Selection Signatures Analysis Reveals Genes Associated with High-Altitude Adaptation in Tibetan Goats from Nagqu, Tibet
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Sample Collection and Genotyping
2.2. Quality Control
2.3. Population Structure Analysis
2.4. Selection Signal Analyses
2.5. Gene Annotation Analysis
2.6. Validation of Selection Signal Results by q-PCR
2.6.1. RNA Extraction
2.6.2. Reverse Transcription and q-PCR Analysis
3. Results and Discussion
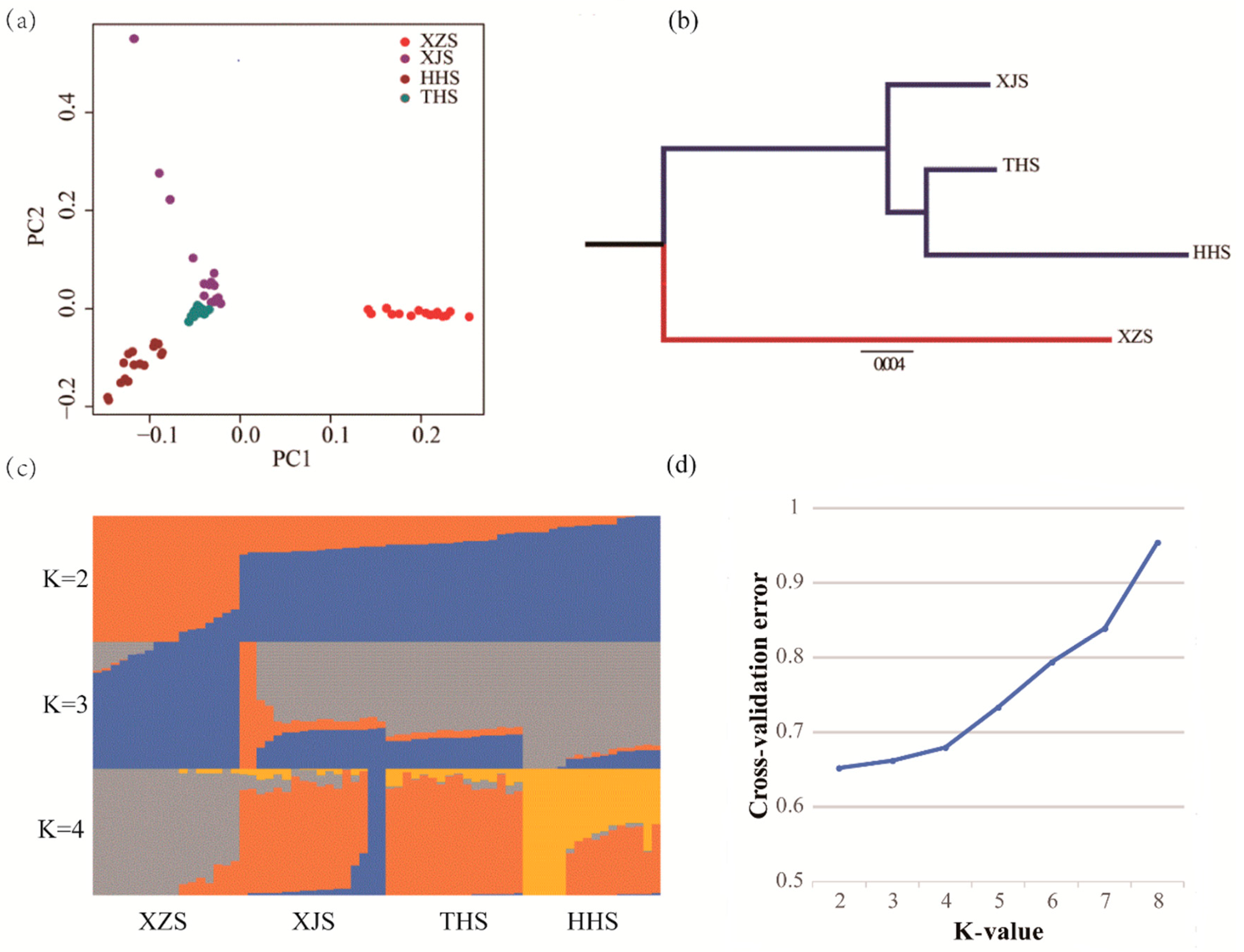
3.1. Genetic Variation and Population Genetic Analysis
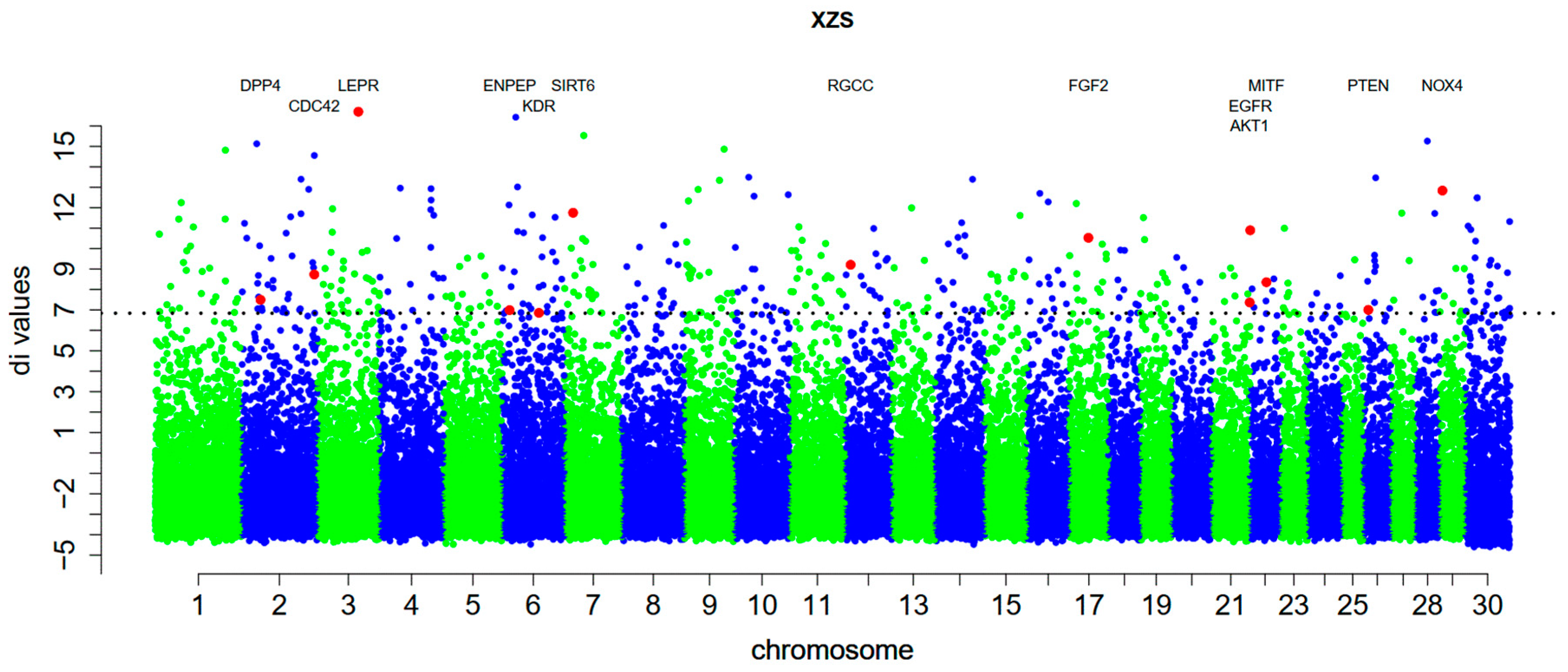
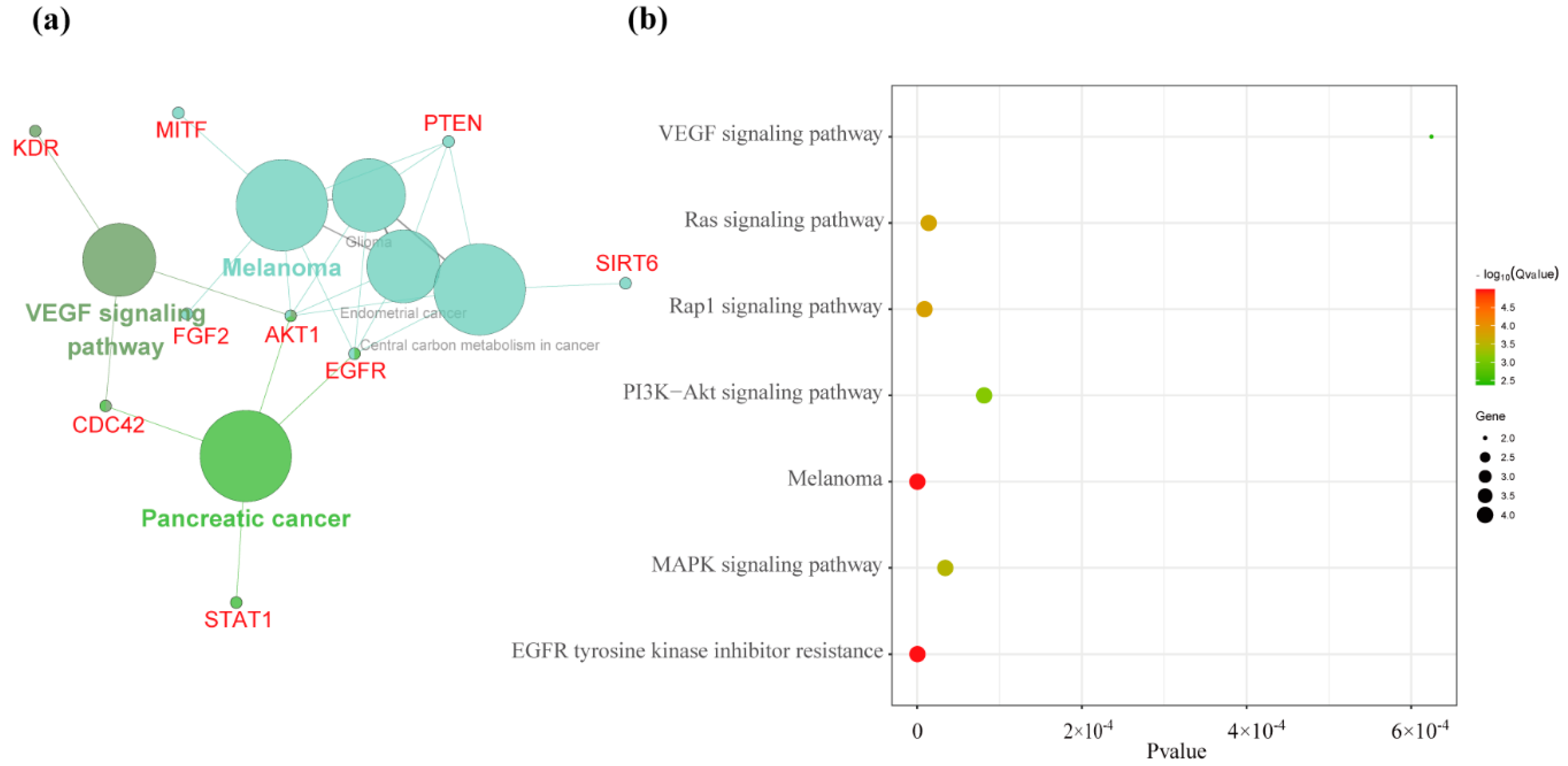
3.2. Detecting Breed Positive Selection Genes
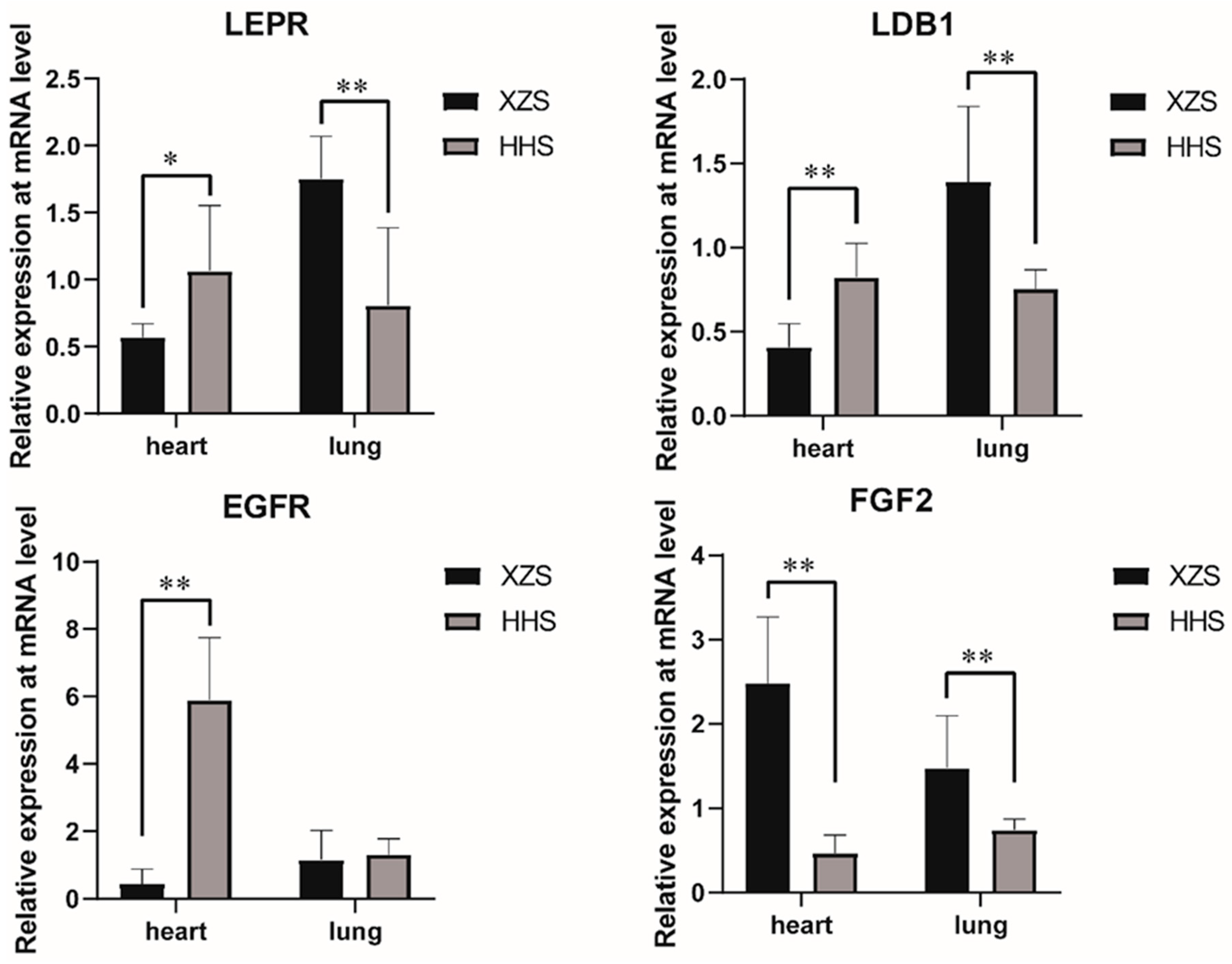
3.3. Validation of Selection Signal Analysis Results by q-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Du, L.X. Animal Genetic Resources in China; China Agriculture Press: Beijing, China, 2011. [Google Scholar]
- Thompson, L.G.; Yao, T.; Mosley-Thompson, E.; Davis, M.E.; Henderson, K.A.; Lin, P. A high-resolution millennial record of the south asian monsoon from himalayan ice cores. Science 2000, 289, 1916–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.; Feng, J.; Li, L.; Zhong, T.; Wang, L.; Guo, J.; Ba, G.; Song, T.; Zhang, H. Polymorphisms, differentiation, and phylogeny of 10 Tibetan goat populations inferred from mitochondrial D-loop sequences. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2018, 29, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Tao, H.; Li, P.; Li, L.; Zhong, T.; Wang, L.; Ma, J.; Chen, X.; Song, T.; Zhang, H. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci. Rep. 2018, 8, 10405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; Yao, N.; Yang, M.; Liu, X.; Dong, K.; Zhao, Q.; Pu, Y.; He, X.; Guan, W.; Yang, N. Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genom. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, C.; Song, S.; Jiang, L.; He, X.; Zhao, Q.; Pu, Y.; Malhi, K.K.; Kamboh, A.A.; Ma, Y. Sequence Characterization of DSG3 Gene to Know Its Role in High-Altitude Hypoxia Adaptation in the Chinese Cashmere Goat. Front. Genet. 2018, 9, 553. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zhou, G.; Guo, J.; Yan, H.; Niu, Y.; Li, Y.; Yuan, C.; Geng, R.; Lan, X.; et al. Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits. Sci. Rep. 2016, 6, 38932. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Lu, J.; Xu, L.; Liu, G.; Wang, Z.; Zhao, F.; Zhang, L.; Han, X.; Du, L.; Liu, C. Genetic structure of Chinese indigenous goats and the special geographical structure in the Southwest China as a geographic barrier driving the fragmentation of a large population. PLoS ONE 2014, 9, e94435. [Google Scholar] [CrossRef]
- Kijas, J.W.; Ortiz, J.S.; McCulloch, R.; James, A.; Brice, B.; Swain, B.; Tosser-Klopp, G.; International Goat Genome, C. Genetic diversity and investigation of polledness in divergent goat populations using 52 088 SNPs. Anim. Genet. 2013, 44, 325–335. [Google Scholar] [CrossRef]
- Yuan, Z.; Liu, E.; Liu, Z.; Kijas, J.W.; Zhu, C.; Hu, S.; Ma, X.; Zhang, L.; Du, L.; Wang, H.; et al. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet. 2017, 48, 55–66. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Rousset, M.R.F. Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar]
- Akey, J.M.; Zhang, G.; Zhang, K.; Jin, L.; Shriver, M.D. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002, 12, 1805–1814. [Google Scholar] [CrossRef] [Green Version]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Akey, J.M.; Ruhe, A.L.; Akey, D.T.; Wong, A.K.; Connelly, C.F.; Madeoy, J.; Nicholas, T.J.; Neff, M.W. Tracking footprints of artificial selection in the dog genome. Proc. Natl. Acad. Sci. USA 2010, 107, 1160–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, C.; Wang, H.; Liu, G.; Zhao, F.; Kijas, J.W.; Ma, Y.; Lu, J.; Zhang, L.; Cao, J.; Wu, M.; et al. Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci. Rep. 2016, 6, 26770. [Google Scholar] [CrossRef] [PubMed]
- Edea, Z.; Dadi, H.; Dessie, T.; Kim, K.S. Genomic signatures of high-altitude adaptation in Ethiopian sheep populations. Genes Genom. 2019, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.; Yang, B.; Li, J.; Xie, X.; Chen, H.; Ren, J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genom. 2014, 15, 834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigham, A.W.; Lee, F.S. Human high-altitude adaptation: Forward genetics meets the HIF pathway. Genes Dev. 2014, 28, 2189–2204. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Chamba, Y.; Shang, P.; Wang, Z.; Ma, J.; Wang, L.; Zhang, H. Comparative transcriptomic and proteomic analyses provide insights into the key genes involved in high-altitude adaptation in the Tibetan pig. Sci. Rep. 2017, 7, 3654. [Google Scholar] [CrossRef] [Green Version]
- Macklin, P.S.; McAuliffe, J.; Pugh, C.W.; Yamamoto, A. Hypoxia and HIF pathway in cancer and the placenta. Placenta 2017, 56, 8–13. [Google Scholar] [CrossRef]
- Huang, D.; Li, C.; Zhang, H. Hypoxia and cancer cell metabolism. Acta Biochim. Biophys. Sin. 2014, 46, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Eichstaedt, C.A.; Antao, T.; Cardona, A.; Pagani, L.; Kivisild, T.; Mormina, M. Genetic and phenotypic differentiation of an Andean intermediate altitude population. Physiol. Rep. 2015, 3. [Google Scholar] [CrossRef]
- An, X.; Jin, Y.; Guo, H.; Foo, S.Y.; Cully, B.L.; Wu, J.; Zeng, H.; Rosenzweig, A.; Li, J. Response gene to complement 32, a novel hypoxia-regulated angiogenic inhibitor. Circulation 2009, 120, 617–627. [Google Scholar] [CrossRef]
- Lendahl, U.; Lee, K.L.; Yang, H.; Poellinger, L. Generating specificity and diversity in the transcriptional response to hypoxia. Nat. Rev. Genet. 2009, 10, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Koo, K.H.; Park, J.W.; Kim, H.J.; Ye, S.K.; Park, J.B.; Park, B.K.; Kim, Y.N. HIF-1 is induced via EGFR activation and mediates resistance to anoikis-like cell death under lipid rafts/caveolae-disrupting stress. Carcinogenesis 2009, 30, 1997–2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pez, F.; Dayan, F.; Durivault, J.; Kaniewski, B.; Aimond, G.; Le Provost, G.S.; Deux, B.; Clézardin, P.; Sommer, P.; Pouysségur, J. The HIF-1–inducible lysyl oxidase activates HIF-1 via the Akt pathway in a positive regulation loop and synergizes with HIF-1 in promoting tumor cell growth. Cancer Res. 2011, 71, 1647–1657. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.-X.; Wang, X.-L.; Tong, M.-M.; Gan, L.; Chen, H.; Wu, S.-S.; Chen, J.-X.; Li, R.-L.; Wu, Y.; Zhang, H.-Y.; et al. SIRT6 protects cardiomyocytes against ischemia/reperfusion injury by augmenting FoxO3α-dependent antioxidant defense mechanisms. Basic Res. Cardiol. 2016, 111, 13. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, N.N.; van Elteren, H.A.; Goos, T.G.; Morales, C.R.; Rivera-Ch, M.; Martin, D.S.; Cabala Peralta, P.; Passano Del Carpio, A.; Aranibar Machaca, S.; Huicho, L.; et al. Pregnancy at high altitude in the Andes leads to increased total vessel density in healthy newborns. J. Appl. Physiol. 2016, 121, 709–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koester-Hegmann, C.; Bengoetxea, H.; Kosenkov, D.; Thiersch, M.; Haider, T.; Gassmann, M.; Schneider Gasser, E.M. High-Altitude Cognitive Impairment Is Prevented by Enriched Environment Including Exercise via VEGF Signaling. Front. Cell. Neurosci. 2018, 12, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brem, H.; Klagsbrun, M. The role of fibroblast growth factors and related oncogenes in tumor growth. Cancer Treat. Res. 1992, 63, 211–231. [Google Scholar]
- Presta, M.; Andrés, G.; Leali, D.; Dell’Era, P.; Ronca, R.J.E.C.N. Inflammatory cells and chemokines sustain FGF2-induced angiogenesis. Eur. Cytokine Netw. 2009, 20, 39. [Google Scholar] [CrossRef] [Green Version]
- Wakayama, Y.; Fukuhara, S.; Ando, K.; Matsuda, M.; Mochizuki, N. Cdc42 mediates Bmp-induced sprouting angiogenesis through Fmnl3-driven assembly of endothelial filopodia in zebrafish. Dev. Cell 2015, 32, 109–122. [Google Scholar] [CrossRef] [Green Version]
- Olszewska-Pazdrak, B.; Hein, T.W.; Olszewska, P.; Carney, D.H. Chronic hypoxia attenuates VEGF signaling and angiogenic responses by downregulation of KDR in human endothelial cells. Am. J. Physiol. Cell Physiol. 2009, 296, C1162–C1170. [Google Scholar] [CrossRef] [Green Version]
- Qian, X.; Li, X.; Shi, Z.; Xia, Y.; Cai, Q.; Xu, D.; Tan, L.; Du, L.; Zheng, Y.; Zhao, D.; et al. PTEN Suppresses Glycolysis by Dephosphorylating and Inhibiting Autophosphorylated PGK1. Mol. Cell 2019, 76, 516–527.e517. [Google Scholar] [CrossRef]
- Caballero Eraso, C.; Shin, M.; Pho, H.; Schwartz, A.; Tang, W.-Y.; Sham, J.; Polotsky, V. Mechanisms and Significance of Leptin-Induced Upregulation of the Hypoxic Ventilatory Response. In D29. Pathophysiology of OSA: Intermittent Hypoxia and Beyond; American Thoracic Society: Baltimore, MD, USA, 2018. [Google Scholar]
- Li, L.; Jothi, R.; Cui, K.; Lee, J.Y.; Cohen, T.; Gorivodsky, M.; Tzchori, I.; Zhao, Y.; Hayes, S.M.; Bresnick, E.H.; et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat. Immunol. 2011, 12, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Ni, T.; Lin, J.; Zhang, C.; Zheng, L.; Luo, M. Long non-coding RNA H19, a negative regulator of microRNA-148b-3p, participates in hypoxia stress in human hepatic sinusoidal endothelial cells via NOX4 and eNOS/NO signaling. Biochimie 2019, 163, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Das, S.S.; Hayashi, H.; Sato, T.; Yamada, R.; Hiratsuka, M.; Hirasawa, N. Regulation of dipeptidyl peptidase 4 production in adipocytes by glucose. Diabetes Metab. Syndr. Obes. 2014, 7, 185–194. [Google Scholar] [PubMed] [Green Version]
- Gunawan, A.; Pramukti, F.; Listyarini, K.; Abuzahra, M.; Jakaria, C.S.; Inounu, I.; Uddin, M. Novel variant in the leptin receptor (LEPR) gene and its association with fat quality, odour, and flavour in sheep. JITAA 2019, 44, 1–9. [Google Scholar] [CrossRef]
- Alim, M.; Hossain, M.; Nusrat, J.; Salimullah, M.; Shu-Hong, Z.; Alam, J. Genetic effects of leptin receptor (LEPR) polymorphism on litter size in a Black Bengal goat population. Anim. Biol. 2019, 69, 411–420. [Google Scholar] [CrossRef]
- Ekegbu, U.J.; Burrows, L.; Amirpour-Najafabadi, H.; Zhou, H.; Hickford, J.G. Gene polymorphisms in PROP1 associated with growth traits in sheep. Gene 2019, 683, 41–46. [Google Scholar] [CrossRef]
- Ma, L.; Qin, Q.; Yang, Q.; Zhang, M.; Zhao, H.; Pan, C.; Lei, C.; Chen, H.; Lan, X. Associations of six SNPs of POU1F1-PROP1-PITX1-SIX3 pathway genes with growth traits in two Chinese indigenous goat breeds. Ann. Anim. Sci. 2017, 17, 399–411. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-S.; Shin, D. Genome-Wide Association Studies Associated with Backfat Thickness in Landrace and Yorkshire Pigs. Genom. Inform. 2018, 16, 59. [Google Scholar] [CrossRef] [Green Version]
- Kowalewska-Luczak, I.; Czerniawska-Piatkowska, E.; Kowalczyk, A. Relationship between polymorphism in the CRH gene and the traits of body conformation of Salers cows. Anim. Biotechnol. 2019, 22, 1–5. [Google Scholar] [CrossRef]
| Breeds | Abbr. | Sample Size | Sex | Breed Characteristics | Location | Living Altitude |
|---|---|---|---|---|---|---|
| Tibetan | XZS | 17 | female | multipurpose, horned, parti-color, small size | Naqu, Tibet, China | above 4500 m |
| Xinjiang | XJS | 17 | female | multipurpose, horned, white color, middle size | Tacheng, Xinjiang, China | about 1100 m |
| Taihang | THS | 16 | female | multipurpose, horned, black color, middle size | Changzhi, Shanxi, China | about 800 m |
| Huanghuai | HHS | 16 | female | meat, horned white color, big size | Zhumadian, Henan, China | about 81 m |
| Breed | XZS | XJS | HHS | Mean Pairwise Fst |
|---|---|---|---|---|
| XZS | 0.049 | |||
| XJS | 0.043 | 0.030 | ||
| HHS | 0.058 | 0.032 | 0.039 | |
| THS | 0.044 | 0.016 | 0.026 | 0.029 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, M.; Lu, J.; Fei, X.; Lu, Z.; Quan, K.; Liu, Y.; Chu, M.; Di, R.; Wei, C.; Wang, H. Selection Signatures Analysis Reveals Genes Associated with High-Altitude Adaptation in Tibetan Goats from Nagqu, Tibet. Animals 2020, 10, 1599. https://doi.org/10.3390/ani10091599
Jin M, Lu J, Fei X, Lu Z, Quan K, Liu Y, Chu M, Di R, Wei C, Wang H. Selection Signatures Analysis Reveals Genes Associated with High-Altitude Adaptation in Tibetan Goats from Nagqu, Tibet. Animals. 2020; 10(9):1599. https://doi.org/10.3390/ani10091599
Chicago/Turabian StyleJin, Meilin, Jian Lu, Xiaojuan Fei, Zengkui Lu, Kai Quan, Yongbin Liu, Mingxing Chu, Ran Di, Caihong Wei, and Huihua Wang. 2020. "Selection Signatures Analysis Reveals Genes Associated with High-Altitude Adaptation in Tibetan Goats from Nagqu, Tibet" Animals 10, no. 9: 1599. https://doi.org/10.3390/ani10091599
APA StyleJin, M., Lu, J., Fei, X., Lu, Z., Quan, K., Liu, Y., Chu, M., Di, R., Wei, C., & Wang, H. (2020). Selection Signatures Analysis Reveals Genes Associated with High-Altitude Adaptation in Tibetan Goats from Nagqu, Tibet. Animals, 10(9), 1599. https://doi.org/10.3390/ani10091599






