Genetic Variability of Methane Production and Concentration Measured in the Breath of Polish Holstein-Friesian Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. CH4 Measurements
3.2. Variance Components
3.3. Heritability Estimates
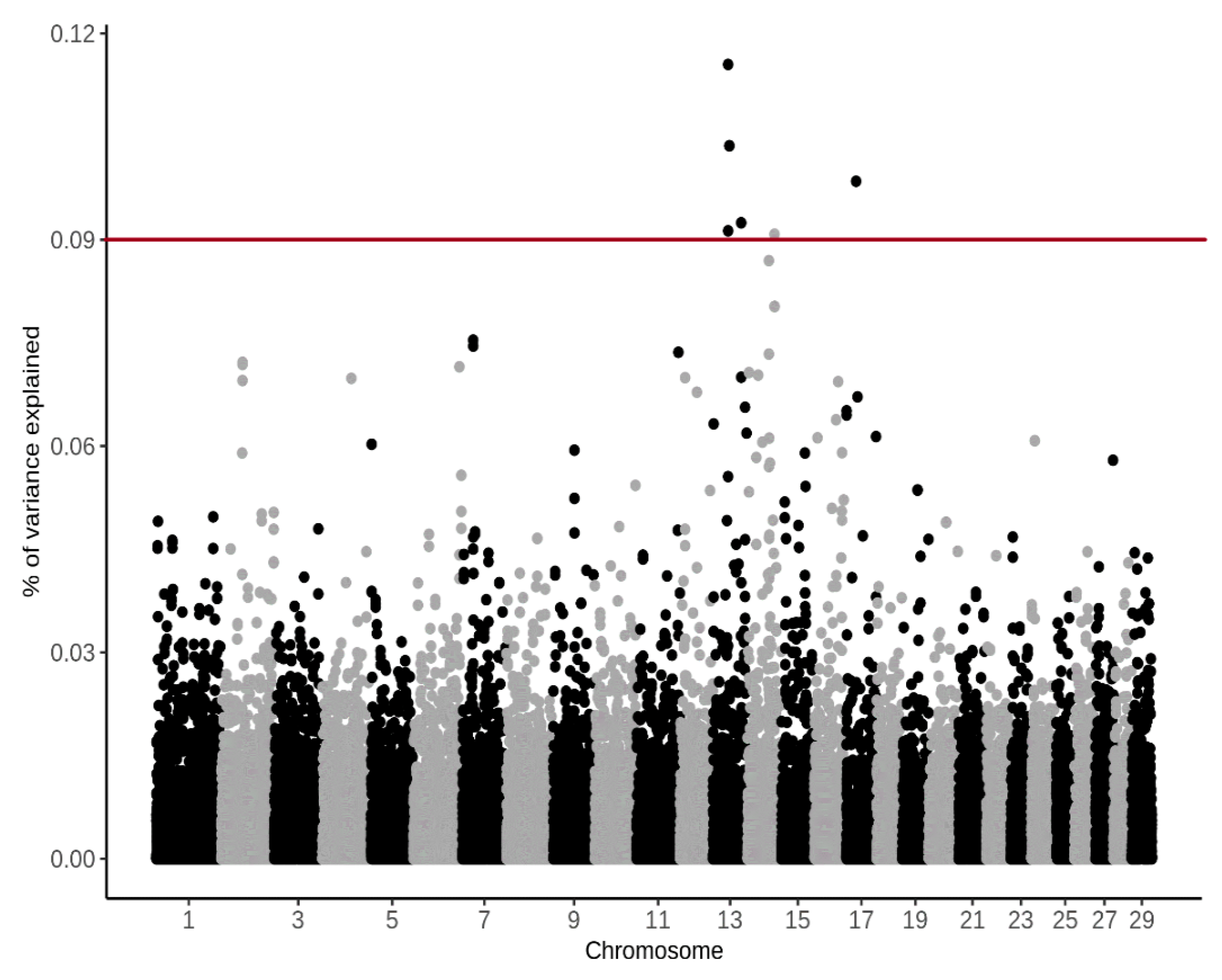
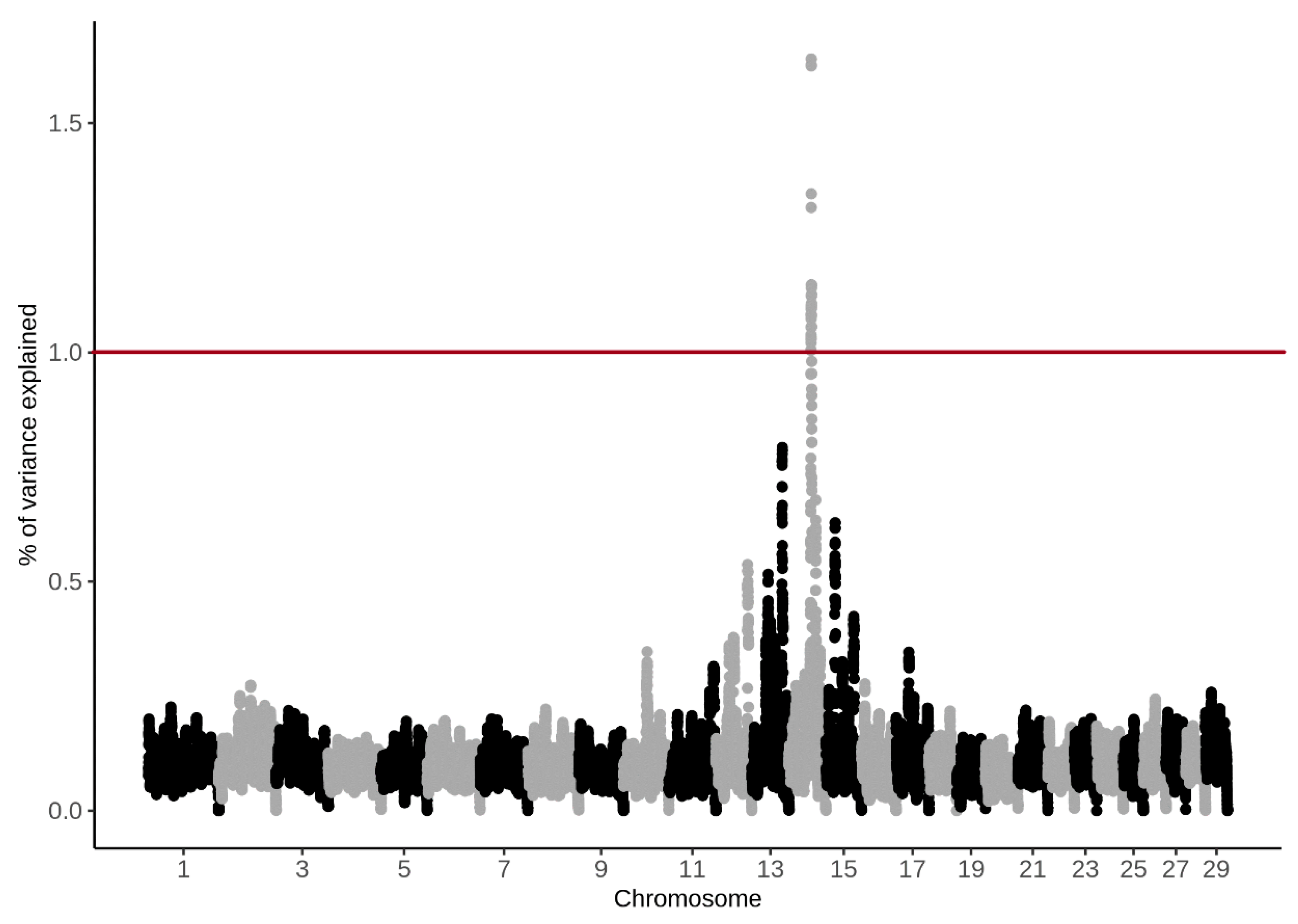
3.4. Genome-Wide Association Study
4. Discussion
4.1. CH4 Measurements
4.2. Variance Components
4.3. Heritability Estimates
4.4. Genome-Wide Association Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myhre, G.D.; Shindell, F.-M.; Bréon, W.; Collins, J.; Fuglestvedt, J.; Huang, D.; Koch, J.-F.; Lamarque, D.; Lee, B.M.; Nakajima, T.A.; et al. Anthropogenic and Natural Radiative Forcing. In Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2013; pp. 659–740. [Google Scholar]
- Steinfeld, H.; Gerber, P.; Wassenaar, T.; Castel, V.; Rosales, M.; de Haan, C. Livestock’s long shadow: Environmental issues and options. Renew. Resour. J. 2006. [Google Scholar] [CrossRef]
- Allen, M.R.; Shine, K.P.; Fuglestvedt, J.S.; Millar, R.J.; Cain, M.; Frame, D.J.; Macey, A.H. A solution to the misrepresentations of CO2-equivalent emissions of short-lived climate pollutants under ambitious mitigation. npj Clim. Atmos. Sci. 2018, 1, 1–8. [Google Scholar] [CrossRef]
- Murray, R.M.; Bryant, A.M.; Leng, R.A. Rates of production of methane in the rumen and large intestine of sheep. Br. J. Nutr. 1976, 36, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garnsworthy, P.C.; Craigon, J.; Hernandez-Medrano, J.H.; Saunders, N. On-farm methane measurements during milking correlate with total methane production by individual dairy cows. J. Dairy Sci. 2012, 95, 3166–3180. [Google Scholar] [CrossRef] [Green Version]
- Szalanski, M.; Kristensen, T.; Difford, G.; Lassen, J.; Buitenhuis, A.J.; Pszczola, M.; Løvendahl, P. Enteric methane emission from Jersey cows during the spring transition from indoor feeding to grazing. J. Dairy Sci. 2019, 102, 6319–6329. [Google Scholar] [CrossRef] [Green Version]
- Pszczola, M.; Rzewuska, K.; Mucha, S.; Strabel, T. Heritability of methane emissions from dairy cows over a lactation measured on commercial farms. J. Anim. Sci. 2017, 95, 4813–4819. [Google Scholar] [CrossRef] [Green Version]
- de Haas, Y.; Pszczola, M.; Soyeurt, H.; Wall, E.; Lassen, J. Invited review: Phenotypes to genetically reduce greenhouse gas emissions in dairying. J. Dairy Sci. 2017, 100, 855–870. [Google Scholar] [CrossRef] [Green Version]
- Pszczola, M.; Strabel, T.; Mucha, S.; Sell-Kubiak, E. Genome-wide association identifies methane production level relation to genetic control of digestive tract development in dairy cows. Sci. Rep. 2018, 8, 15164. [Google Scholar] [CrossRef] [Green Version]
- Difford, G.F.; Plichta, D.R.; Løvendahl, P.; Lassen, J.; Noel, S.J.; Højberg, O.; Wright, A.D.G.; Zhu, Z.; Kristensen, L.; Nielsen, H.B.; et al. Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows. PLoS Genet. 2018, 14, e1007580. [Google Scholar] [CrossRef] [Green Version]
- Madsen, J.; Bjerg, B.S.; Hvelplund, T.; Weisbjerg, M.R.; Lund, P. Methane and carbon dioxide ratio in excreted air for quantification of the methane production from ruminants. Livest. Sci. 2010, 129, 223–227. [Google Scholar] [CrossRef]
- Chwalibog, A. Husdyrernæring: Bestemmelse af næringsværdi og næringsbehov; Faculty of Life Sciences, University of Copenhagen, DSR forlag.: Copenhagen, Denmark, 1991. [Google Scholar]
- Sorg, D.; Difford, G.F.; Mühlbach, S.; Kuhla, B.; Swalve, H.H.; Lassen, J.; Strabel, T.; Pszczola, M. Comparison of a laser methane detector with the GreenFeed and two breath analysers for on-farm measurements of methane emissions from dairy cows. Comput. Electron. Agric. 2018, 153, 285–294. [Google Scholar] [CrossRef]
- Difford, G.F.; Olijhoek, D.W.; Hellwing, A.L.F.; Lund, P.; Bjerring, M.A.; de Haas, Y.; Lassen, J.; Løvendahl, P. Ranking cows’ methane emissions under commercial conditions with sniffers versus respiration chambers. Acta Agric. Scand. Anim. Sci. 2018, 68, 25–32. [Google Scholar] [CrossRef] [Green Version]
- van Engelen, S.; Bovenhuis, H.; van der Tol, P.P.J.; Visker, M.H.P.W. Genetic background of methane emission by Dutch Holstein Friesian cows measured with infrared sensors in automatic milking systems. J. Dairy Sci. 2018, 101, 2226–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Engelen, S.; Bovenhuis, H.; Dijkstra, J.; van Arendonk, J.A.M.; Visker, M.H.P.W. Short communication: Genetic study of methane production predicted from milk fat composition in dairy cows. J. Dairy Sci. 2015, 98, 8223–8226. [Google Scholar] [CrossRef]
- Manzanilla-Pech, C.I.V.; De Haas, Y.; Hayes, B.J.; Veerkamp, R.F.; Khansefid, M.; Donoghue, K.A.; Arthur, P.F.; Pryce, J.E. Genomewide association study of methane emissions in angus beef cattle with validation in dairy cattle. J. Anim. Sci. 2016, 94, 4151–4166. [Google Scholar] [CrossRef] [Green Version]
- Løvendahl, P.; Bjerring, M.A. Detection of carryover in automated milk sampling equipment. J. Dairy Sci. 2006, 89, 3645–3652. [Google Scholar] [CrossRef]
- Lassen, J.; Løvendahl, P.; Madsen, J. Accuracy of noninvasive breath methane measurements using Fourier transform infrared methods on individual cows. J. Dairy Sci. 2012, 95, 890–898. [Google Scholar] [CrossRef] [Green Version]
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- R Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 1 October 2021).
- Aguilar, I.; Tsuruta, S.; Masuda, Y.; Lourenco, D.A.L.; Legarra, A.; Misztal, I. BLUPF90 suite of programs for animal breeding with focus on genomics. In Proceedings of the 11th World Congress on Genetics Applied to Livestock Production, Auckland, New Zeeland, 11–16 February 2018; pp. 11–16. [Google Scholar]
- Kirkpatrick, M.; Lofsvold, D.; Bulmer, M. Analysis of the inheritance, selection and evolution of growth trajectories. Genetics 1990, 124, 979–993. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93. [Google Scholar] [CrossRef]
- Legarra, A.; Christensen, O.F.; Aguilar, I.; Misztal, I. Single Step, a general approach for genomic selection. Livest. Sci. 2014, 166. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. BLUPF90 and related programs (BGF90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; 2002; p. 33. [Google Scholar]
- Aguilar, I.; Legarra, A.; Cardoso, F.; Masuda, Y.; Lourenco, D.; Misztal, I. Frequentist p-values for large-scale-single step genome-wide association, with an application to birth weight in American Angus cattle. Genet. Sel. Evol. 2019, 51, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gualdrón Duarte, J.L.; Cantet, R.J.C.; Bates, R.O.; Ernst, C.W.; Raney, N.E.; Steibel, J.P. Rapid screening for phenotype-genotype associations by linear transformations of genomic evaluations. BMC Bioinform. 2014, 15, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatkar, M.S.; Zenger, K.R.; Hobbs, M.; Hawken, R.J.; Cavanagh, J.A.L.; Barris, W.; McClintock, A.E.; McClintock, S.; Thomson, P.C.; Tier, B.; et al. A primary assembly of a bovine haplotype block map based on a 15,036-single-nucleotide polymorphism panel genotyped in Holstein-Friesian cattle. Genetics 2007, 176. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef] [Green Version]
- Huhtanen, P.; Cabezas-Garcia, E.H.; Utsumi, S.; Zimmerman, S. Comparison of methods to determine methane emissions from dairy cows in farm conditions. J. Dairy Sci. 2015, 98. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Koerkamp, P.W.G.G.; Ogink, N. Uncertainty assessment of the breath methane concentration method to determine methane production of dairy cows. J. Dairy Sci. 2018, 101. [Google Scholar] [CrossRef] [Green Version]
- Lassen, J.; Løvendahl, P. Heritability estimates for enteric methane emissions from Holstein cattle measured using noninvasive methods. J. Dairy Sci. 2016, 99, 1959–1967. [Google Scholar] [CrossRef] [Green Version]
- Mancin, E.; Lourenco, D.; Bermann, M.; Mantovani, R.; Misztal, I. Accounting for population structure and phenotypes from relatives in association mapping for farm animals: A simulation study. Front. Genet. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- George, E.I.; McCulloch, R.E. Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 1993, 88. [Google Scholar] [CrossRef]
- Dikmen, S.; Cole, J.B.; Null, D.J.; Hansen, P.J. Genome-Wide association mapping for identification of quantitative trait loci for rectal temperature during heat stress in Holstein cattle. PLoS ONE 2013, 8, e69202. [Google Scholar] [CrossRef] [Green Version]
- Tiezzil, F.; Parker-Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. A genome-wide association study for clinical mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS ONE 2015, 10, e114919. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Peñagaricano, F. Unravelling the genomic architecture of bull fertility in Holstein cattle. BMC Genet. 2016, 17, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legarra, A.; Ricard, A.; Varona, L. GWAS by GBLUP: Single and multimarker EMMAX and bayes factors, with an example in detection of a major gene for horse gait. G3 Genes Genomes Genet. 2018, 8, 2301–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]



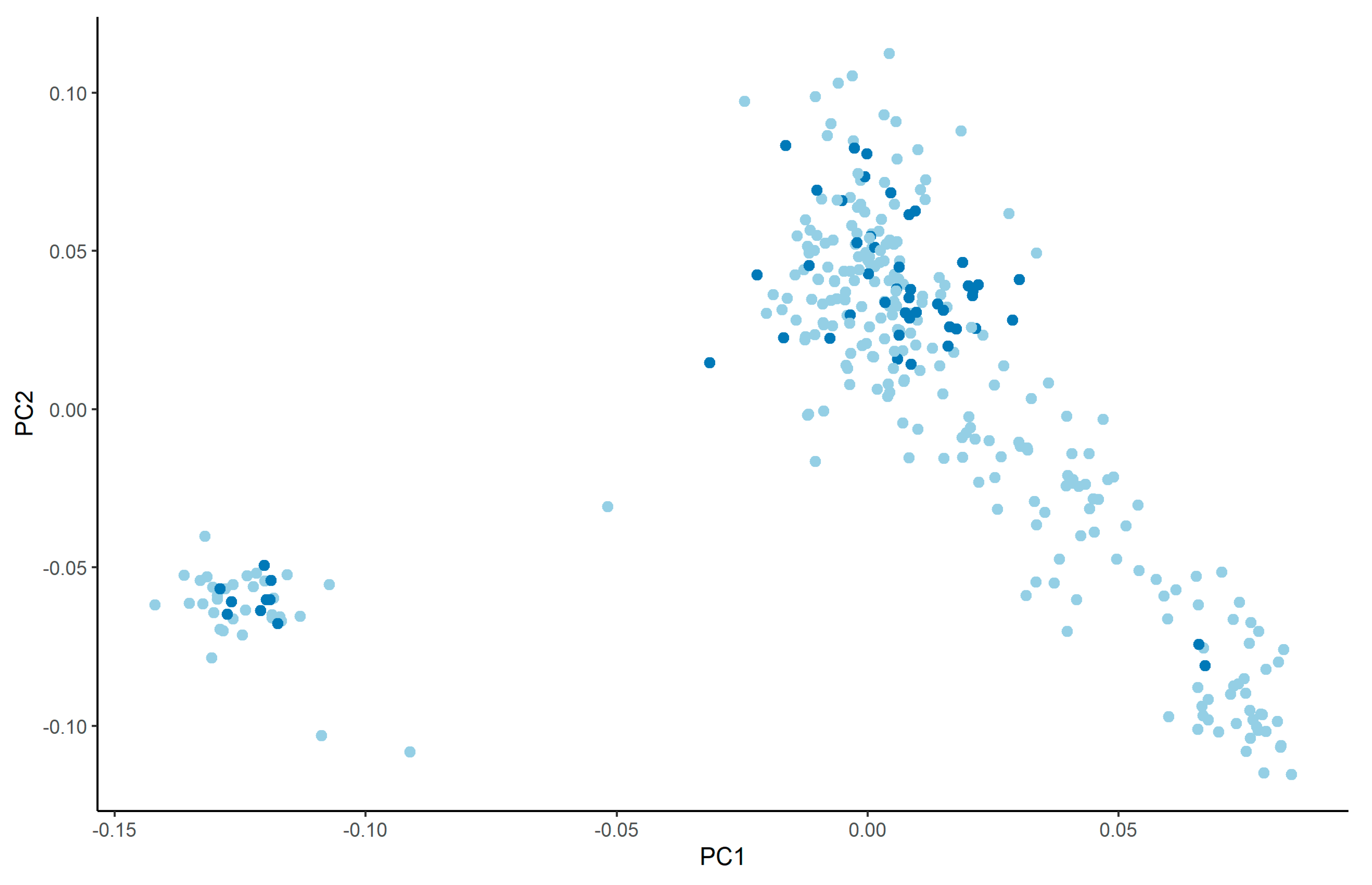
| Farm 1 | Farm 2 | |||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| CH4 ppm/day | 505 | 190 | 517 | 166 |
| CH4 g/day | 396 | 59.70 | 503 | 93.20 |
| Live weight (kg) | 544 | 115 | 669 | 73 |
| FPCM (kg) | 33.70 | 6.82 | 36.60 | 8.79 |
| No. of records | 31,179 | 3180 | ||
| BTA | SNP Name | Position (bp) | (%) CH4 g/d | (%) CH4 ppm/d |
|---|---|---|---|---|
| 14 | BTB-00568855 | 48890778 | 1.62 | 1.19 |
| 14 | Hapmap57707-rs29024913 | 48858864 | 1.31 | 1.01 |
| 14 | UA-IFASA-9208 | 48726666 | 1.05 | 0.93 |
| 14 | ARS-BFGL-NGS-45675 | 48679326 | 1.09 | 0.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sypniewski, M.; Strabel, T.; Pszczola, M. Genetic Variability of Methane Production and Concentration Measured in the Breath of Polish Holstein-Friesian Cattle. Animals 2021, 11, 3175. https://doi.org/10.3390/ani11113175
Sypniewski M, Strabel T, Pszczola M. Genetic Variability of Methane Production and Concentration Measured in the Breath of Polish Holstein-Friesian Cattle. Animals. 2021; 11(11):3175. https://doi.org/10.3390/ani11113175
Chicago/Turabian StyleSypniewski, Mateusz, Tomasz Strabel, and Marcin Pszczola. 2021. "Genetic Variability of Methane Production and Concentration Measured in the Breath of Polish Holstein-Friesian Cattle" Animals 11, no. 11: 3175. https://doi.org/10.3390/ani11113175






