Simple Summary
This study examined the impact of whole sesame seeds (WSS), rich in both linoleic acid and lignans, on the innate immunity of goats. WSS were incorporated in the concentrates of the control group at 5 and 10% respectively, by partial substitution of both soybean meal and corn grain. The highest supplementation level of WSS resulted in a significant down-regulation in the expression levels of several pro-inflammatory genes in the neutrophils of goats. In conclusion, the dietary supplementation of goats with WSS might be a good nutritional strategy to improve their innate immunity.
Abstract
Whole sesame seeds (WSS) are rich in both linoleic acid (LA) and lignans. However, their impact on the innate immunity of goats is not well studied. Twenty-four goats were divided into three homogeneous sub-groups; comprise one control (CON) and two treated (WWS5 and WWS10). In the treated groups, WSS were incorporated in the concentrates of the CON at 5 (WSS5) and 10% (WSS10) respectively, by partial substitution of both soybean meal and corn grain. The expression levels of MAPK1, IL6, TRIF, IFNG, TRAF3, and JUND genes in the neutrophils of WSS10 fed goats were reduced significantly compared with the CON. The same was found for the expression levels of IFNG and TRAF3 genes in the neutrophils of WSS5 fed goats. Both treated groups primarily affected the MYD88-independent pathway. The dietary supplementation of goats with WSS might be a good nutritional strategy to improve their innate immunity.
1. Introduction
N-3 polyunsaturated fatty acids (PUFA) in humans have anti-inflammatory role [1] since resolve inflammation [2] and eliminate pain in inflammatory circumstances [3]. On the other hand, increased consumption of linoleic acid (LA), the main fatty acid (FA) of the n-6 PUFA group, might be related with inflammatory diseases due to its metabolization in LA-derived pro-inflammatory lipoxins and arachidonic acid, which further leads to pro-inflammatory eicosanoids and prostaglandins production [4]. An enhancement in the concentrations of pro-inflammatory leukotriene and prostaglandins [5] was found in rats fed with high LA diets. Significantly higher tumor necrosis factor a (TNFA), and interleukin-7 concentrations in the liver of pregnant rat, consumed a high compared with low LA diet, was observed, without the cytokines content in their blood to be affected [6]. Similarly, excessive dietary LA consumption increased significantly the TNFA content in plasma and nuclear factor-kappa B (NF-KB) expression in rats’ aortas [7]. On the other hand, recent reviews and meta-analysis studies provide evidence that LA intake decreases [8] or has no effect on cardiovascular diseases [9,10] disputing its role in chronic diseases involving inflammatory process.
So far, to the best of our knowledge, no information exists on the impact of LA in the innate immunity of productive animals, and particularly in goats. Thus, whole sesame seeds (WSS), due to their high LA (44%) content [11], can be used in goats’ diets to test this hypothesis. Moreover, WSS contain lignans such as sesamin and sesamolin, which might have several beneficial effects in immunity [12]. The anti-inflammatory properties of sesame in rats’ models through in vitro and in vivo trials have been reviewed recently [13]. Sesamin down-regulates the expression of Toll-like receptor 4 (TLR4) gene in lipopolysaccharide (LPS) stimulated BV-2 microglial cell line of rats, in a dose dependent matter in vitro [14]. Accordingly, 50 μM of sesamin suppressed the activation of p38 mitogen-activated protein kinase (MAPK) signaling pathway after its stimulation with LPS [15]. A significant decline in the expression of interleukin 1 Beta (IL1B), interleukin-2 (IL2) and TNFA genes in mouse senescence-accelerated brain cells was found, when fed with sesaminol [16]. So far, the impact of dietary inclusion of WSS in the immune system of produced animals has not been studied.
The immune system is broken down into innate and adaptive [17]. Neutrophils comprise one of the main cellular component of the innate immune system and the first line of defense against pathogens [18]. The innate immune system employs special receptors known as pattern-recognition receptors (PRRs) such as NOD-like receptors (NLRs) that recognize pathogen- or damage-associated molecular patterns (PAMPs and DAMPs, respectively) [19]. Among the PRRs, Toll-like receptors (TLRs) enact the induction of immune response [20]. All TLR signaling pathways end up in activation of the transcription factor nuclear factor-kappa B (NF-KB) and interferons (IRFs), which regulate the outcome of innate immune responses [19]. In addition, a core element of the NF-KB cascade is the IκB kinase (IKK) complex or conserved helix-loop-helix ubiquitous kinase (CHUK) which is encoded by the CHUK gene [21]. TLR activation stimulates the release of various inflammatory cytokines (TNFA, interferon IFNG, interleukins (IL1B, IL2 and IL6) and immune modulators such as IL8, C-C motif chemokine ligand 5 (CCL5) and chemokine (C-X-C motif) ligand 16 (CXCL16) [22,23,24]. After that, IL6 induces downstream signaling of the signal transducer and activator of transcription 3 (STAT3) [25]. Upon PAMPs and DAMPs recognition, TLRs recruit Toll-interleukin-1 receptor TIR domains, which transmit downstream signals via adaptor molecules such as myeloid differentiation primary response gene 88 (MΥD88) and the TIR (Toll/Interleukin-1 Receptor) domain-containing adaptor protein inducing interferon beta (TRIF) [26,27]. The MYD88-dependent pathway activates the IRF5 gene expression [28] and the pathway involving mitogen-activated protein kinases (MAPKs). TNF Receptor-associated Factor 3 (TRAF3) is incorporated into both MYD88 and TRIF complex, activating MYD88-dependent signaling and suppressing TRIF-dependent pathway [29]. TRAF3 mediates activation of IRF3 [30]. The extracellular signal regulated kinase (ERK)–mitogen-activated protein kinase pathway determines the regulation of JUND gene expression [31]. Finally, Heme Oxygenase-1 (HO1) gene has the ability to modulate immune responses [32].
Taking into account all the above, the objective of this study was to investigate the effects of dietary inclusion of WSS at two different levels (5 and 10%) on the expression of selected key-genes (NLRC3, TLR4, MYD88, NF-KB, MAPK1, IL1A, IL1B, TNFA, TNFB, IL2, IL6, IL10, STAT3, TRIF, IRF3, IFNG, TRAF3, IRF5, CCL5, IL8, CXCL16, HO1, JUND and CHUK) involved in the innate immunity of dairy goats.
2. Materials and Methods
2.1. Animals and Diets
Animal handling procedures were performed in accordance with protocols approved by the Agricultural University of Athens Ethical Committee of the Faculty of Animal Sciences. Twenty-four goats were divided into three homogenous subgroups (n = 8) according to their fat-corrected milk yield (1.00 ± 0.22 kg/day) and body weight (44.9 ± 5.4 kg). The goats were fed on a group basis with a basal diet consisted of alfalfa hay, wheat straw and concentrates (Forage/Concentrate ratio = 50/50), for a seven-day adaptation period. The forages were provided separately from the concentrates while they were both offered to the animals twice a day (in two equal parts at 08:00 and 18:30 h) after milking. After the adaptation period the control goats continued to consume the basal diet, in the concentrates of which hulled sesame seeds were not included (CON). On the other hand, in the concentrates of the two other groups whole sesame seeds at 5 (WSS5) and 10% (WSS10) respectively, were incorporated by partial substitution of both soybean meal and corn grain (Table 1), in order the dietary treatments to be iso-energetic and iso-protein, and to meet the animals’ average maintenance and lactation requirements [33]. The quantities of food offered to the animals were adjusted every two weeks, according to their average requirements, based on their body weight and milk fat-corrected yield. Diet selectivity did not occur, and no refusals of forage and/or concentrates were observed. The mineral and vitamin premix of both concentrates contained the following (per kg as mixed): 150 g Ca, 100 g P, 100 g Na, 100 mg Co, 300 mg I, 5000 mg Fe, 10,000 mg Mn, 20,000 mg Zn, 100,000 mg Se, 5,000,000 IU retinol, 500,000 IU cholecalciferol and 15,000 mg α-tocopherol. The experimental period, lasted 100 days and all the animals had free access to fresh water.

Table 1.
Nutrients and fatty acids intake from forages and concentrates, and the total antioxidant capacity and phenolic content of concentrates only.
2.2. Feed Sample Analyses
Samples of the alfalfa hay, wheat straw and concentrate were analyzed for organic matter (OM; Official Method 7.009), dry matter (DM; Official Method 7.007) and crude protein (CP; Official Method 7.016) according to the AOAC (1984) and for neutral detergent fiber (NDF) and acid detergent fiber (ADF)-expressed exclusive of residual ash-according to the methods of Van Soest et al. [34].
2.3. Blood Samples
2.3.1. Blood Sample Collection for Neutrophil Isolation
Blood samples were taken at the 30th, 60th and 90th day from the beginning of the experiment for neutrophil isolation from the jugular vein into 17 Units/mL heparine-containing tubes (BD Vacutainer, Plymouth, UK).
2.3.2. Cell Isolation
Cell isolation was performed according to Tsiplakou et al. [35]. More specifically, isolation of neutrophils is carried out using density gradient centrifugation Histopaque 1077 (Sigma-Aldrich, St. Louis, MO, USA). Analytically, whole blood mixed with an equal volume of Hanks’ balanced salt solution and three parts diluted blood layered on two parts Histopaque. Samples were centrifuged for 40 min at 500× g, 4 °C with the minimum acceleration and deceleration. After centrifugation, the upper phases were rejected. Neutrophils cells, which remained in the red cell layer, were lysed with the addition of endotoxin-free ultrapure water, and were vigorously shaken. NaCl was then added to resuspend cells in an isotonic solution (0.9% NaCl). These cells were washed several times and centrifuged for 5 min at 1000 g and 4 °C until a white and consistent cell pellet was clearly visible at the bottom of the tube. In the sequel, the final cell suspensions were cultured in 1 mL of growth medium RPMI (Sigma-Aldrich, St. Louis, MO, USA) which is incubated at 37 °C and then centrifuged at 1000 g for 5 min at 4 °C. Finally, the resulting cell pellets were again washed at least twice in 0.5 mL of phosphate-buffered saline (PBS) and centrifuged at 700 rpm for 1 min at 4 °C.
2.3.3. RNA Extraction
The isolated cells were homogenized with TRIzol™ (Invitrogen, Carlsbad, CA, USA) and after centrifugation with 24:23:1 phenol: chloroform: isoamyl alcohol solution, three distinct layers were obtained. The upper clear aqueous phase containing the RNA was transferred carefully into a new tube, without disturbing the interphase. RNA pellet was precipitated with 70% ethanol and then was dissolved in milli-Q water. The quantity and quality of the extracted RNA were evaluated by ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA); the quantity was measured in ng/μL, and its purity was determined based on the A260/A280 and A260/A23 ratios. In addition, RNA integrity was assessed by electrophoresis on an agarose gel. As defined, the isolated RNA was treated with Turbo™ DNase I (2U/μL, commercially available kit: Invitrogen, Carlsbad, CA, USA), accordingly to the manufacturer’s instructions. Absence of genomic DNA contamination was confirmed by PCR, using glycer aldehyde3-phosphatedehydro genase (GAPDH; housekeeping gene) Then, RNA samples were further purified by using phenol: chloroform and ethanol precipitation. The quantity and quality of the pure RNA samples were again confirmed by spectrophotometry (NanoDropND-1000) and by agarose gel (0.7%) electrophoresis.
2.3.4. cDNA Synthesis
Approximately 500 ng of RNA was used per cDNA synthesis by using the Prime Script First Strand cDNA Synthesis Kit (Takara, Shiga, Japan) according to the manufacturer’s protocol using a mix of random hexamers and oligo-dT primers.
2.3.5. Primers
To derive primers sequences the ARS1 goat annotation was used. A pair of primers specific for each target gene (Table 2) were used by previous studies [35,36] designed to be specific for Capra hircus by using Primer Express Software (version 3.0) and verified using the Geneious Software (Biomatters, Auckland, New Zealand) and were tested against genomic DNAs to confirm that a single amplicon of 70 bp would result from quantitative real-time PCR (qPCR). In addition, dissociation curves were generated, and the amplification products were subjected to agarose gel electrophoresis to confirm the production of a single amplicon per reaction.

Table 2.
Primers used for real-time qPCR and the mean PCR efficiency for each gene as calculated by LinRegPCR software [37].
2.3.6. Real-Time Quantitative PCR
The expression levels of genes were estimated by a Step One Plus ™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR Select Master Mix (Applied Biosystems, Austin, TX, USA), gene-specific primers at a final concentration of 0.2 μM each (forward and reverse) and 1 μL of each cDNA as template. Thermal cycling was started with denaturation at 95 °C for 15 min, followed by 40 cycles at 95 °C for 15 s and 62 °C for 10 s. GAPDH and YWHAZ were used as housekeeping genes to normalize the cDNA template concentrations. The choice of housekeeping genes was based on a study by Vorachek et al. [38].
2.3.7. Normalization
The expression levels of the genes were calculated as (1 + E)−ΔCt, where ΔCt is the difference between the geometric mean of the two housekeeping genes’ Cts and the Ct of the target gene, and the primer efficiency is the mean of each amplicon’s efficiency per primer, which was calculated by employing the linear regression method on the log (fluorescence) per cycle number (ΔRn) using the LinReg PCR software [37].
2.3.8. Statistical Analysis
Experimental data are presented as least squares means ± standard errors and were analyzed using a general linear model (GLM) for repeated measures, considering the sampling time (T) as the repeated measure, with fixed effects of dietary treatments (D) (CON, WSS5, WSS10), sampling time (T) (30th, 60th, 90th experimental day) and the interactions among them (D × T) according to the model:
where Υijk is the dependent variable, μ the overall mean, Di the effect of dietary treatment (I = 1, 2, 3), Tj the effect of sampling time (j = 1, 2, 3), (D × T) ij the interaction between dietary treatments and sampling time, Ak the animal’s random effect and eijk the residual error. Post hoc analyses were performed when appropriate using Duncan’s multiple range test. Kolmogorov-Smirnov test revealed that all variables followed a normal distribution. Pearson’s correlation coefficients were used to determine the relationships between gene expression in neutrophils using heat map chart. For all tests, the significance was set at 0.05. Graphs were drawn using SPSS software (version 20.0, IBM, Armonk, NY, USA), and the error bars represent the standard error of the mean (SEM). Statistical analysis was performed using the statistical packages SPSS software (version 20.0, IBM, Armonk, NY, USA).
Yijk = μ + Di + Tj + (D × T) ij + Ak + eijk
3. Results
A significant reduction in the expression levels of MAPK1, IL6, TRIF, IFNG, TRAF3 and JUND genes in the neutrophils of WSS10 fed goats compared with the CON was found (Figure 1).
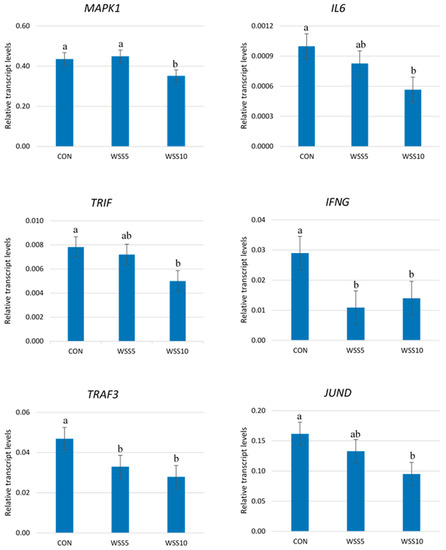
Figure 1.
The Transcript abundance of several genes in the neutrophils of goats. Bars represent means ± SEM of each (n = 8) of the three dietary treatments; CON: control, basal diet; WSS5: basal diet + 5% whole sesame seed; WSS10: basal diet + 10% whole sesame seed in goats. For each gene, bars with different superscripts (a, b) between the three dietary treatments (CON, WSS5, WSS10) differ significantly (p ≤ 0.05), according to the analysis of variance (ANOVA) using a general linear model (GLM) for repeated measures. Post hoc analysis was performed using Duncan’s multiple range test.
The same trend was found for the IFNG and TRAF3 genes in the neutrophils of SS5 fed goats (Figure 1). No differences were found in the expression levels of the above genes between the treated groups (Supplementary Figure S1).
A significant reduction in the expression levels of NF-KB, MYD88, MAPK1, TNFA, and STAT3 genes in the neutrophils of goats throughout the experimental period was observed (Supplementary Table S1). The opposite happened in the relative abundance transcripts of IRF5 and HO1 genes (Supplementary Table S1). The highest expression levels of NLRC3, TNFB and TRIF genes were indicated in the 60th experimental day while in this day the TLR4 gene showed the lowest expression levels (Supplementary Table S1). A significant decline in the expression levels of IL1A, ΙL-2 and CCL5 genes was found in the 90th compared with the 30th and 60th experimental period, while the opposite trend was observed for the IL10 and CXXL-16 genes (Supplementary Table S1).
Significantly positive correlations between the expression levels of; TLR4 with NF-KB, MYD88, and IL1B, MYD88 with NF-KB and IRF3, STAT3 with MYD88 and NF-KB, JUND with HO1, NF-KB with IRF3 genes respectively, as well as between MAPK1 with MYD88 and TLR4 and NLRC3 with IL6 and TNFB genes respectively, were found (Figure 2).
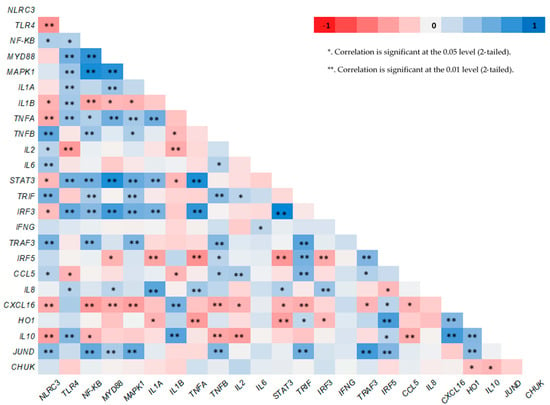
Figure 2.
Pearson’s heat map correlations between the expression level of several genes in neutrophils of goats.
4. Discussion
The impact of sesame seeds in the innate immunity of ruminants has rarely been investigated to the best of our knowledge. Neutrophils are involved in initiation of the inflammatory response [39], through the expression of several families of PRRs such as NLRs and TLRs [40,41] which can identify either microbial pathogens or components of host’s cells that are released during cell damage or death. Significantly higher expression levels of TLR4 gene have been found in blood neutrophils of ketotic cows [42]. Moreover, Roldan-Montes et al. [43] found in milk, a significant association between the identified polymorphisms of the TLR4 gene and the somatic cell score of water buffaloes. A significant down-regulation in the expression of TLR4 gene in LPS-stimulated BV-2 microglial cell line of rats was observed in vitro [14]. Thus, the results of this study referring to NLRC3 and TLR4 genes (Supplementary Figure S1) might show not only absence of any inflammation (clinical or subclinical) but also the protective role of sesame seeds in goats’ cells survival.
TLR4 gene regulates the NF-KB pathway through the MYD88 gene [44] which affects the MAPKs cascade [45]. Indeed, significantly positive correlations between the expression levels of TLR4 with NF-KB, TLR4 with MYD88 and MYD88 with NF-KB genes, were found (Figure 2). It was observed that sesamin, one of the main antioxidant compounds of sesame seeds, reduces the activation of NF-KB (measured by ELISA) and P38 MAPK kinase (measured by Western blot) in mice microglia cells treated with LPS [15]. The same has been shown for the expression of TLR4 gene (measured by flow cytometry) in hepatic tissue of mice [46]. Sesamin inclusion in RPMI-8226 cells [47] and sesame oil aqueous extract in RAW 264.7 macrophages of mice treated with LPS [48] down regulate the expression of NF-KB gene in vitro. Additionally, a significant up-regulation in expression levels of TLR4 and MYD88 genes in goats’ mammary epithelial cells [49], and in human endometrial cells [50] when stimulated in vitro by LPS have been found. Moreover, significantly higher expression levels of MYD88 gene have been observed in bovine mastitis tissue [51]. Thus, the results of this study, concerning the NF-KB, MYD88 and MAPK1 genes, not only show no pathogens, stress or endogenous inflammatory factors in goats’ organisms but also indicate a positive effect in their innate immunity when the animals were fed with the higher supplementation level (10%) of WSS.
NLRs and TLRs stimulate the MAPKs cascade thought the MYD88 pathway [52] and trigger the cytokines production [53,54]. The significantly positive correlations between the expression levels of MAPK1 with MYD88 and TLR4 genes respectively, confirm this close relationship (Figure 2). Moreover, the positive correlations between the expression levels of TLR4 and IL1B genes, as well as between the expression levels of NLRC3 with IL6 and TNFB genes respectively (Figure 2) show that both NLRs and TLRs regulate the cytokines expression. IL2 has anti-inflammatory properties [55] while IL6 is elevated in most cases of inflammation and have been recognized as target for therapeutic intervention [56]. Indeed, it has been found recently that elevated IL6 levels in blood plasma resulted a STAT3 hyperactivation in tumor cells [25]. However, sesamin has the ability to suppress the STAT3 signaling pathway (IL6/JAK/STAT3) in human hepatocellular carcinoma cell line HepG2 [57]. The anti-cancer effects of sesamin have been attributed to its ability to reduce significantly the expression of NF-KΒ, IL6 and transcriptional target of STAT3 [58]. In accordance with our findings, sesamin inhibits the expression levels of IL6 gene, in a dose depend matter in vitro [15]. The anti-inflammatory activities of sesamin have been shown also, in influenza H1N1-induced peripheral blood mononuclear cells of humans by either the reduction in the expression levels of both IL1B and TNFA genes or the increase in the expression of IL2 gene [59]. A significant reduction in the expression levels of IL2 gene was observed in cows infected with malignant catarrhal fever [60,61]. Furthermore, a significant down-regulation in the expression levels of pro-inflammatory (IL1A, IL1B, TNFA) genes including IL6 was found in the liver of mice fed with a sesame oil rich diet [62]. Sesamin, reversed the inflammation which caused by the consumption of a high fat diet in rats by reducing the expression of IL6 and TNFA genes [63]. Thus, the results of this study, as the IL2, IL6, STAT3 and cytokines genes expression is concerned could high light as well the idea of an improvement of goats’ innate immunity especially, when they were fed with the higher inclusion level of sesame seeds.
Although MYD88 is a common adaptor for all the TLRs except TLR3, TRIF, is an adaptor for TLR3 and TLR4 which promotes an alternative pathway that leads to the activation of IRF3 for induction of type IFN [19,64]. Our results referring to the TRIF, IRF3 and IFNG genes, show that the highest dietary inclusion level (10%) of WSS affected also the MYD88 independent pathway. This is further supported by the changes in the expression levels of TRAF gene since both MYD88 and TRIF pathways are controlled by TRAF regulators such as TRAF3 [65,66]. Similar to our findings, a significant down-regulation in the expression levels of IFNG gene in cultured mononuclear cells of experimental autoimmune encephalomyelitis mice, fed with sesame oil, has been found [67]. Additionally, sesame oil reduces significantly the concentrations of IFNG in multiple sclerosis patients [68]. On the other hand, the expression level of IRF3 gene increased significantly in goats’ mammary epithelial cells after 3 h incubation in vitro with both toxins from LPS and gram-positive lipoteichoic acid bacterial [49]. The same was observed in bovine mammary epithelial cells when stimulated either with Escherichia coli or Staphylococcus aureus [69]. Furthermore, the pro-inflammatory role of IRF3 has been indicated also in mice macrophages, through the activation of TLR4-TRIF metabolic pathway which regulates the production of pro-inflammatory cytokines [70].
Chemokines such as CCL5, IL8 and CXCL16 can be produced by many cells including neutrophils [71] after proper stimulation [72]. So far, significant higher expression levels of CCL5 gene have been observed in infected blood macrophages with Mycobacterium in vitro [73]. The same trend has been indicated in goats’ mammary epithelial cells after incubation with gram-negative and/or gram-positive bacteria cell wall components in vitro [49]. The expression of IL8 gene enhanced significantly in blood neutrophils of calving cows with clinical mastitis [71]. A positive correlation between IL8 gene expression and the incidence of severe mastitis has been also shown [74]. However, chemokines such as IL8 can be also released from the cells as response to the reactive oxygen species (ROS) [75]. Additionally, CXCL16 chemokine can have a scavenger role for the uptake of oxidase molecules such as the low-density lipoproteins [76]. It has been shown that various antioxidant compounds can protect low-density lipoprotein (LDL) from oxidation in vitro [77]. Indeed, a delay in the oxidation of lipoproteins in the blood plasma of mice fed with sesame oil has been found due to its sesamin and sesamone content which was accompanied by a significant reduction in the CXCL16 blood plasma content [62]. Thus, the results of this study, concerning the expressions of chemokines (CCL5, IL8 and CXCL16), further support the use of sesame seeds as a nutritional tool for the improvement of goats both innate immunity and antioxidant status.
HO1 is a highly inducible gene well known for its anti-inflammatory, immunomodulatory and antioxidants functions [78]. Similar with our findings, sesamin did not modify the expression levels of HO1 gene in rats in vitro [79]. On the contrary, a significant up-regulation in the expression levels of HO1 gene has been found in the liver of bovine and mice, infected by Fachiola hepatica [80]. The same was observed for both HO1 and IL10 genes in LPS-stimulated macrophages of mice [81]. Although the metabolic pathway which regulates HO1 gene expression in not clear, activation of STAT3 by IL10 cytokine has repeatedly been suggested. The positive relationship between the expression levels of HO1 and IL10 genes (p < 0.01) supports this suggestion while the negative relation between the expression levels of STAT3 and IL10 genes (p < 0.01), which was found in this study (Figure 2), needs further investigation in order to clarify the role of STAT3 gene in this metabolic pathway. Thus, referring to the results on the expression levels of HO1, IL10 and STAT3 genes of this study, an enhancement of the innate immune responses with the higher supplementation level of WSS could be claimed.
JUND gene might have also an involvement in the HO1 gene expression. It has been found that JUND protein repressed HO1 gene expression in human renal epithelial cells [82]. The relationship between the expression levels of HO1 and JUND genes supports this link (Figure 2). Moreover, the expression levels of JUND gene followed the same trend with the expression level of MAPK1 gene (Supplementary Table S1). JUND gene has a fundamental role in the defense against oxidative stress [83]. Thus, its sharpest down-regulation with the highest supplementation level of sesame seeds might show that the SS10 goats had a sufficient pool of antioxidants compounds in their organism such as sesamin, sesaminol, etc. which enhance their innate immunity.
So far, in the innate immunity, little attention has been given in inflammatory mediators such as the IkappaB kinase (IKK). Τhe role of IKK-α subunit (CHUK) in inflammation is not well known. However, CHUK gene is required for the activation of the “alternative” NF-KB pathway which is activated by the TNF family cytokines [84]. Moreover, CHUK has anti-inflammatory role through the regulation of SUMO (small ubiquitin-related modifier) ligase activity of protein inhibitor of activated STAT1 (PIAS) [85]. In accordance with our findings sesamin had no effect on the expression of CHUK gene in various human cells lines in vitro [47]. More research is needed in order to clarify the role of CHUK gene in the innate immunity.
5. Conclusions
Overall, our study provides new evidence regarding the impact of dietary supplementation with WSS in the innate immunity of dairy goats. The highest inclusion level (WSS10) seems the best modulator of goats’ innate immunity, as demonstrated by the sharpest decline in the expression levels of genes (MAPK1, IL6, TRIF, IFNG, TRAF3, and JUND) involved with inflammatory metabolic pathways. The topmost intake of WSS also regulates both MYD88 dependent (MAPK1) and independent (TRIF, TRAF3, IFNG) pathway, while this of WSS5 the independent one only. The above findings are very important in animal husbandry since inflammation should be limited as much as possible, and animals’ innate immunity should be activated only when is needed in order to be stronger and more effective. Finally, lignans can eliminate the pro-inflammatory compound, which is produced by LA’s metabolism, making WSS one of the best way to administer LA in goats’ diet.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/11/2/468/s1, Figure S1: The Transcript abundance of several genes in the neutrophils of goats. Bars represent means ± SEM of each (n = 8) of the three dietary treatments; CON: control, basal diet; WSS5: basal diet + 5% whole sesame seed; WSS10: basal diet + 10% whole sesame seed in goats. The analysis of variance (ANOVA) using a general linear model (GLM) for repeated measures revealed that for these genes there was not significant difference between the three dietary treatments (p > 0.05), Table S1: Transcript abundance of several genes in the neutrophils of goats: NOD-like receptor (NLRC3), Toll-like receptors 4 (TLR4), Nuclear factor kappa B (NF-KB), Myeloid-Differentiation-primary response gene 88 (MYD88), Mitogen-Activated Protein Kinase-1 (MAPK1), Interleukin 1 Alpha (IL1A), Interleukin 1 Beta (IL1B), Tumor necrosis factor Alpha (TNFA), Τumor necrosis factor Beta (TNFB), Interleukin 2 (IL2), Interleukin 6 (IL6), Signal Transducer and Activator of Transcription 3 (STAT3), TIR (Toll/Interleukin-1 Receptor) domain-containing adaptor protein inducing interferon beta (TRIF), Interferon Regulatory Factor 3 (IRF3), Interferon gamma (IFNG), TNF Receptor-associated Factor 3 (TRAF3), Interferon Regulatory Factor 5 (IRF5), C-C motif chemokine ligand 5 (CCL5), Interleukin 8 (IL8), Chemokine (C-X-C motif) ligand 16 (CXCL16), Heme Oxygenase-1 (HO1), Interleukin 10 (IL10), Transcription factor JunD (JUND) and Conserved Helix-Loop-Helix-Ubiquitous Kinase (CHUK) or IKKA relative to the geometrical mean of the references genes (Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) and Tyrosine 3-monoxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ)).
Author Contributions
Conceptualization, E.T.; methodology, E.T., E.F. and C.M.; software, K.S and D.S.; validation, E.T., K.S., E.F., D.S. and C.M.; formal analysis, C.M.; investigation, E.T.; data curation, E.T., K.S. and C.M.; writing—original draft preparation, E.T. and C.M.; writing—review and editing, E.T. and E.F.; visualization, C.M.; supervision, E.T.; project administration, E.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research is co-financed by Greece and the European Union (European Social Fund-ESF) through the Operational Programme «Human Resources Development, Education and Lifelong Learning» in the context of the project “Strengthening Human Resources Research Potential via Doctorate Research” (MIS-5000432), implemented by the State Scholarships Foundation (ΙΚΥ)».
Institutional Review Board Statement
The study was conducted according to the guidelines of Ethical Committee guidelines of Faculty of Animal Science of Agricultural University of Athens (026/10022017).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Calder, P.C.; Bosco, N.; Bourdet-Sicard, R.; Capuron, L.; Delzenne, N.; Doré, J.; Franceschi, C.; Lehtinen, M.J.; Recker, T.; Salvioli, S.; et al. Health relevance of the modification of low grade inflammation in ageing (inflammageing) and the role of nutrition. Ageing Res. Rev. 2017, 40, 95–119. [Google Scholar] [CrossRef] [PubMed]
- Bannenberg, G.; Serhan, C.N. Specialized pro-resolving lipid mediators in the inflammatory response: An update. Biochim. Biophys. Acta. 2010, 1801, 1260–1273. [Google Scholar] [CrossRef]
- Senftleber, N.K.; Nielsen, S.M.; Andersen, J.R.; Bliddal, H.; Tarp, S.; Lauritzen, L.; Furst, D.E.; Suarez-Almazor, M.E.; Lyddiatt, A.; Christensen, R. Marine Oil Supplements for Arthritis Pain: A Systematic Review and Meta-Analysis of Randomized Trials. Nutrients 2017, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Ramsden, C.E.; Ringel, A.; Feldstein, A.E.; Taha, A.Y.; MacIntosh, B.A.; Hibbeln, J.R.; Majchrzak-Hong, S.F.; Faurot, K.R.; Rapoport, S.I.; Cheon, Y.; et al. Lowering dietary linoleic acid reduces bioactive oxidized linoleic acid metabolites in humans. Prostaglandins Leukot. Essent Fatty Acids 2012, 87, 135–141. [Google Scholar] [CrossRef]
- Ilich, J.Z.; Kelly, O.J.; Kim, Y.; Spicer, M.T. Low-grade chronic inflammation perpetuated by modern diet as a promoter of obesity and osteoporosis. Arh. Hig. Rada Toksikol. 2014, 65, 139–148. [Google Scholar] [CrossRef]
- Shrestha, N.; Cuffe, J.S.; Holland, O.J.; Bulmer, A.C.; Hill, M.; Perkins, A.V.; Muhlhausler, B.S.; McAinch, A.J.; Hryciw, D.H. Elevated maternal linoleic acid reduces circulating leptin concentrations, cholesterol levels and male fetal survival in rat model. J. Physiol. 2019, 597, 3349–3361. [Google Scholar] [CrossRef] [PubMed]
- Marchix, J.; Choque, B.; Kouba, M.; Fautrel, A.; Catheline, D.; Legrand, P. Excessive dietary linoleic acid induces proinflammatory markers in rats. J. Nutr. Biochem. 2015, 12, 1434–1441. [Google Scholar] [CrossRef] [PubMed]
- Froyen, E.; Burns-Whitmore, B. The Effects of Linoleic Acid Consumption on Lipid Risk Markers for Cardiovascular Disease in Healthy Individuals: A Review of Human Intervention Trials. Nutrients 2020, 12, 2329. [Google Scholar] [CrossRef]
- Chowdhury, R.; Warnakula, S.; Kunutsor, S.; Crowe, F.; Ward, H.A.; Johnson, L.; Franco, O.H.; Butterworth, A.S.; Forouhi, N.G.; Thompson, S.G.; et al. Association of dietary, circulating, and supplement fatty acids with coronary risk: A systematic review and meta-analysis. Ann. Intern. Med. 2014, 160, 398–406. [Google Scholar] [CrossRef]
- Hooper, L.; Al-Khudairy, L.; Abdelhamid, A.S.; Rees, K.; Brainard, J.S.; Brown, T.J.; Ajabnoor, S.M.; O’Brien, A.T.; Winstanley, L.E.; Donaldson, D.H.; et al. Omega-6 fats for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018, 7. [Google Scholar] [CrossRef]
- Hansen, R. Sesame Profile. Available online: http://www.agmrc.org/commodities__products/grains__oilseeds/sesame_profile (accessed on 19 August 2011).
- Khorrami, S.; Daneshmandi, S.; Mosayeb, G. Sesame seeds essential oil and Sesamol modulate the pro-inflammatory function of macrophages and dendritic cells and promote Th2 response. Med. J. Islam Repub. Iran. 2018, 32, 566–573. [Google Scholar] [CrossRef]
- Wu, M.S.; Aquino, L.; Barbaza, M.; Hsieh, C.L.; Castro-Cruz, K.A.; Yang, L.L.; Tsai, P.W. Anti-Inflammatory and Anticancer Properties of Bioactive Compounds from Sesamum indicum L.-A Review. Molecules 2019, 24, 4426. [Google Scholar] [CrossRef]
- Udomruk, S.; Kaewmool, C.; Pothacharoen, P.; Phitak, T.; Kongtawelert, P. Sesamin suppresses LPS-induced microglial activation via regulation of TLR4 expression. J. Funct. Foods 2018, 49, 32–43. [Google Scholar] [CrossRef]
- Jeng, K.C.; Hou, R.C.; Wang, J.C.; Ping, L.I. Sesamin inhibits lipopolysaccharide-induced cytokine production by suppression of p38 mitogen-activated protein kinase and nuclear factor-κB. Immunol. Lett. 2005, 97, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Sugiyama, H.; Kushimoto, S.; Uchiyama, Y.; Hirano, M.; Nakamura, S. Effects of Sesaminol Feeding on Brain Aβ Accumulation in a Senescence-Accelerated Mouse-Prone 8. J. Agric. Food Chem. 2016, 64, 4908–4913. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, I.; Estep, R.; Robinson, B.; Wong, S.W. Nonhuman Primate Models of Human Immunology. Antioxid. Redox Signal. 2011, 14, 261–273. [Google Scholar] [CrossRef]
- Rosales, C.; Demaurex, N.; Lowell, C.A.; Uribe-Querol, E. Neutrophils: Their Role in Innate and Adaptive Immunity. J. Immunol. Res. 2016, 2016, 1469780. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Majewska, M.; Szczepanik, M. The role of Toll-like receptors (TLR) in innate and adaptive immune responses and their function in immune response regulation. Postepy. Hig. Med. Dosw. 2006, 60, 52–63. [Google Scholar]
- Huang, W.; Hung, M. Beyond NF-κB activation: Nuclear functions of IκB kinase α. J. Biomed. Sci. 2013, 20. [Google Scholar] [CrossRef]
- Roman-Blas, J.A.; Jimenez, S.A. NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthr. Cartil. 2006, 14, 839–848. [Google Scholar] [CrossRef]
- Cho, J.W.; Lee, K.S.; Kim, C.W. Curcumin attenuates the expression of IL-1beta, IL-6, and TNF-alpha as well as cyclin E in TNF-alpha-treated HaCaT cells; NF-κB and MAPKs as potential upstream targets. Int. J. Mol. Med. 2007, 19, 469–474. [Google Scholar] [CrossRef]
- Wong, S.W.; Kwon, M.J.; Choi, A.M.; Kim, H.P.; Nakahira, K.; Hwang, D.H. Fatty acids modulate toll-like receptor 4 activation through regulation of receptor dimerization and recruitment into lipid rafts in a reactive oxygen species-dependent manner. J. Biol. Chem. 2009, 284, 27384–27392. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. Toll-like receptors in innate immunity. Int. Immunol. 2005, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, A.; Yanai, H.; Kondo, S.; Duncan, G.; Negishi, H.; Mizutani, T.; Kano, S.-I.; Honda, K.; Ohba, Y.; Mak, T.W.; et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 2005, 434, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Barton, G.M.; Kagan, J.C. A cell biological view of Toll-like receptor function: Regulation through compartmentalization. Nat. Rev. Immunol. 2009, 9, 535–542. [Google Scholar] [CrossRef]
- Xie, X.; Jin, J.; Zhu, L.; Jie, Z.; Li, Y.; Zhao, B.; Cheng, X.; Li, P.; Sun, S.-C. Cell type-specific function of TRAF2 and TRAF3 in regulating type I IFN induction. Cell Biosci. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Floyd, D.H.; Weilbaecher, K.N.; Green, P.L.; Boris-Lawrie, K. Multiple facets of junD gene expression are atypical among AP-1 family members. Oncogene 2008, 27, 4757–4767. [Google Scholar] [CrossRef]
- Schumacher, A.; Zenclussen, A.C. Effects of heme oxygenase-1 on innate and adaptive immune responses promoting pregnancy success and allograft tolerance. Front. Pharmacol. 2015, 5, 288. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Small Ruminants: Sheep, Goats, Cervids, and New World Camelids; The National Academies Press: Washington, DC, USA, 2007. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Tsiplakou, E.; Mavrommatis, A.; Skliros, D.; Sotirakoglou, K.; Flemetakis, E.; Zervas, G. The effects of dietary supplementation with rumen-protected amino acids on the expression of several genes involved in the immune system of dairy sheep. J. Anim. Physiol. Anim. Nutr. 2018, 102, 1437–1449. [Google Scholar] [CrossRef]
- Tsiplakou, E.; Mavrommatis, A.; Skliros, D.; Righi, F.; Flemetakis, E. The impact of rumen-protected amino acids on the expression of key- genes involved in the innate immunity of dairy sheep. PLoS ONE 2020, 15, e0233192. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Vorachek, W.; Hugejiletu Bobe, G.; Hall, J. Reference gene selection for quantitative PCR studies in sheep neutrophils. Int. J. Mol. Sci. 2013, 14, 11484–11495. [Google Scholar] [CrossRef]
- Rosales, C.; Lowell, C.A.; Schnoor, M.; Uribe-Querol, E. Neutrophils: Their Role in Innate and Adaptive Immunity. J. Immunol. Res. 2017, 2017. [Google Scholar] [CrossRef]
- Wu, B.; Peisley, A.; Tetrault, D. Molecular imprinting as a signal activation mechanism of the viral RNA sensor RIG-I. Mol. Cell 2014, 55, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Sun, S.C. Ubiquitin signaling immune responses. Cell Res. 2016, 26, 457–483. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Zhang, H.; Zhao, Z.; Peng, Z.; Wang, Z.; Liu, G.; Li, X. Non-Esterified Fatty Acids Over-Activate the TLR2/4-NF-Κb Signaling Pathway to Increase Inflammatory Cytokine Synthesis in Neutrophils from Ketotic Cows. Cell Physiol. Biochem. 2018, 48, 827–837. [Google Scholar] [CrossRef]
- Roldan-Montes, V.; Cardoso, D.F.; Hurtado-Lugo, N.A.; do Nascimento, A.V.; Santos, D.; Scalez, D.; de Freitas, A.C.; Herrera, A.C.; Albuquerque, L.G.; de Camargo, G.; et al. Polymorphisms in TLR4 Gene Associated with Somatic Cell Score in Water Buffaloes (Bubalus bubalis). Front. Vet. Sci. 2020, 7, 568249. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Fatty acids Long-chain fatty acids and inflammation. Proc. Nutr. Soc. 2012, 71, 274–289. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Ma, L.; Gong, X.; Kuang, G.; Jiang, R.; Chen, R.; Wan, J. Sesamin ameliorates lipopolysaccharide/d-galactosamine-induced fulminant hepatic failure by suppression of Toll-like receptor 4 signaling in mice. Biochem. Biophys. Res. Commun. 2015, 461, 230–236. [Google Scholar] [CrossRef]
- Harikumar, K.B.; Sung, B.; Tharakan, S.T.; Pandey, M.K.; Joy, B.; Guha, S.; Krishnan, S.; Aggarwal, B.B. Sesamin manifests chemopreventive effects through the suppression of NF-kappa B-regulated cell survival, proliferation, invasion, and angiogenic gene products. Mol. Cancer Res. 2010, 8, 751–761. [Google Scholar] [CrossRef]
- Selvarajan, K.; Narasimhulu, C.A.; Bapputty, R.; Parthasarathy, S. Anti-inflammatory and antioxidant activities of the nonlipid (aqueous) components of sesame oil: Potential use in atherosclerosis. J. Med. Food 2015, 18, 393–402. [Google Scholar] [CrossRef]
- Bulgari, O.; Dong, X.; Roca, A.L.; Caroli, A.M.; Loor, J.J. Innate immune responses induced by lipopolysaccharide and lipoteichoic acid in primary goat mammary epithelial cells. J. Anim. Sci. Biotechnol. 2017, 8, 29–38. [Google Scholar] [CrossRef]
- Rashidi, N.; Mirahmadian, M.; Jeddi-Tehrani, M.; Rezania, S.; Ghasemi, J.; Kazemnejad, S. Lipopolysaccharide and lipoteichoic acid-mediated pro-inflammatory cytokine production and modulation of TLR2, TLR4 and MyD88 expression in human endometrial cells. J. Reprod. Infertil. 2005, 16, 72–81. [Google Scholar]
- Wu, Z.; Li, F.; Liu, D.; Xue, H.; Zhao, X. Novel Type XII Staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase CcrC2. Antimicrob. Agents Chemother. 2015, 59, 7597–7601. [Google Scholar] [CrossRef]
- Qian, C.; Cao, X. Regulation of Toll-like receptor signaling pathways in innate immune responses. Ann. N. Y. Acad. Sci. 2012, 1283, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Dong, C. MAP kinases in immune responses. Cell Mol. Immunol. 2005, 2, 20–27. [Google Scholar]
- Lim, M.X.; Png, C.W.; Tay, C.Y.; Teo, J.D.; Jiao, H.; Lehming, N.; Tan, K.S.; Zhang, Y. Differential regulation of proinflammatory cytokine expression by mitogen-activated protein kinases in macrophages in response to intestinal parasite infection. Infect. Immun. 2014, 82, 4789–4801. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Wolint, P.; Walton, S.; Schwarz, K.; Oxenius, A. Differential role of IL-2R signaling for CD8+ T cell responses in acute and chronic viral infections. Eur. J. Immunol. 2007, 37, 1502–1512. [Google Scholar] [CrossRef]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro-and anti-inflammatory properties of the cytokine inteleukin-6. Biochim. Biophys. Acta 2011, 1813, 878–888. [Google Scholar] [CrossRef]
- Deng, P.; Wang, C.; Chen, L.; Wang, C.; Du, Y.; Yan, X.; Chen, M.; Yang, G.; He, G. Sesamin induces cell cycle arrest and apoptosis through the inhibition of signal transducer and activator of transcription 3 signalling in human hepatocellular carcinoma cell line HepG2. Biol. Pharm. Bull. 2013, 36, 1540–1548. [Google Scholar] [CrossRef]
- Kong, X.; Ma, M.Z.; Zhang, Y.; Weng, M.Z.; Gong, W.; Guo, L.Q.; Zhang, J.X.; Wang, G.D.; Su, Q.; Quan, Z.W.; et al. Differentiation therapy: Sesamin as an effective agent in targeting cancer stem-like side population cells of human gallbladder carcinoma. BMC Complement. Altern Med. 2014, 14, 254. [Google Scholar] [CrossRef]
- Fanhchaksai, K.; Kodchakorn, K.; Pothacharoen, P.; Kongtawelert, P. Effect of sesamin against cytokine production from influenza type A H1N1-induced peripheral blood mononuclear cells: Computational and experimental studies. In Vitro Cell Dev. Biol. Anim. 2015, 52, 107–119. [Google Scholar] [CrossRef]
- Meier-Trummer, C.S.; Rehrauer, H.; Franchini, M.; Patrignani, A.; Wagner, U.; Ackermann, M. Malignant catarrhal fever of cattle is associated with low abundance of IL-2 transcript and a predominantly latent profile of ovine Herpesvirus 2 gene expression. PLoS ONE 2009, 4, 6265. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.C.; Benavides, J.; Grant, D.M.; Todd, H.; Thomson, J.; Puri, V.; Nath, M.; Haig, D.M. Host gene expression changes in cattle infected with Alcelaphine herpesvirus 1. Virus Res. 2012, 169, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Narasimhulu, C.A.; Selvarajan, K.; Litvinov, D.; Parthasarathy, S. Anti-atherosclerotic and anti-inflammatory actions of sesame oil. J. Med. Food 2015, 18, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Yu, Y.; Hu, S.; Zhang, J.; Yang, H.; Han, B.; Cheng, Y.; Luo, X. Sesamin ameliorates hepatic steatosis and inflammation in rats on a high-fat diet via LXRα and PPARα. Nutr. Res. 2016, 36, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Luo, J.; Chen, D.; Xu, H.; Shi, H.; Jing, X.; Zang, W. CD36 regulates lipopolysaccharide-induced signalling pathways and mediates the internalization of Escherichia coli in cooperation with TLR4 in goat mammary gland epithelial cells. Sci. Rep. 2016, 6, 23132. [Google Scholar] [CrossRef] [PubMed]
- Hacker, H.; Tseng, P.H.; Karin, M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 2011, 7, 457–468. [Google Scholar] [CrossRef]
- Yang, X.D.; Sun, S.C. Targeting signaling factors for degradation an emerging mechanism for TRAF functions. Immunol. Rev. 2015, 266, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Javan, M.R.; Zamani, M.R.; Aslani, S.; Dargahi Abbasabad, G.; Beirami Khalaj, M.; Serati-Nouri, H. Cytokine Modulatory Effects of Sesamum Indicum Seeds Oil Ameliorate Mice with Experimental Autoimmune Encephalomyelitis. Arch. Asthma Allergy Immunol. 2017, 1, 86–93. [Google Scholar] [CrossRef]
- Faraji, F.; Hashemi, M.; Ghiasabadi, A.; Davoudian, S.; Talaie, A.; Ganji, A.; Mosayebi, G. Combination therapy with interferon beta-1a and sesame oil in multiple sclerosis. Complement. Ther. Med. 2019, 45, 275–279. [Google Scholar] [CrossRef]
- Gilbert, F.B.; Cunha, P.; Jensen, K.; Glass, E.J.; Foucras, G.; Robert-Granie, C.; Rupp, R.; Rainard, P. Differential response of bovine mammary epithelial cells to Staphylococcus aureus or Escherichia coli agonists of the innate immune system. Vet. Res. 2013, 44, 40. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Urban, J.F.; Anthony, R.M.; Sun, R.; Stiltz, J.; van Rooijen, N.; Wynn, T.A.; Gause, W.C.; Shea-Donohue, T. Th2 Cytokine-Induced Alterations in Intestinal Smooth Muscle Function Depend on Alternatively Activated Macrophages. Gastroenterology 2008, 135, 217–225. [Google Scholar] [CrossRef]
- Alhussien, M.; Manjari, P.; Sheikh, A.A.; Mohammed Seman, M.; Reddi, S.; Mohanty, A.K.; Dang, A.K. Immunological attributes of blood and milk neutrophils isolated from crossbred cows during different physiological conditions. Czech J. Anim. Sci. 2016, 61, 223–231. [Google Scholar] [CrossRef]
- Jarczak, J.; Kaba, J.; Reczyńska, D.; Bagnicka, E. Impaired expression of cytokines as a result of viral infections with an emphasis on small ruminant lentivirus infection in goats. Viruses 2016, 8, 186. [Google Scholar] [CrossRef] [PubMed]
- Machugh, D.E.; Taraktsoglou, M.; Killick, K.E.; Nalpas, N.C.; Browne, J.A.; De Park, S.; Magee, D.A. Pan-genomic analysis of bovine monocyte-derived macrophage gene expression in response to in vitro infection with Mycobacterium avium subspecies paratuberculosis. Vet. Res. 2012, 43, 25. [Google Scholar] [CrossRef] [PubMed]
- Galvao, K.N.; Pighetti, G.M.; Cheong, S.H.; Nydam, D.V.; Gilbert, R.O. Association between interleukin-8 receptor-α (CXCR1) polymorphism and disease incidence, production, reproduction, and survival in Holstein cows. J. Dairy Sci. 2011, 94, 2083–2091. [Google Scholar] [CrossRef] [PubMed]
- DeForge, L.E.; Preston, A.M.; Takeuchi, E.; Kenney, J.; Boxer, L.A.; Remick, D.G. Regulation of interleukin 8 gene expression by oxidant stress. J. Biol. Res. 1993, 268, 25568–25576. [Google Scholar]
- Lehrke, M.; Millington, S.C.; Lefterova, M.; Cumaranatunge, R.G.; Szapary, P.; Wilensky, R.; Rader, D.J.; Lazar, M.A.; Reilly, M.P. CXCL16 Is a Marker of Inflammation, Atherosclerosis, and Acute Coronary Syndromes in Humans. J. Am. Coll. Cardiol. 2007, 49, 442–449. [Google Scholar] [CrossRef]
- Shariat, S.Z.A.S.; Mostafavi, S.A.; Khakpour, F. Antioxidant effects of vitamins c and e on the low-density lipoprotein oxidation mediated by myeloperoxidase. Iran. Biomed. J. 2013, 17, 22–28. [Google Scholar] [CrossRef]
- Paine, A.; Eiz-Vesper, B.; Blasczyk, R.; Immenschuh, S. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochem. Pharmacol. 2010, 80, 1895–1903. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, M.; Ohnishi, M.; Shiratsuchi, A.; Kawakami, T.; Takahashi, M.; Motomura, M.; Egusa, K.; Urasaki, T.; Inoue, A. Sesamin increases heme oxygenase-1 protein in RAW 264.7 macrophages through inhibiting its ubiquitination process. Eur. J. Pharmacol. 2014, 741, 214–221. [Google Scholar] [CrossRef]
- Carasi, P.; Racedo, S.M.; Jacquot, C.; Elie, A.M.; Serradell, M.L.; Urdaci, M.C. Enterococcus durans EP1 a Promising Anti-inflammatory Probiotic Able to Stimulate sIgA and to Increase Faecalibacterium prausnitzii Abundance. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-S.; Chau, L.-Y. Heme oxygenase-1 mediates the anti-inflammatory effect of interleukin-10 in mice. Nat. Med. 2002, 8, 240–246. [Google Scholar] [CrossRef]
- Hock, T.D.; Liby, K.; Wright, M.M.; McConnell, S.; Schorpp-Kistner, M.; Ryan, T.M.; Agarwal, A. JunB and JunD regulate human heme oxygenase-1 gene expression in renal epithelial cells. J. Biol. Chem. 2007, 282, 6875–6886. [Google Scholar] [CrossRef]
- Raghunath, A.; Nagarajan, R.; Sundarraj, K.; Panneerselvam, L.; Perumal, E. Genome-wide identification and analysis of Nrf2 binding sites—Antioxidant response elements in zebrafish. Toxicol. Appl. Pharmacol. 2018, 360, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T. The Nuclear Factor NF- B Pathway in Inflammation. Cold Spring Harbor. Perspect. Biol. 2009, 1. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yang, Y.; Chernishof, V.; Loo, R.R.O.; Jang, H.; Tahk, S.; Yang, R.; Mink, S.; Shultz, D.; Bellone, C.J.; et al. Proinflammatory Stimuli Induce IKKα-Mediated Phosphorylation of PIAS1 to Restrict Inflammation and Immunity. Cell 2007, 129, 903–914. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).