Glycolysis Combined with Core Pluripotency Factors to Promote the Formation of Chicken Induced Pluripotent Stem Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Cell Culture
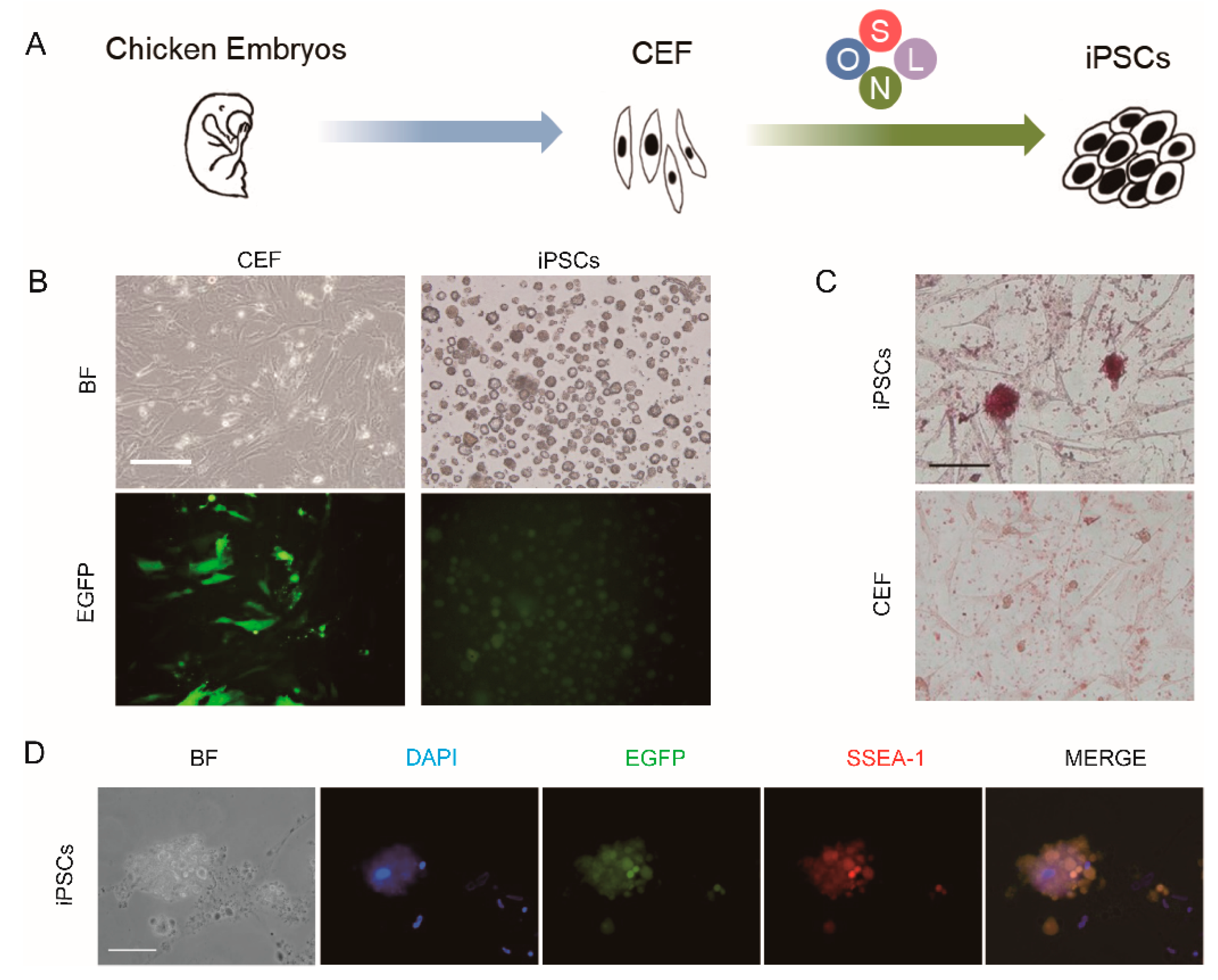
2.3. Generation of Chicken iPSCs from CEF Cultures
2.4. Alkaline Phosphatase Staining
2.5. Immunofluorescence Staining
2.6. Flow Cytometry Analysis
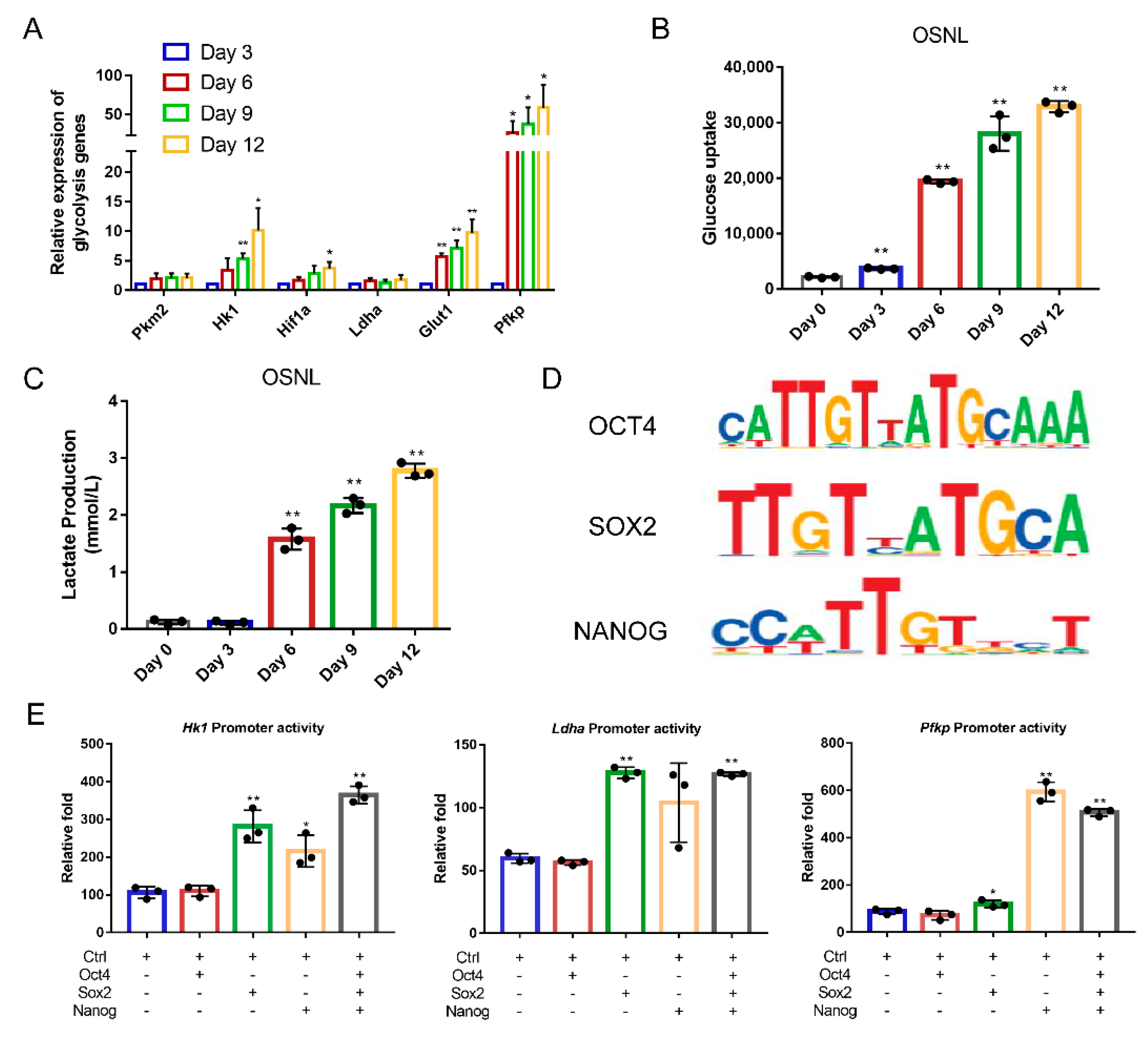
2.7. Glucose Uptake Assay
2.8. Lactate Production Assay
2.9. JC-1 Mitochondrial Membrane Potential Detection
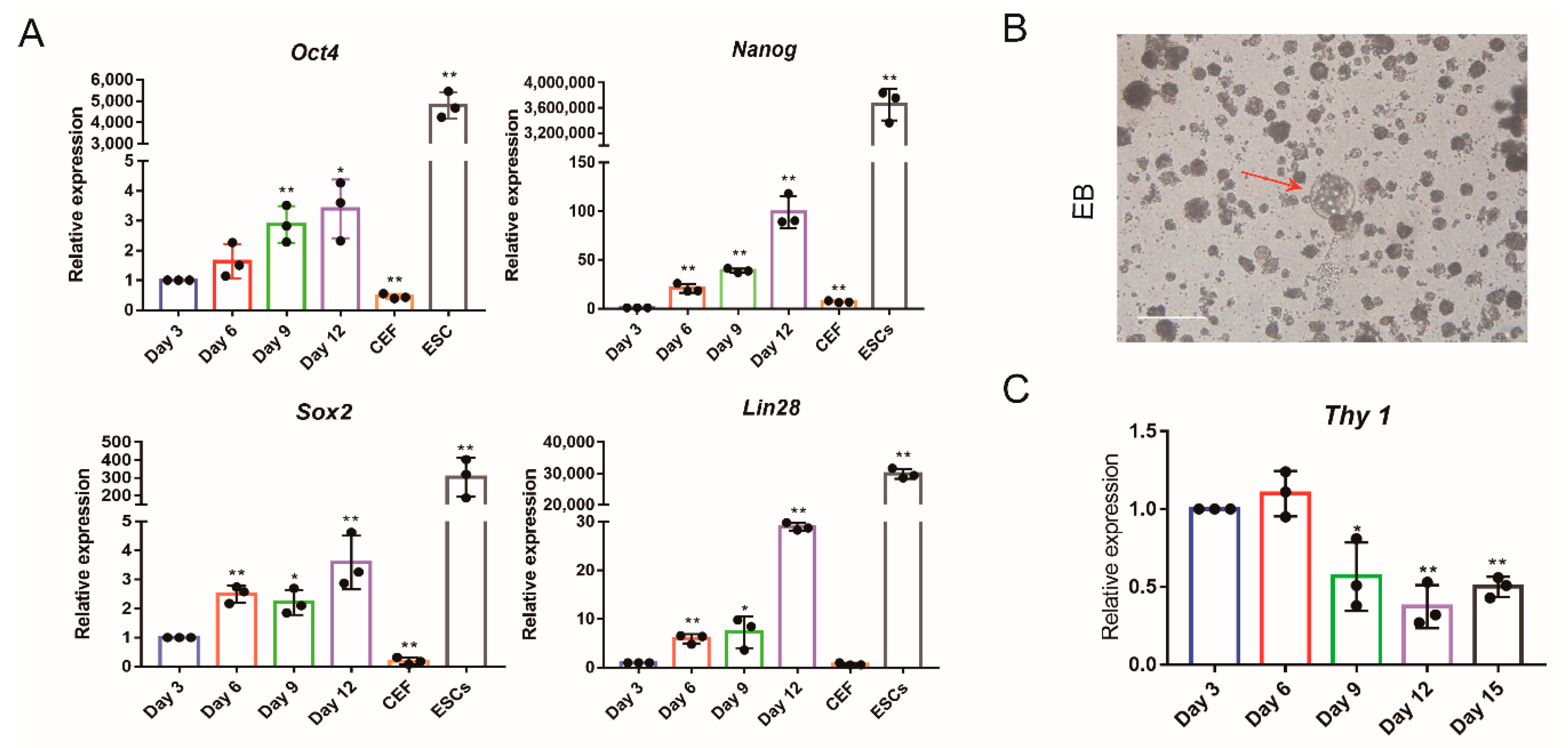
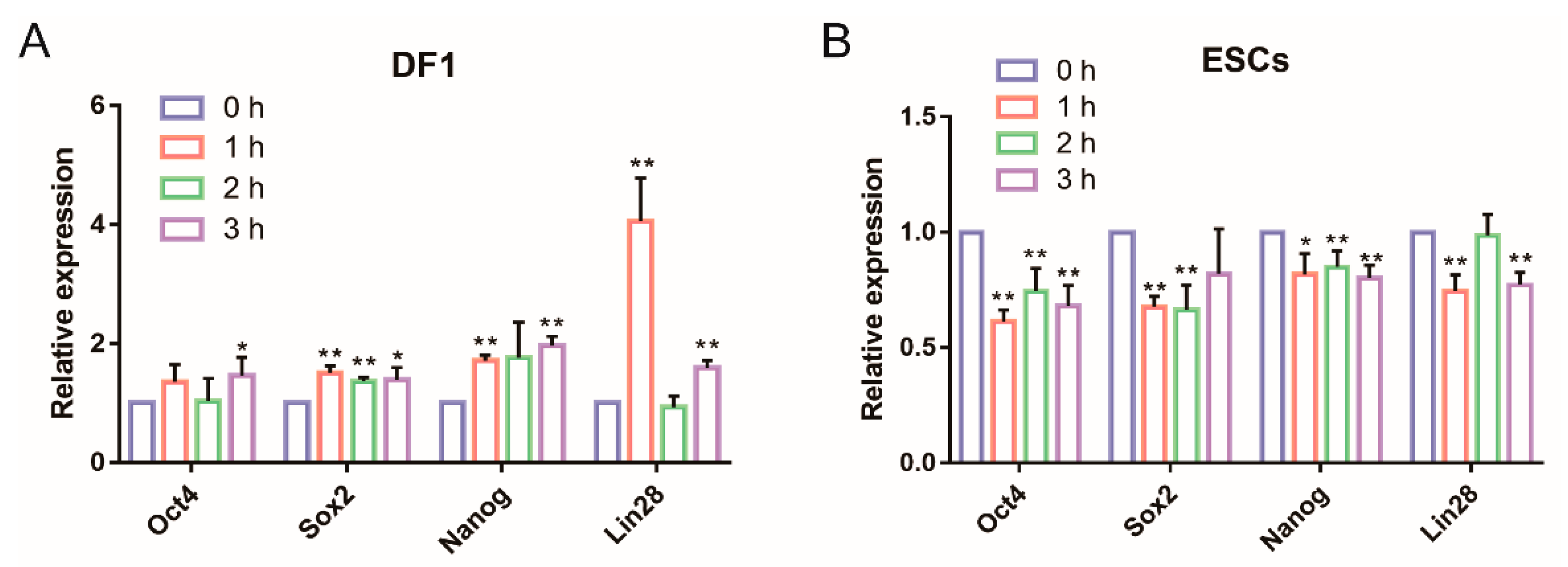
2.10. qRT-PCR
2.11. Dual Luciferase Reporter Assay
2.12. Embryoid Bodies Formation In Vitro
2.13. Statistical Analysis
3. Results
3.1. Exogenous OSNL Factors Activate Glycolysis
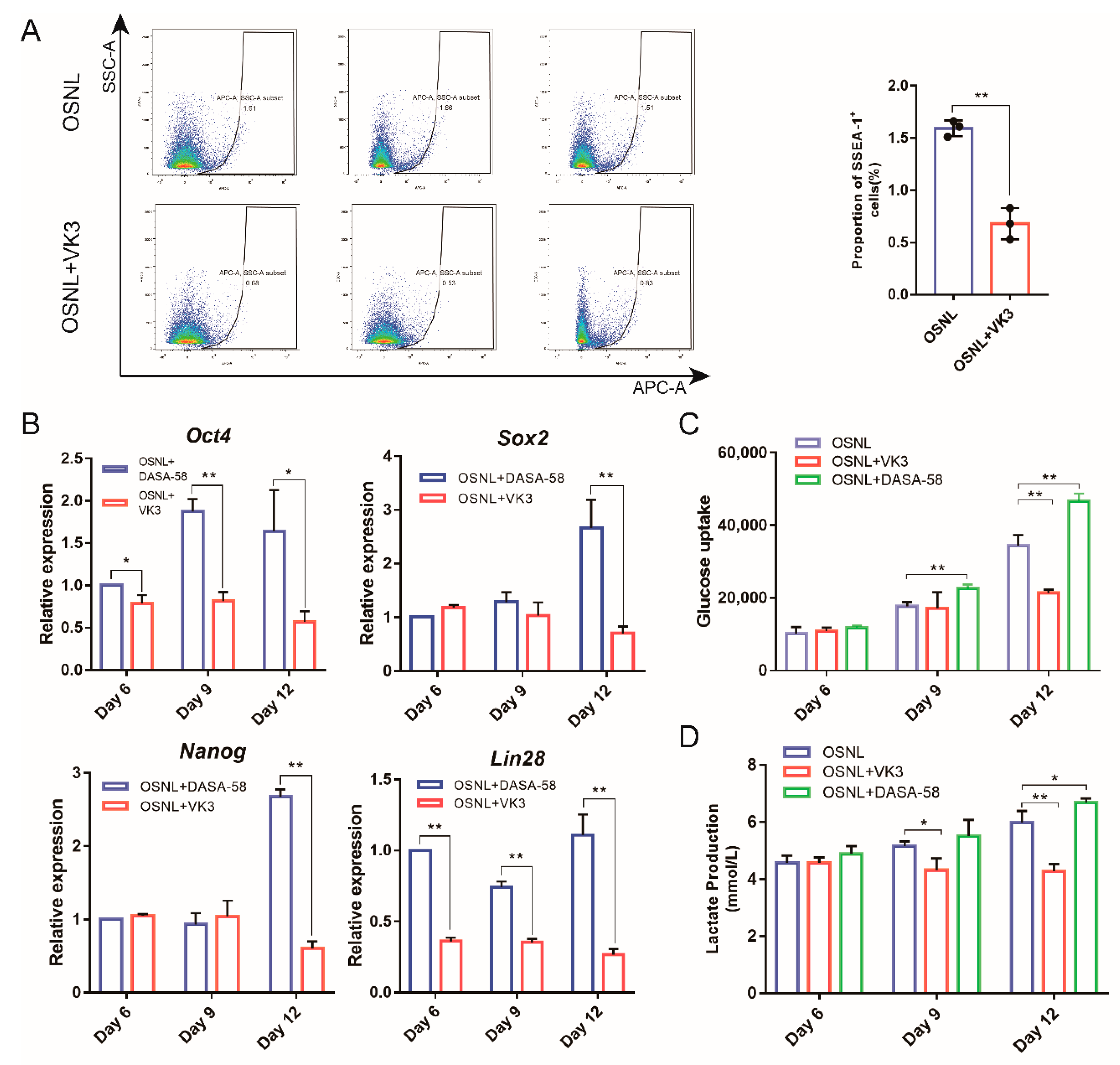
3.2. Glycolysis Activates the Expression of Endogenous Pluripotent Genes to Promote the Formation of Chicken iPSCs
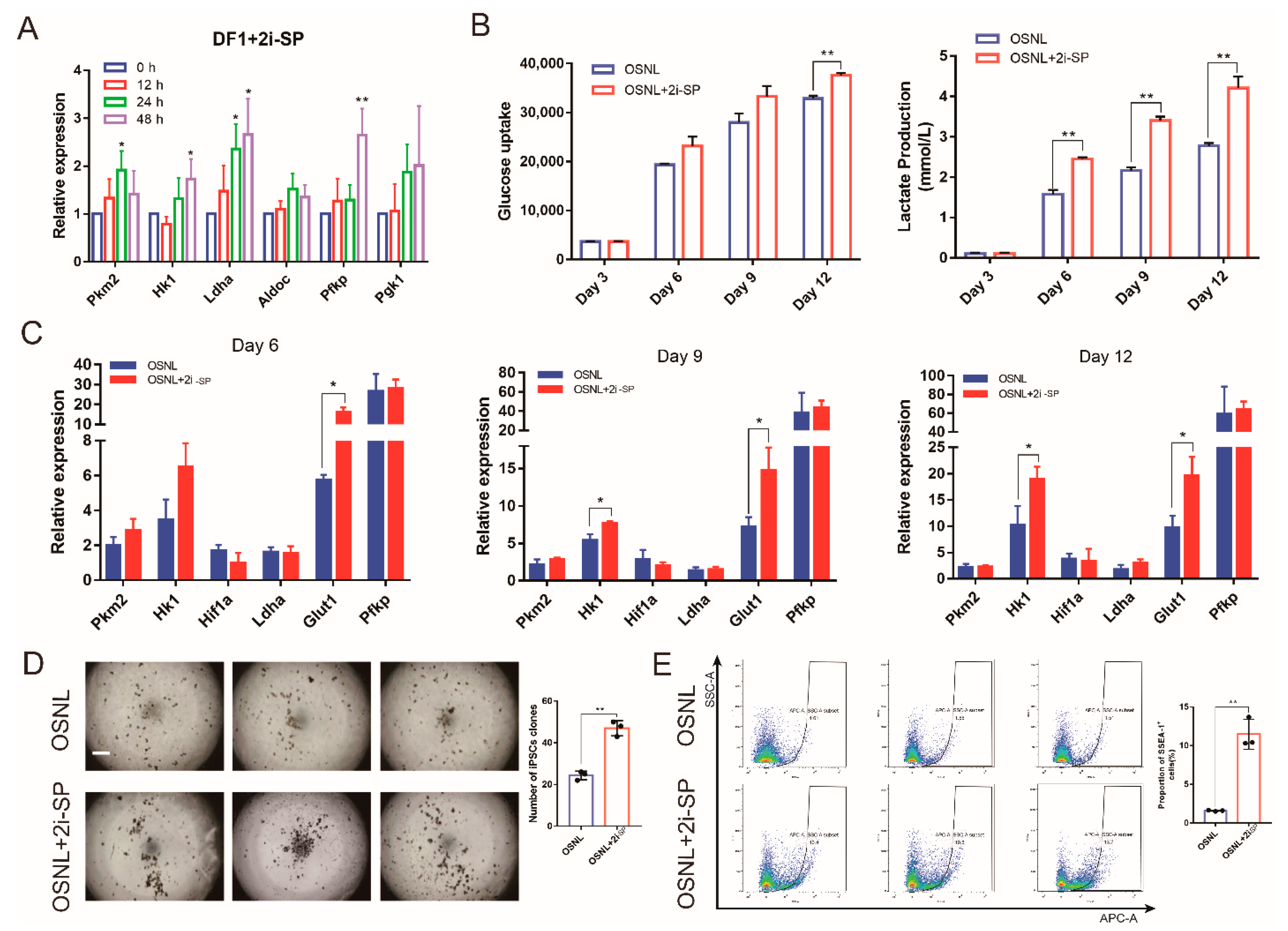
3.3. 2i-SP Increases Glycolysis Levels to Promote Chicken iPSC Formation
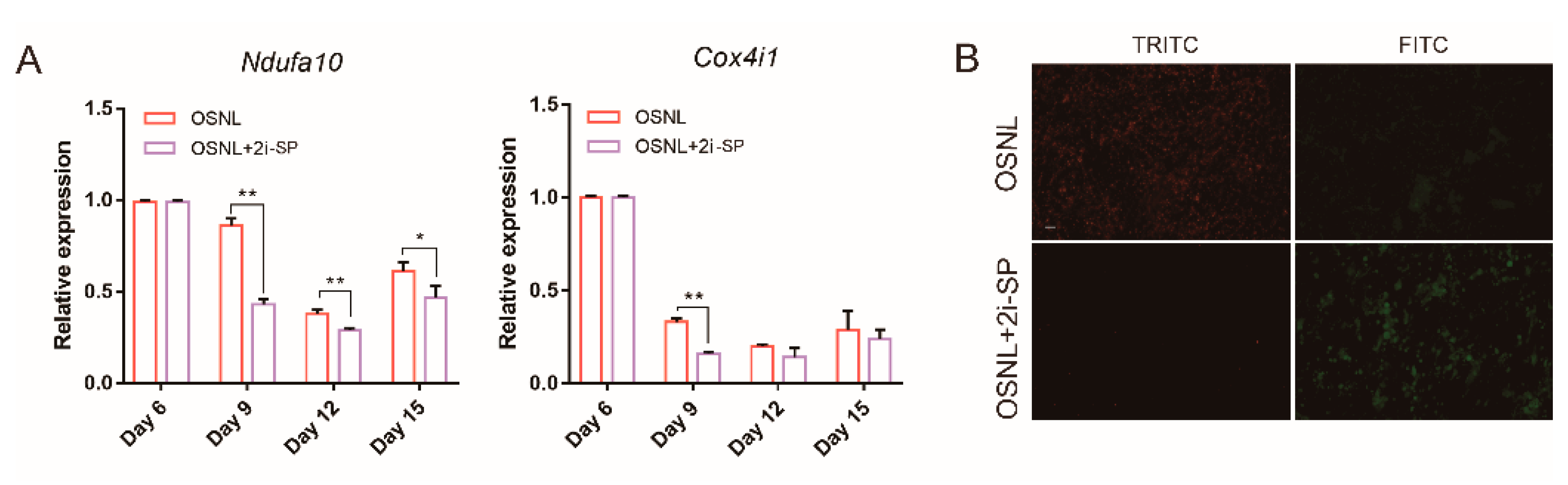
3.4. 2i-SP Inhibits Oxidative Phosphorylation to Promote Chicken iPSC Formation
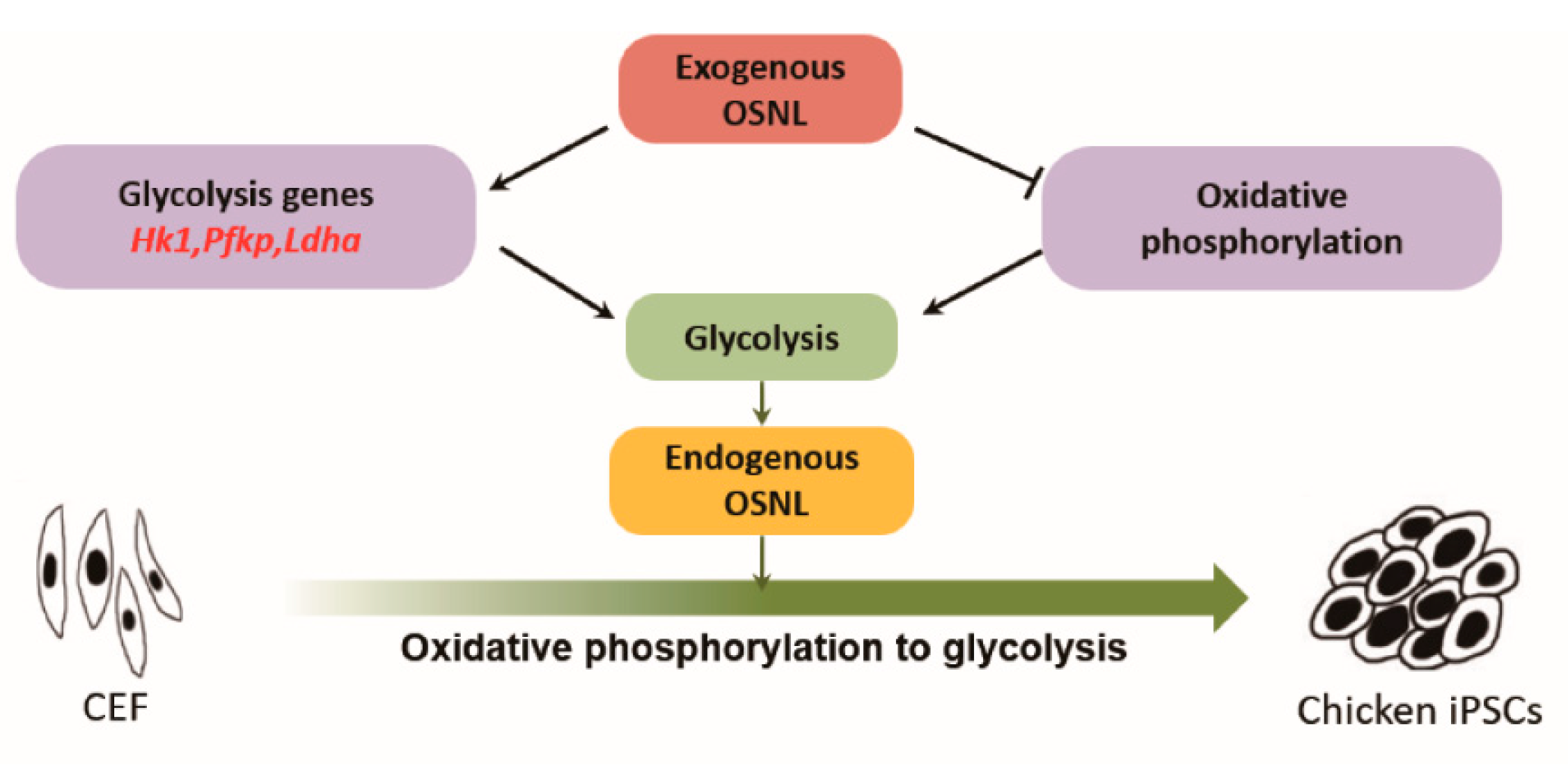
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reynaud, G. Capacités reproductrices et descendance de poulets ayant subi un transfert de cellules germinales primordiales durant la vie embryonnaire. Dev. Genes Evol. 1976, 179, 85–110. [Google Scholar] [CrossRef]
- Wentworth, B.C.; Tsai, H.; Hallett, J.H.; Gonzales, D.S.; Rajcic-Spasojevic, G. Manipulation of Avian Primordial Germ Cells and Gonadal Differentiation. Poult. Sci. 1989, 68, 999–1010. [Google Scholar] [CrossRef]
- Chang, I.; Jeong, D.K.; Hong, Y.H.; Park, T.S.; Moon, Y.K.; Ohno, T.; Han, J.Y. Production of germline chimeric chickens by transfer of cultured primordial germ cells. Cell Biol. Int. 1997, 21, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Chang, I.; Yoshiki, A.; Kusakabe, M.; Tajima, A.; Chikamune, T.; Naito, M.; Ohno, T. Germ line chimera produced by transfer of cultured chick primordial germ cells. Cell Biol. Int. 1995, 19, 569–576. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; West, F.D.; Jordan, B.J.; Mumaw, J.L.; Jordan, E.T.; Gallegos-Cardenas, A.; Beckstead, R.B.; Stice, S.L. Avian-induced pluripotent stem cells derived using human reprogramming factors. Stem Cells Dev. 2012, 21, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R. Establishment the System of Chicken CEF Reprogramming Induce to PGC and Research of its Migration Homing Fuction. Master’s Thesis, Yangzhou University, Yangzhou, China, 2019. [Google Scholar]
- Varum, S.; Rodrigues, A.S.; Moura, M.B.; Momcilovic, O.; Iv, C.A.E.; Ramalho-Santos, J.; van Houten, B.; Schatten, G. Energy Metabolism in Human Pluripotent Stem Cells and Their Differentiated Counterparts. PLoS ONE 2011, 6, e20914. [Google Scholar] [CrossRef]
- Panopoulos, A.D.; Yanes, O.; Ruiz, S.; Kida, Y.S.; Diep, D.; Tautenhahn, R.; Herrerías, A.; Batchelder, E.M.; Plongthongkum, N.; Lutz, M.; et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2011, 22, 168–177. [Google Scholar] [CrossRef]
- Folmes, C.D.L.; Nelson, T.J.; Martinez-Fernandez, A.; Arrell, D.K.; Lindor, J.Z.; Dzeja, P.P.; Ikeda, Y.; Perez-Terzic, C.; Terzic, A. Somatic Oxidative Bioenergetics Transitions into Pluripotency-Dependent Glycolysis to Facilitate Nuclear Reprogramming. Cell Metab. 2011, 14, 264–271. [Google Scholar] [CrossRef]
- Goldman, A. Isolation of Fibroblasts from Chicken Embryos. Cold Spring Harb. Protoc. 2006, 2006, pdb.prot4475. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, H.; Shi, Q.; Zhang, Z.; Zheng, M.; Wang, D.; Li, B. Isolation of chicken embryonic stem cells and exploration of their chimeric preparation conditions. China J. Anim. Husb. 2012, 48, 4. [Google Scholar]
- Trosko, J.E. Induction of iPS cells and of cancer stem cells: The stem cell or reprogramming hypothesis of cancer? Anat. Rec. 2014, 297, 1611–1673. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Xia, W.; Liu, J.-C.; Yang, J.-Y.; Lee, D.-F.; Xia, J.; Bartholomeusz, G.; Li, Y.; Pan, Y.; Li, Z.; et al. Erk Associates with and Primes GSK-3β for Its Inactivation Resulting in Upregulation of β-Catenin. Mol. Cell 2005, 19, 159–170. [Google Scholar] [CrossRef]
- Yu, D.; Wang, Y.; Gao, Y.; Liu, C. Effect of Akt/GSK-3β/Snail signaling pathway on EMT in A549/DDP cells mediated by TGF-β1. Chin. J. Pathophysiol. 2018, 34, 1124–1128. [Google Scholar]
- Liu, C.-C.; Wang, H.; Wang, W.-D.; Wang, L.; Liu, W.-J.; Wang, J.-H.; Geng, Q.-R.; Lu, Y. ENO2 Promotes Cell Proliferation, Glycolysis, and Glucocorticoid-Resistance in Acute Lymphoblastic Leukemia. Cell. Physiol. Biochem. 2018, 46, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Chambers, I.; Pollard, S.; Smith, A. Nanog promotes transfer of pluripotency after cell fusion. Nat. Cell Biol. 2006, 441, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Rosner, M.H.; Vigano, M.A.; Ozato, K.; Timmons, P.M.; Poirie, F.; Rigby, P.W.J.; Staudt, L.M. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nat. Cell Biol. 1990, 345, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Rodda, D.J.; Chew, J.-L.; Lim, L.-H.; Loh, Y.-H.; Wang, B.; Ng, H.-H.; Robson, P. Transcriptional Regulation of Nanog by OCT4 and SOX2. J. Biol. Chem. 2005, 280, 24731–24737. [Google Scholar] [CrossRef]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef]
- Li, Y.; McClintick, J.; Zhong, L.; Edenberg, H.J.; Yoder, M.C.; Chan, R.J. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood 2005, 105, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional Expression Cloning of Nanog, a Pluripotency Sustaining Factor in Embryonic Stem Cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef]
- Richards, M.; Tan, S.; Tan, J.H.; Chan, W.K.; Bongso, A. The Transcriptome Profile of Human Embryonic Stem Cells as Defined by SAGE. Stem Cells 2004, 22, 51–64. [Google Scholar] [CrossRef]
- Hayashi, K.; Saitou, M. Generation of Functional Primordial Germ Cells from Pluripotent Stem Cells. J. Mamm. Ova Res. 2012, 29, 2–10. [Google Scholar] [CrossRef]
- Kim, H.; Jang, H.; Kim, T.W.; Kang, B.-H.; Lee, S.E.; Jeon, Y.K.; Chung, D.H.; Choi, J.; Shin, J.; Cho, E.-J.; et al. Core Pluripotency Factors Directly Regulate Metabolism in Embryonic Stem Cell to Maintain Pluripotency. Stem Cells 2015, 33, 2699–2711. [Google Scholar] [CrossRef] [PubMed]
- Liu, X. Functions of Mitochondrial Biogenesis and Metabolism Related Genes in Mouse iPSCs Induction and Insight into Mechanism. Master’s Thesis, University of Science and Technology of China, Hefei, China, 2011. [Google Scholar]
- Li, X.-B.; Gu, J.-D.; Zhou, Q.-H. Review of aerobic glycolysis and its key enzymes—New targets for lung cancer therapy. Thorac. Cancer 2015, 6, 17–24. [Google Scholar] [CrossRef]
- Esteban, M.A.; Wang, T.; Qin, B.; Yang, J.; Qin, D.; Cai, J.; Li, W.; Weng, Z.; Chen, J.; Ni, S.; et al. Vitamin C Enhances the Generation of Mouse and Human Induced Pluripotent Stem Cells. Cell Stem Cell 2010, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Snitow, M.; Chen, A.E.; Melton, D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008, 26, 795–797. [Google Scholar] [CrossRef]
- Lin, T.; Ambasudhan, R.; Yuan, X.; Li, W.; Hilcove, S.; Abujarour, R.; Lin, X.; Hahm, H.S.; Hao, E.; Hayek, A.; et al. A chemical platform for improved induction of human iPSCs. Nat. Methods 2009, 6, 805–808. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, Y.; Gordon, A.; Wang, Z.Z.; Qian, Z.; Wu, W.-S. Efficient Generation of Fully Reprogrammed Human iPS Cells via Polycistronic Retroviral Vector and a New Cocktail of Chemical Compounds. PLoS ONE 2011, 6, e26592. [Google Scholar] [CrossRef]
- Ichida, J.K.; Blanchard, J.; Lam, K.; Son, E.Y.; Chung, J.E.; Egli, D.; Loh, K.M.; Carter, A.C.; Di Giorgio, F.P.; Koszka, K.; et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in re-programming by inducing Nanog. Cell Stem Cell 2009, 5, 491–503. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward Primers (5′–3′) | Reverse Primers (5′–3′) |
|---|---|---|
| Oct4 UTR | CGGGATCTCCATGAACAACAG | CTGGCCCCAGGCAGGTAA |
| Sox2 UTR | CATATGTAAGACAAAGGGG | AAGGTCCAGAATTTCTAATAA |
| Nanog UTR | GTATGCAACCAGCTCACC | TAGTAGTGTCCGCACCTAAC |
| Lin28 UTR | AAAGCCAATGCCAAGTGA | CAAACAAACCCAAAGATACG |
| Oct4 | TGCAATGCAGAGCAAGTGCTGG | ACTGGGCTTCACACATTTGCGG |
| Sox2 | ACTCGGCCGCGAACAACCAG | GCCCCGAGCCGTTTGCTGAT |
| Nanog | TGCACACCAGGCTTACAGCAGTG | TGCTGGGTGTTGCAGCTTGTTC |
| Lin28 | ACACCCGTCTGGGCAACGAC | CCCGCTGGATGCGCTTCATC |
| Thy1 | GACTGCCGCTATGAGAACA | GGAGACGCTGATGGTGCT |
| Hk1 | CCTCTTGGCTTCACATTC | TTCACAGTTTGGGTCTTCAT |
| Pkm2 | GGCACCCACGAGTATCAT | CATTGTCCAGCGTCACTTT |
| Pfkp | GCCACAACAAACCTATAACA | ATCAAAGGCAGACGAACA |
| Ldha | TGGGCATCCATCCTCTGA | CCTGCTTGTGAACCTCCT |
| Glut1 | TGTTTGGCTTGGACTTGAT | TCTTGAGGACGCTCTTGG |
| Hif1a | TTGACAAGGCATCCATTA | TCCTCAGAAAGCACCATA |
| Aldoc | CTGACGACGGCACTCCTTT | GACAGCCCATCCAGACCCT |
| Pgk1 | AGGGCTGCATCACCATTA | CCACCTCCAGTGCTAACG |
| Ndufa10 | TGAGCGAGACCAGTGAAA | GGAATGAACCGAGGGATA |
| Cox4i1 | TTTCAGCCATCCAGCATA | GAAGAGCACTCCACCAAG |
| β-actin | CAGCCATCTTTCTTGGGTAT | CTGTGATCTCCTTCTGCATCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Zhang, C.; Zhao, R.; Jiang, J.; Shi, X.; Zhang, M.; Sun, H.; Zuo, Q.; Zhang, Y.; Song, J.; et al. Glycolysis Combined with Core Pluripotency Factors to Promote the Formation of Chicken Induced Pluripotent Stem Cells. Animals 2021, 11, 425. https://doi.org/10.3390/ani11020425
Yuan X, Zhang C, Zhao R, Jiang J, Shi X, Zhang M, Sun H, Zuo Q, Zhang Y, Song J, et al. Glycolysis Combined with Core Pluripotency Factors to Promote the Formation of Chicken Induced Pluripotent Stem Cells. Animals. 2021; 11(2):425. https://doi.org/10.3390/ani11020425
Chicago/Turabian StyleYuan, Xia, Chen Zhang, Ruifeng Zhao, Jingyi Jiang, Xiang Shi, Ming Zhang, Hongyan Sun, Qisheng Zuo, Yani Zhang, Jiuzhou Song, and et al. 2021. "Glycolysis Combined with Core Pluripotency Factors to Promote the Formation of Chicken Induced Pluripotent Stem Cells" Animals 11, no. 2: 425. https://doi.org/10.3390/ani11020425
APA StyleYuan, X., Zhang, C., Zhao, R., Jiang, J., Shi, X., Zhang, M., Sun, H., Zuo, Q., Zhang, Y., Song, J., Chen, G., & Li, B. (2021). Glycolysis Combined with Core Pluripotency Factors to Promote the Formation of Chicken Induced Pluripotent Stem Cells. Animals, 11(2), 425. https://doi.org/10.3390/ani11020425






