Comparative Transcriptome Analysis Identifies Candidate Genes Related to Black-Spotted Pattern Formation in Spotted Scat (Scatophagus argus)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statements
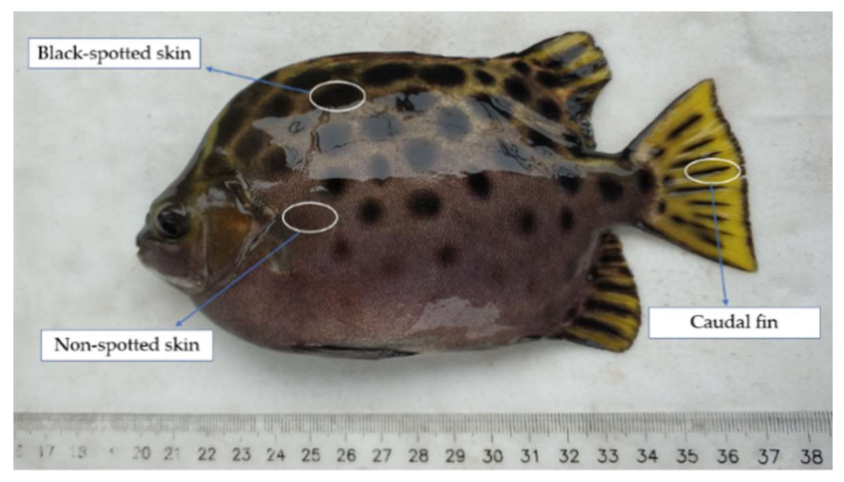
2.2. Sample Preparation
2.3. RNA Extraction and Illumina Library Preparation
2.4. Data Filtering, Reads Mapping and Differential Expression Analysis
2.5. Validation of DEGs by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
3. Results
3.1. Raw Sequencing Reads and Quality Statistics
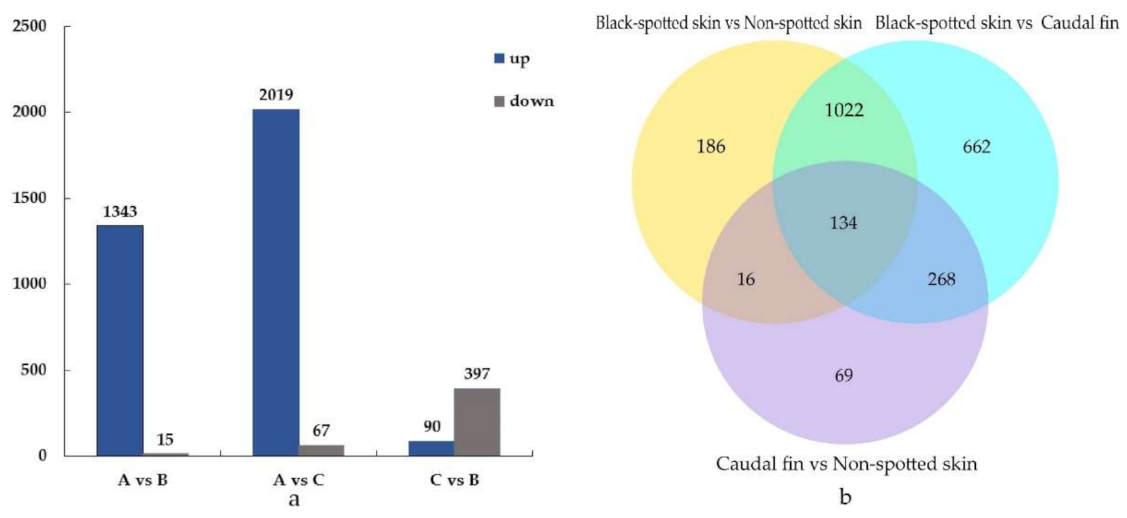
3.2. Comparative Analysis of DEGs
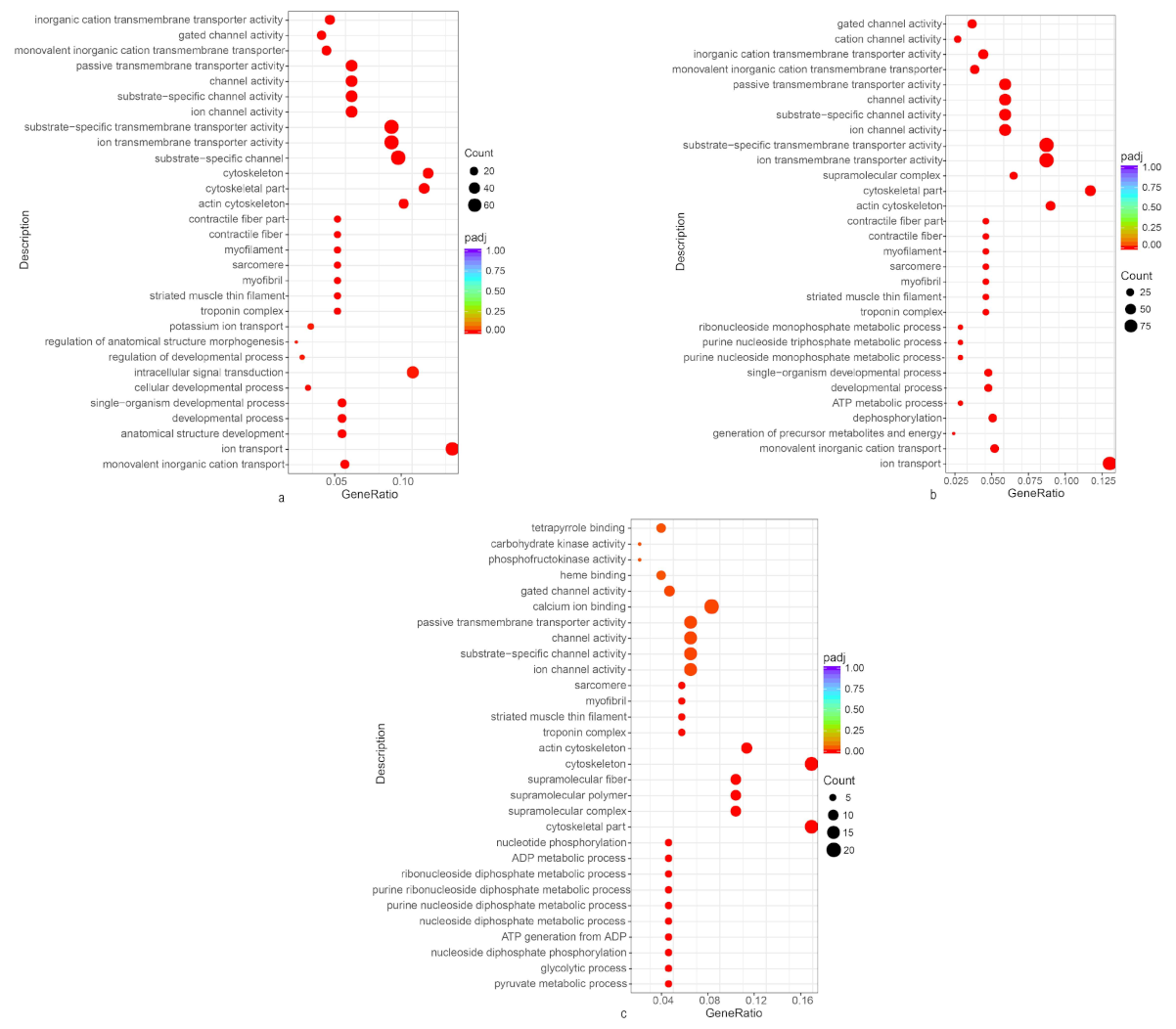
3.3. GO and KEGG Pathway Enrichment Analysis
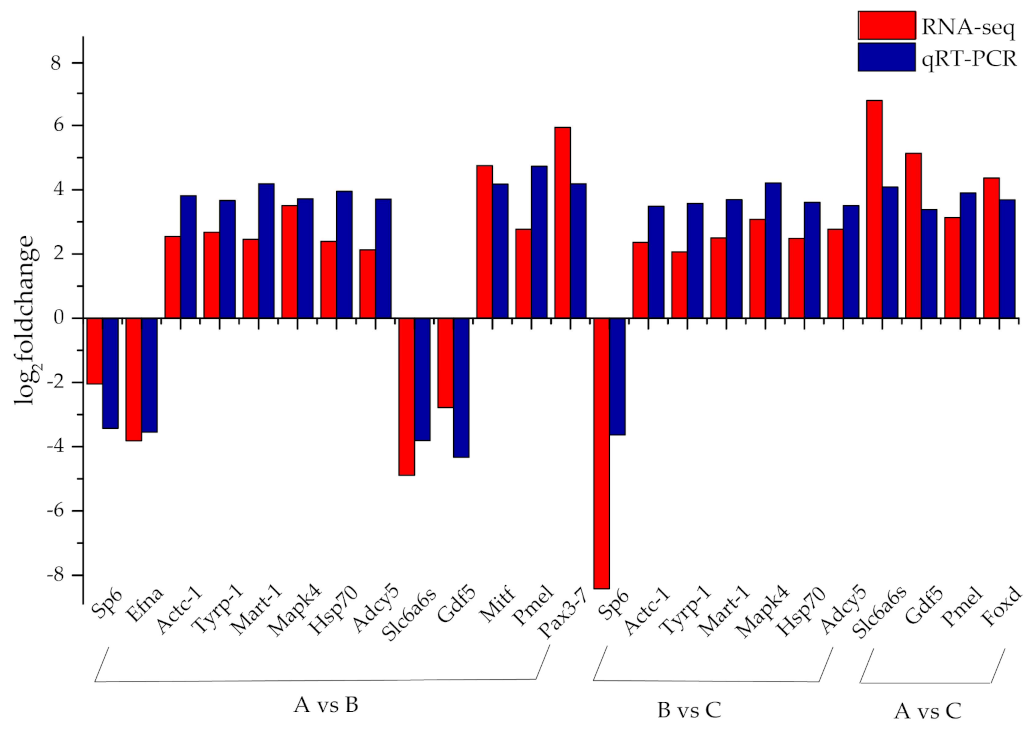
3.4. Validation of Gene Expression Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hubbard, J.K.; Uy, J.A.; Hauber, M.E.; Hoekstra, H.E.; Safran, R.J. Vertebrate pigmentation: From underlying genes to adaptive function. Trends Genet. 2010, 26, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Protas, M.E.; Patel, N.H. Evolution of coloration patterns. Annu. Rev. Cell Dev. Biol. 2008, 24, 425–446. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, G.M.; Kelley, J.L.; Morrell, L.J. Colour change and assortment in the western rainbowfish. Anim. Behav. 2010, 79, 1025–1030. [Google Scholar] [CrossRef]
- Jiang, Y. Comparative transcriptome analysis reveals the genetic basis of skin color variation in Common Carp. PLoS ONE 2014, 9, e108200. [Google Scholar] [CrossRef]
- Bagnara, J.T.; Fernandez, P.J.; Fujii, R. On the blue coloration of vertebrates. Pigment Cell Res. 2007, 20, 14–26. [Google Scholar] [CrossRef]
- Braasch, I.; Brunet, F.; Volff, J.N.; Schartl, M. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol. Evol. 2009, 1, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Kelsh, R.N.; Brand, M.; Jiang, Y.J.; Heisenberg, C.P.; Lin, S.; Haffter, P.; Odenthal, J.; Mullins, M.C.; van Eeden, F.J.; Furutani-Seiki, M.; et al. Zebrafish pigmentation mutations and the processes of neural crest development. Development 1996, 123, 369–389. [Google Scholar] [PubMed]
- Parichy, D.M. Evolution of danio pigment pattern development. Heredity 2006, 97, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Bagnara, T.J.; Matsumoto, J. Comparative anatomy and physiology of pigment cells in nonmammalian tissues. In The Pigmentary System: Physiology and Pathophysiology, 2nd ed.; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2007; pp. 11–59. [Google Scholar]
- Kelsh, R.N.; Inoue, C.; Momoi, A.; Kondoh, H.; Furutani-Seiki, M.; Ozato, K.; Wakamatsu, Y. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech. Dev. 2004, 121, 841–859. [Google Scholar] [CrossRef] [PubMed]
- Mellgren, E.M.; Johnson, S.L. The evolution of morphological complexity in zebrafish stripes. Trends Genet. 2002, 18, 128–134. [Google Scholar] [CrossRef]
- Braasch, I.; Schartl, M.; Volff, J.N. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol. Biol. 2007, 7, 74. [Google Scholar] [CrossRef]
- Li, S.; Wang, C.; Yu, W.; Zhao, S.; Gong, Y. Identification of genes related to white and black plumage formation by RNA-Seq from white and black feather bulbs in ducks. PLoS ONE 2012, 7, e36592. [Google Scholar] [CrossRef] [PubMed]
- Logan, D.W.; Burn, S.F.; Jackson, I.J. Regulation of pigmentation in zebrafish melanophores. Pigment Cell Res. 2006, 19, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Liedtke, D.; Volff, J.N.; Schartl, M. Pigmentary function and evolution of tyrp1 gene duplicates in fish. Pigment Cell Melanoma Res. 2009, 22, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Altschmied, J.; Delfgaauw, J.; Wilde, B.; Wilde, B.; Duschl, J.; Bouneau, L.; Bouneau, L.; Schartl, M. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics 2002, 161, 259–267. [Google Scholar] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SAR Tools: A DESeq2- and edge R-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Djurdjevič, I.; Kreft, M.E.; Sušnik, B.S. Comparison of pigment cell ultrastructure and organisation in the dermis of marble trout and brown trout, and first description of erythrophore ultrastructure in salmonids. J. Anat. 2015, 227, 583–595. [Google Scholar] [CrossRef]
- Rad, H.H.; Yamashita, T.; Jin, H.Y.; Hirosaki, K.; Wakamatsu, K.; Ito, S.; Jimbow, K. Tyrosinase-related proteins suppress tyrosinase-mediated cell death of melanocytes and melanoma cells. Exp. Cell Res. 2004, 298, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Sarangarajan, R.; Boissy, R.E. Tyrp1 and oculocutaneous albinism type 3. Pigment Cell Res. 2001, 14, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Hearing, V.J. Direct interaction of tyrosinase with Tyrp1 to form heterodimeric complexes in vivo. J. Cell Sci. 2007, 120, 4261–4268. [Google Scholar] [CrossRef]
- Shibahara, S.; Tomita, Y.; Yoshizawa, M.; Shibata, K.; Tagami, H. Identification of mutations in the pigment cell-specific gene located at the brown locus in mouse. Pigment Cell Res. Suppl. 2010, 3, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, S.M.; Berryere, T.G.; Goldfinch, A.D. TYRP1 and MC1R genotypes and their effects on coat color in dogs. Mamm. Genome 2002, 13, 380–387. [Google Scholar] [CrossRef]
- Schmidt-Kuntzel, A.; Eizirik, E.; O’Brien, S.J.; Menotti-Raymond, M. Tyrosinase and tyrosinase related protein 1 alleles specify domestic cat coat color phenotypes of the albino and brown loci. J. Hered. 2005, 96, 289–301. [Google Scholar] [CrossRef]
- Berryere, T.G.; Schmutz, S.M.; Schimpf, R.J.; Cowan, C.M.; Potter, J. TYRP1 is associated with dun coat colour in Dexter cattle or how now brown cow? Anim. Genet. 2003, 34, 169–175. [Google Scholar] [CrossRef]
- Gratten, J.; Beraldi, D.; Lowder, B.V.; McRae, A.F.; Visscher, P.M. Compelling evidence that a single nucleotide substitution in TYRP1 is responsible for coat-colour polymorphism in a free-living population of Soay sheep. Proc. Biol. Sci. 2007, 274, 619–626. [Google Scholar] [CrossRef]
- Mollaaghababa, R.; Pavan, W.J. The importance of having your SOX on: Role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene 2003, 22, 3024–3034. [Google Scholar] [CrossRef]
- Hou, L.; Arnheiter, H.; Pavan, W.J. Interspecies difference in the regulation of melanocyte development by SOX10 and MITF. Proc. Natl. Acad. Sci. USA 2006, 103, 9081–9085. [Google Scholar] [CrossRef]
- Newton, R.A.; Cook, A.L.; Roberts, D.W.; Leonard, J.H.; Sturm, R.A. Post-transcriptional regulation of melanin biosynthetic enzymes by cAMP and resveratrol in human melanocytes. J. Investig. Dermatol. 2007, 127, 2216–2227. [Google Scholar] [CrossRef]
- Oren, M.; Bartek, J. The sunny side of p53. Cell 2007, 128, 826–828. [Google Scholar] [CrossRef]
- Fadeev, A.; Krauss, J.; Frohnhöfer, H.G.; Irion, U.; Nüsslein-Volhard, C. Tight junction protein 1a regulates pigment cell organisation during zebrafish colour patterning. eLife 2015, 4, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, J.; Takai, Y. Structural and functional associations of apical junctions with cytoskeleton. Biochim. Biophys. Acta Biomembr. 2008, 1778, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Hellstrom, A.R.; Watt, B.; Fard, S.S.; Tenza, D.; Mannstrom, P.; Narfstrōm, K.; Ekesten, B.; Ito, S.; Wakamatsu, K.; Larsson, J.; et al. Inactivation of Pmel alters melanosome shape but has only a subtle effect on visible pigmentation. PLoS Genet. 2011, 7, e1002285. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, A.C.; Kerje, S.; Hallbook, F.; Jensen, P. The dominant white mutation in the PMEL17 gene does not cause visual impairment in chickens. Vet. Ophthalmol. 2009, 12, 292–298. [Google Scholar] [CrossRef]
- Brunberg, E.; Andersson, L.; Cothran, G.; Sandberg, K.; Mikko, S.; Lindgren, G. A missense mutation in PMEL17 is associated with the Silver coat color in the horse. BMC Genet. 2006, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Shyy, J.Y.; Chien, S. Role of integrins in endothelial mechanosensing of shear stress. Circ. Res. 2002, 91, 769–775. [Google Scholar] [CrossRef] [PubMed]
| Group | Raw Reads | Clean Reads | Clean Bases (G) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|
| Black-spotted skin (A) | |||||
| A1 | 58,647,296 | 57,844,474 | 8.68 | 93.16 | 46.97 |
| A2 | 54,843,424 | 53,929,484 | 8.09 | 92.84 | 46.67 |
| A3 | 61,189,662 | 60,096,498 | 9.01 | 94.12 | 46.56 |
| Non-spotted skin (B) | |||||
| B1 | 55,148,934 | 54,250,656 | 8.14 | 92.72 | 45.94 |
| B2 | 53,000,358 | 52,199,276 | 7.83 | 92.69 | 46.70 |
| B3 | 56,077,298 | 55,093,088 | 8.26 | 92.64 | 46.07 |
| Caudal fin (C) | |||||
| C1 | 46,402,098 | 45,867,418 | 6.88 | 92.16 | 46.15 |
| C2 | 50,405,758 | 49,913,528 | 7.49 | 92.47 | 46.31 |
| C3 | 49,958,956 | 48,909,024 | 7.34 | 94.57 | 46.88 |
| Gene | Gene ID | Note | Log2 Fold Change |
|---|---|---|---|
| Black-spotted skin vs. Non-spotted skin | |||
| Tyrp1 | EVM0000142 | Tyrosinase-related protein 1 | 2.68 |
| Wnt2 | EVM0010869 | Wingless-type MMTV integration site family, member 2 | 8.04 |
| Camk2 | EVM0018979 | Calcium/calmodulin-dependent protein kinase | 2.75 |
| Adcy5 | EVM0008013 | Adenylate cyclase 5 | 2.12 |
| Adcy6 | EVM0004537 | Adenylate cyclase 6 | 2.76 |
| Wnt11 | EVM0010741 | Wingless-type MMTV integration site family, member 11 | 5.96 |
| Foxd | EVM0009699 | Forkhead box protein D | 4.67 |
| Mapk4 | EVM0001218 | Mitogen-activated protein kinase 4 | 3.51 |
| Adcy9 | EVM0003371 | Adenylate cyclase 9 | 2.29 |
| Hgf | EVM0021385 | Hepatocyte growth factor | 2.29 |
| Tgfb3 | EVM0004513 | Transforming growth factor beta-3 | 2.96 |
| Rasgrf1 | EVM0005858 | Ras-specific guanine nucleotide-releasing factor 1 | 4.77 |
| Efna | EVM0012046 | Ephrin-A | −3.81 |
| Mart-1 | EVM0011519 | Melanoma antigen recognized by T-cells 1 | 2.45 |
| Tjp1 | EVM0021943 | Tight junction protein 1 | 2.12 |
| Hsp70 | EVM0013804 | Heat shock 70 kDa protein 12A isoform X1 | 2.39 |
| Black-spotted skin vs. Caudal fin | |||
| Pmel | EVM0007408 | Premelanosome protein | 2.23 |
| Slc7a2 | EVM0021987 | Solute carrier family 7 member 2 | 4.29 |
| Pax3 | EVM0000393 | Paired box protein 3 | 2.45 |
| Pdgfrb | EVM0022774 | Platelet-derived growth factor receptor beta | 3.89 |
| Flt1 | EVM0007297 | FMS-like tyrosine kinase 1 | 3.04 |
| Dusp | EVM0020294 | Dual specificity MAP kinase phosphatase | 2.04 |
| Hspb1 | EVM0002084 | Heat shock protein beta-1 | 4.37 |
| Kdr | EVM0020836 | Kinase insert domain protein receptor | 2.96 |
| Ntrk2 | EVM0017245 | Neurotrophic tyrosine kinase receptor type 2 | 4.74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Tian, C.; Huang, Y.; Shi, H.; Li, G. Comparative Transcriptome Analysis Identifies Candidate Genes Related to Black-Spotted Pattern Formation in Spotted Scat (Scatophagus argus). Animals 2021, 11, 765. https://doi.org/10.3390/ani11030765
Lin X, Tian C, Huang Y, Shi H, Li G. Comparative Transcriptome Analysis Identifies Candidate Genes Related to Black-Spotted Pattern Formation in Spotted Scat (Scatophagus argus). Animals. 2021; 11(3):765. https://doi.org/10.3390/ani11030765
Chicago/Turabian StyleLin, Xiaozhan, Changxu Tian, Yang Huang, Hongjuan Shi, and Guangli Li. 2021. "Comparative Transcriptome Analysis Identifies Candidate Genes Related to Black-Spotted Pattern Formation in Spotted Scat (Scatophagus argus)" Animals 11, no. 3: 765. https://doi.org/10.3390/ani11030765
APA StyleLin, X., Tian, C., Huang, Y., Shi, H., & Li, G. (2021). Comparative Transcriptome Analysis Identifies Candidate Genes Related to Black-Spotted Pattern Formation in Spotted Scat (Scatophagus argus). Animals, 11(3), 765. https://doi.org/10.3390/ani11030765







