Fibromodulin Modulates Chicken Skeletal Muscle Development via the Transforming Growth Factor-β Signaling Pathway
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Cultures
2.3. Fmod Knockdown and Overexpression
2.4. RNA Extration and Real-Time Quantitative PCR
2.5. Western Blot
2.6. Immunofluorescence Analysis
2.7. Lentiviral Intramuscular Injections
2.8. Histomorphological Observation
2.9. Transcriptome Analysis
2.10. Statistical Analysis
3. Results
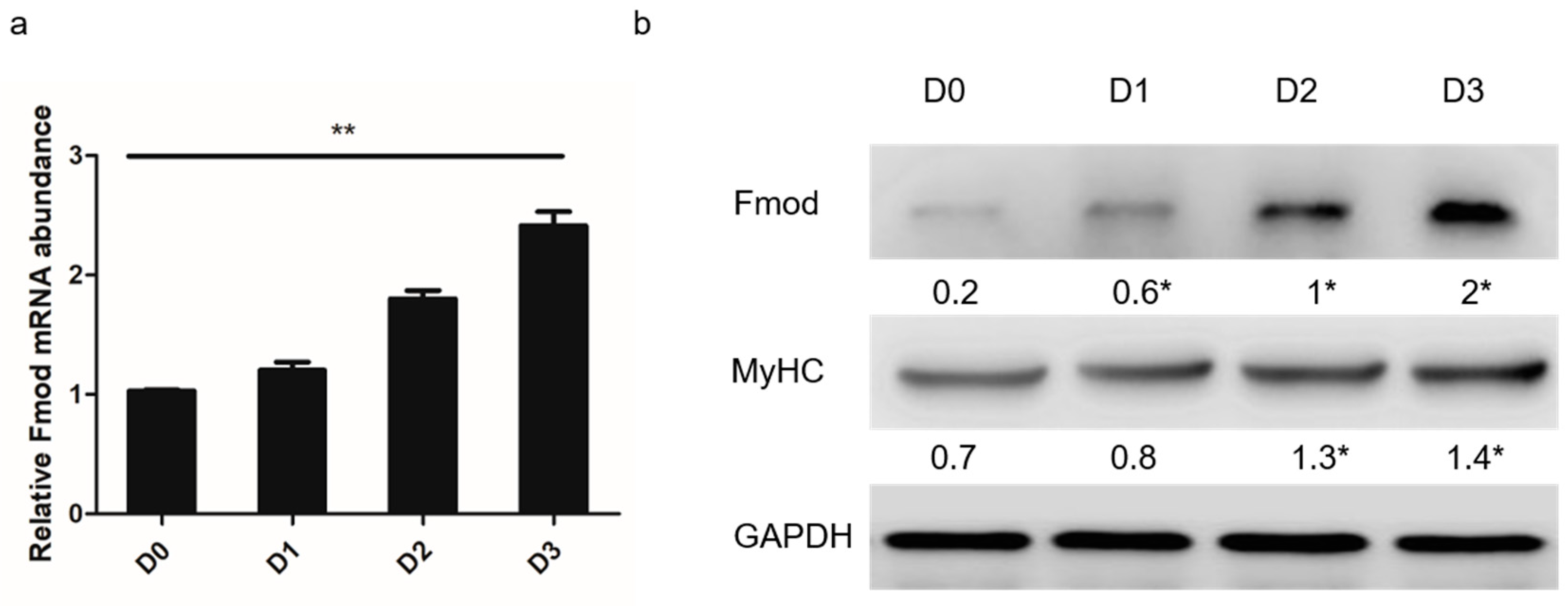
3.1. Fmod Expression in Differentiation of Myoblasts
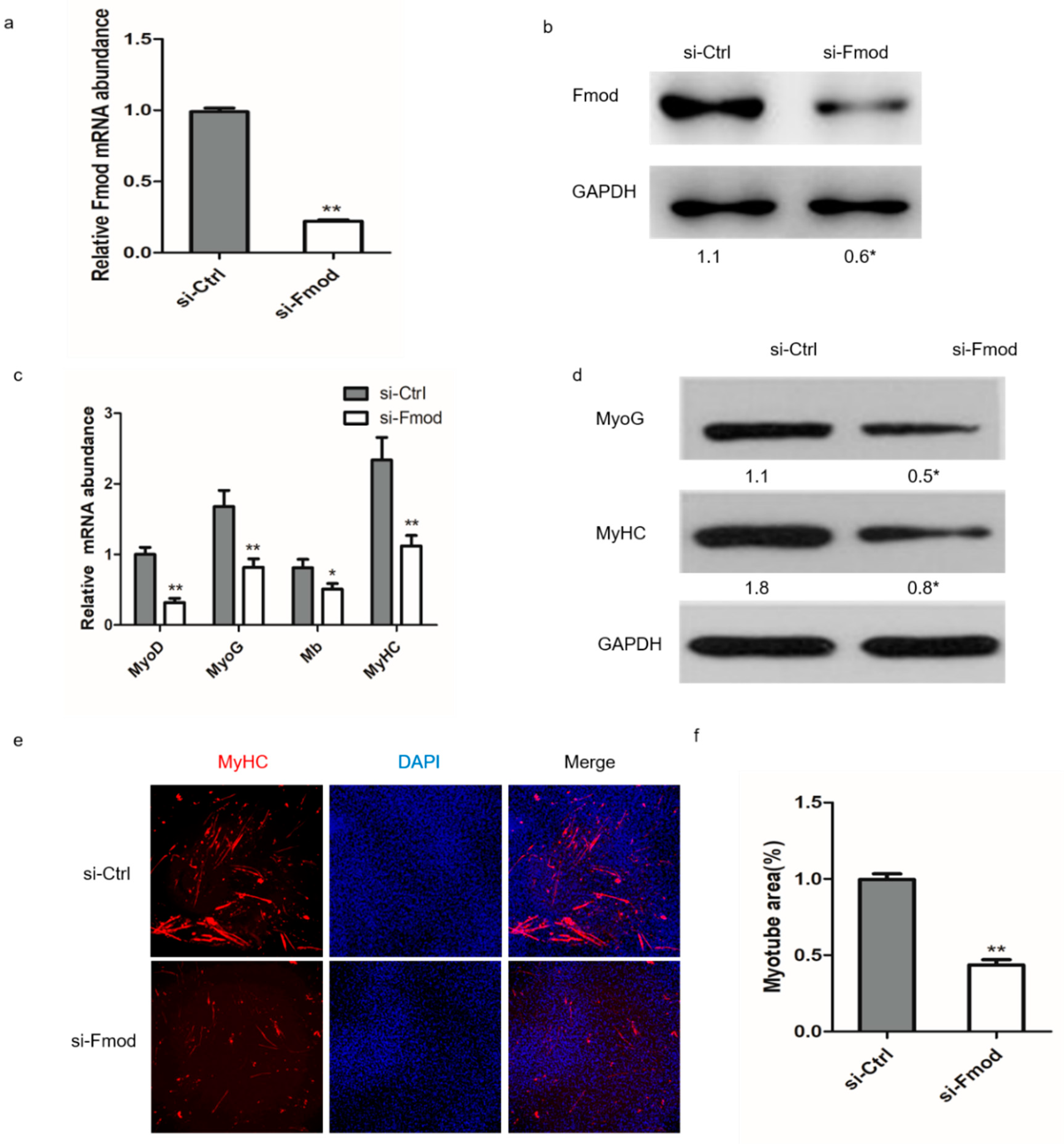
3.2. Interference with Fmod Restricts Formation of Myotubes
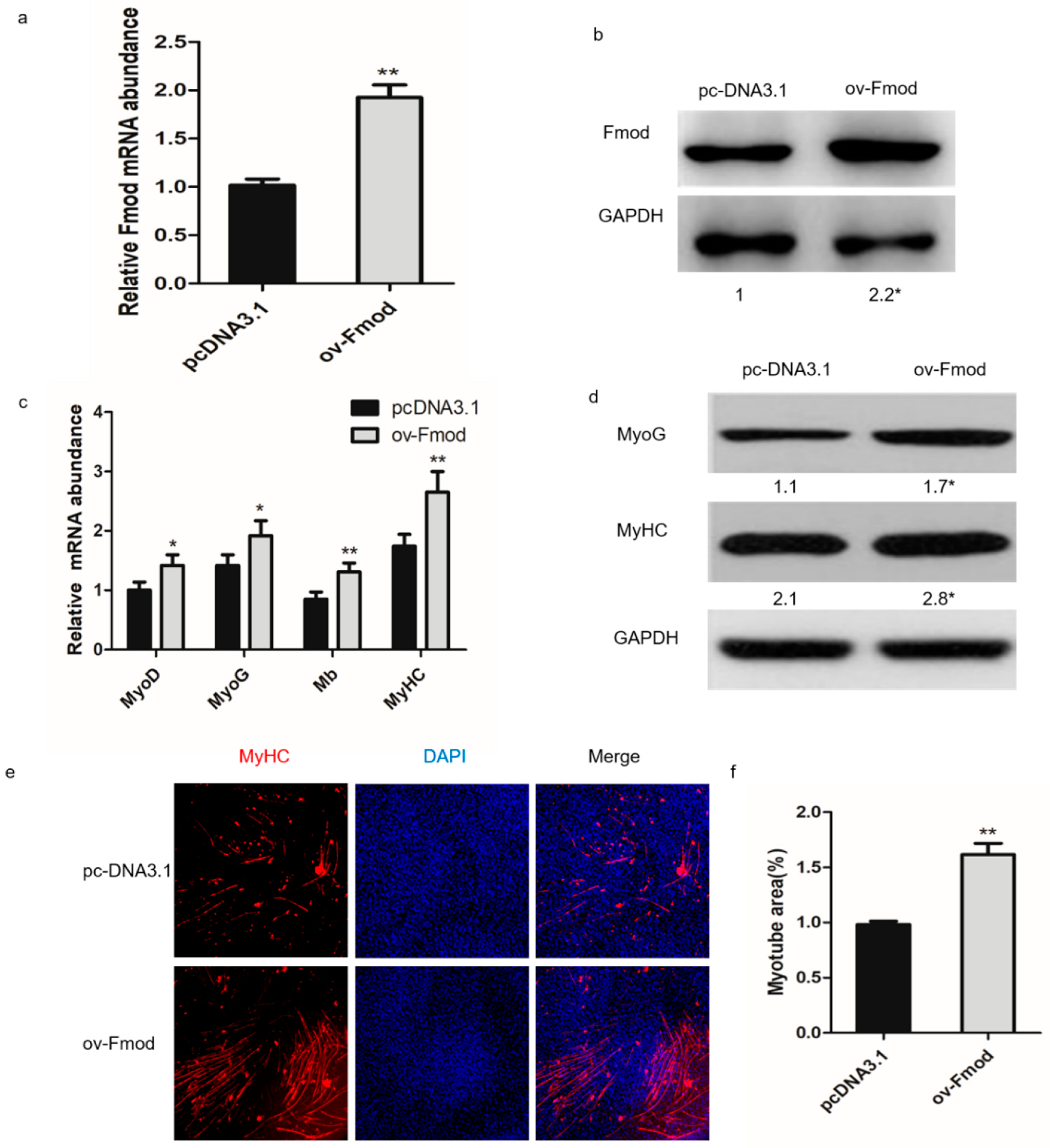
3.3. Overexpression of Fmod Promotes Formation of Myotubes
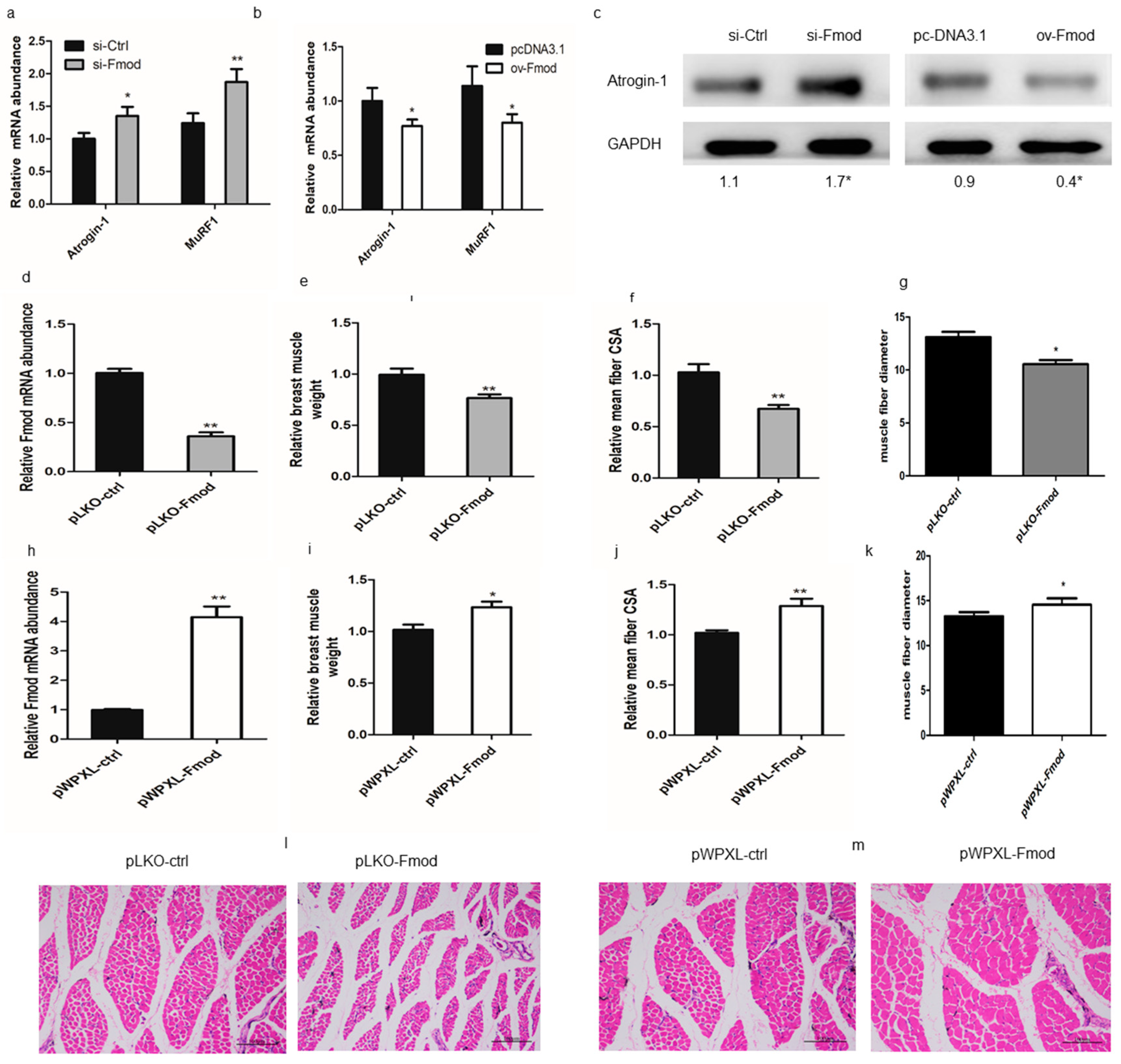
3.4. Fmod is Involved in Skeletal Muscle Atrophy in Chickens
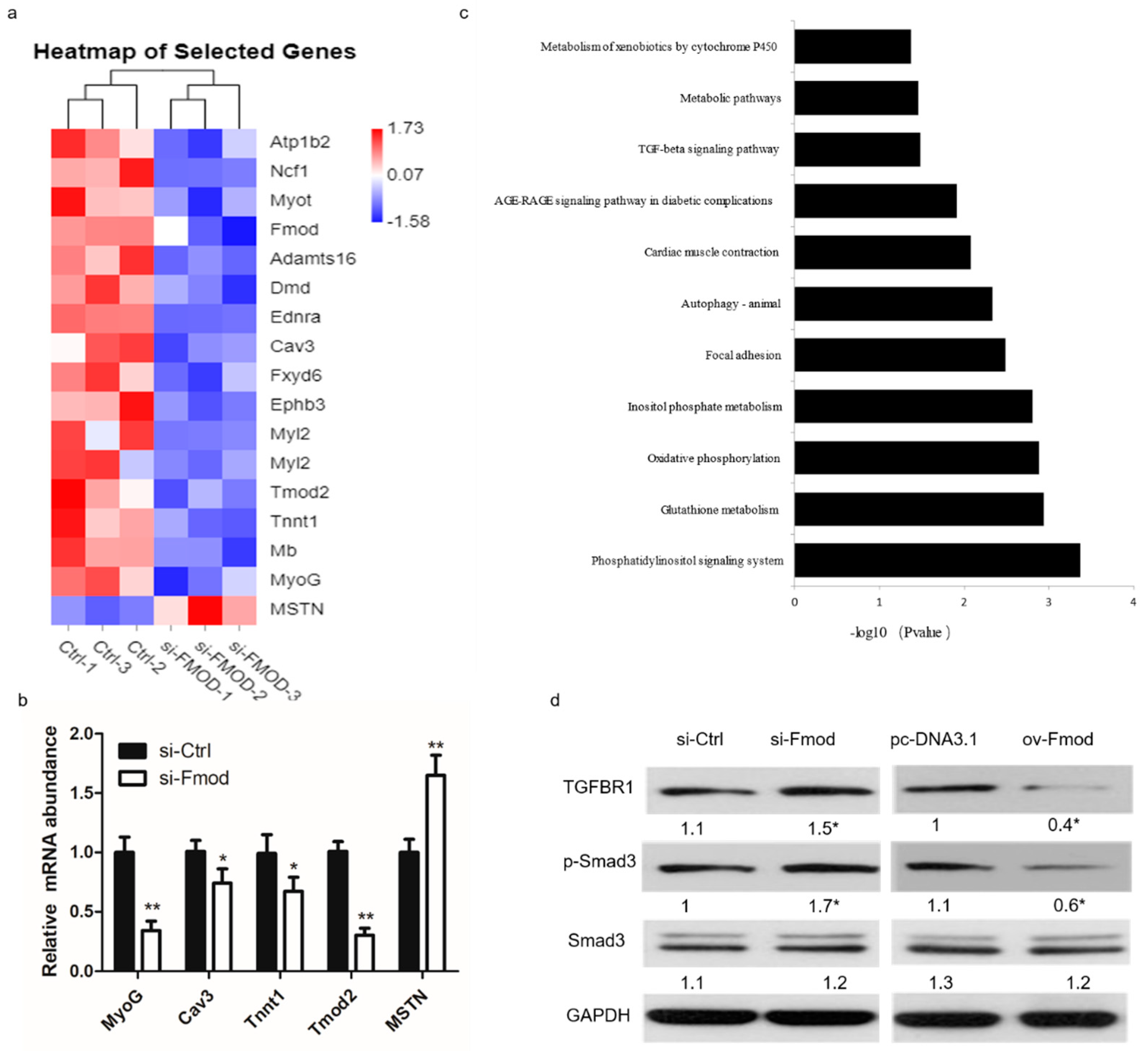
3.5. Fmod Regulates Differentiation of Myoblasts via TGF-β Signaling Pathway
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rattray, B.; Thompson, M.; Ruell, P.; Caillaud, C. Specific training improves skeletal muscle mitochondrial calcium homeostasis after eccentric exercise. Graefe’s Arch. Clin. Exp. Ophthalmol. 2012, 113, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Schmalbruch, H. Growth and denervation response of skeletal muscle fibers of newborn rats. Muscle Nerve 1990, 13, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Boil. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, A.; Romarís, M.; Rasmussen, L.M.; Heinegård, D.; Twardzik, D.R.; Border, W.A.; Ruoslahti, E. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor β. Biochem. J. 1994, 302, 527–534. [Google Scholar] [CrossRef]
- Antonsson, P.; Heinegård, D.; Oldberg, A. Posttranslational modifications of fibromodulin. J. Boil. Chem. 1991, 266, 16859–16861. [Google Scholar]
- Ao, Z.; Yu, S.; Qian, P.; Gao, W.; Guo, R.; Dong, X.; Xu, J.; Zhang, R.; Jiang, C.; Ji, F.-Y.; et al. Tumor angiogenesis of SCLC inhibited by decreased expression of FMOD via downregulating angiogenic factors of endothelial cells. Biomed. Pharmacother. 2017, 87, 539–547. [Google Scholar] [CrossRef]
- Zheng, Z.; Nguyen, C.; Zhang, X.; Khorasani, H.; Wang, J.Z.; Zara, J.N.; Chu, F.; Yin, W.; Pang, S.; Le, A.; et al. Delayed wound closure in fibromodulin-deficient mice is associated with increased TGF-β3 signaling. J. Investig. Dermatol. 2010, 131, 769–778. [Google Scholar] [CrossRef]
- Choudhury, A.; Derkow, K.; Daneshmanesh, A.H.; Mikaelsson, E.; Kiaii, S.; Kokhaei, P.; Österborg, A.; Mellstedt, H. Silencing of ROR1 and FMOD with siRNA results in apoptosis of CLL cells. Br. J. Haematol. 2010, 151, 327–335. [Google Scholar] [CrossRef]
- Dürrenfeld, P.; Iacocca, E.; Åkerman, J.; Muduli, P.K. Modulation-mediated unlocking of a parametrically phase-locked spin torque oscillator. Appl. Phys. Lett. 2014, 105, 252404. [Google Scholar] [CrossRef]
- Mayr, C.; Bund, D.; Schlee, M.; Moosmann, A.; Kofler, D.M.; Hallek, M.; Wendtner, C.M. Fibromodulin as a novel tumor-associated antigen (TAA) in chronic lymphocytic leukemia (CLL), which allows expansion of specific CD8+ autologous T lymphocytes. Blood 2005, 105, 1566–1573. [Google Scholar] [CrossRef]
- Meriane, M.; Charrasse, S.; Comunale, F.; Gauthier-Rouvière, C. Transforming growth factor β activates Rac1 and Cdc42Hs GTPases and the JNK pathway in skeletal muscle cells. Boil. Cell 2002, 94, 535–543. [Google Scholar] [CrossRef]
- Zheng, Z.; Jian, J.; Zhang, X.; Zara, J.N.; Yin, W.; Chiang, M.; Liu, Y.; Wang, J.; Pang, S.; Ting, K.; et al. Reprogramming of human fibroblasts into multipotent cells with a single ECM proteoglycan, fibromodulin. Biomaterials 2012, 33, 5821–5831. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Cui, C.; He, H.; Shen, X.; Chen, Y.; Wang, Y.; Li, D.; Zhu, Q.; Yin, H. FHL1 regulates myoblast differentiation and autophagy through its interaction with LC3. J. Cell. Physiol. 2019, 235, 4667–4678. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Cui, C.; Wang, Y.; He, H.; Liu, Z.; Shen, X.; Chen, Y.; Li, D.; Zhu, Q.; Yin, H. Knockdown of CSRP3 inhibits differentiation of chicken satellite cells by promoting TGF-β/Smad3 signaling. Gene 2019, 707, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Mansukhani, A.; Ambrosetti, D.; Holmes, G.; Cornivelli, L.; Basilico, C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J. Cell Boil. 2005, 168, 1065–1076. [Google Scholar] [CrossRef]
- Luo, W.; Chen, J.; Li, L.; Ren, X.; Cheng, T.; Lu, S.; Lawal, R.A.; Nie, Q.; Zhang, X.; Hanotte, O. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs. Cell Death Differ. 2018, 26, 426–442. [Google Scholar] [CrossRef]
- Hu, H.; Li, S.; Li, J.; Huang, C.; Zhou, F.; Zhao, L.; Yu, W.; Qin, X. Knockdown of fibromodulin inhibits proliferation and migration of RPE cell via the VEGFR2-AKT pathway. J. Ophthalmol. 2018, 2018, 1–8. [Google Scholar] [CrossRef]
- Mondal, B.; Patil, V.; Shwetha, S.D.; Sravani, K.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Kanduri, M.; Somasundaram, K. Integrative functional genomic analysis identifies epigenetically regulated fibromodulin as an essential gene for glioma cell migration. Oncogene 2016, 36, 71–83. [Google Scholar] [CrossRef]
- Lee, E.J.; Jan, A.T.; Baig, M.H.; Ashraf, J.M.; Nahm, S.-S.; Kim, Y.W.; Park, S.-Y.; Choi, I. Fibromodulin: A master regulator of myostatin controlling progression of satellite cells through a myogenic program. FASEB J. 2016, 30, 2708–2719. [Google Scholar] [CrossRef]
- Berti, F.; Nogueira, J.M.; Wöhrle, S.; Sobreira, D.R.; Hawrot, K.; Dietrich, S. Time course and side-by-side analysis of mesodermal, pre-myogenic, myogenic and differentiated cell markers in the chicken model for skeletal muscle formation. J. Anat. 2015, 227, 361–382. [Google Scholar] [CrossRef]
- Deligiannis, A. Exercise rehabilitation and skeletal muscle benefits in hemodialysis patients. Clin. Nephrol. 2004, 61, S46. [Google Scholar] [PubMed]
- Dasarathy, S.; Dodig, M.; Muc, S.M.; Kalhan, S.C.; McCullough, A.J. Skeletal muscle atrophy is associated with an increased expression of myostatin and impaired satellite cell function in the portacaval anastamosis rat. Am. J. Physiol. Liver Physiol. 2004, 287, G1124–G1130. [Google Scholar] [CrossRef] [PubMed]
- Sukhanov, S.; Yoshida, T.; Tabony, A.M.; Higashi, Y.; Galvez, S.; Delafontaine, P.; Semprun-Prieto, L. Angiotensin II, oxidative stress and skeletal muscle wasting. Am. J. Med. Sci. 2011, 342, 143–147. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, P.C.D.; Wang, X.; Goldman, D. Myogenin regulates denervation-dependent muscle atrophy in mouse soleus muscle. J. Cell. Biochem. 2011, 112, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Al-Qattan, M.M.; Al-Qattan, A.M. Fibromodulin: Structure, physiological functions, and an emphasis on its potential clinical applications in various diseases. J. Coll. Physicians Surg. Pak. 2018, 28, 783–790. [Google Scholar] [PubMed]
- Massagué, J.; Chen, Y.G. Controlling TGF-beta signaling. Genes Dev. 2000, 14, 627. [Google Scholar]
- Witte, D.; Zeeh, F.; Gädeken, T.; Gieseler, F.; Rauch, B.H.; Settmacher, U.; Kaufmann, R.; Lehnert, H.; Ungefroren, H. Proteinase-activated receptor 2 is a novel regulator of TGF-β signaling in pancreatic cancer. J. Clin. Med. 2016, 5, 111. [Google Scholar] [CrossRef]
- Bierie, B.; Moses, H.L. Transforming growth factor beta (TGF-β) and inflammation in cancer. Cytokine Growth Factor Rev. 2010, 21, 49–59. [Google Scholar] [CrossRef]
- Srivastava, V.; Khanna, M.; Sharma, S.; Kumar, B. Resolution of immune response by recombinant transforming growth factor-beta (rTGF-β) during influenza A virus infection. Indian J. Med Res. 2012, 136, 641–648. [Google Scholar]
- Lijnen, P.J.; Petrov, V.V.; Fagard, R.H. Stimulation of collagen production by TGF-beta 1 during differenatiation of cardiac fibroblasts to myofibro-blasts. Am. J. Hypertens. 2002, 15, A99. [Google Scholar] [CrossRef]
- Casey, T.; Eneman, J.D.; Crocker, A.; White, J.; Tessitore, J.; Stanley, M.; Harlow, S.P.; Bunn, J.Y.; Weaver, D.L.; Muss, H.; et al. Cancer associated fibroblasts stimulated by transforming growth factor beta1 (TGF-β1) increase invasion rate of tumor cells: A population study. Breast Cancer Res. Treat. 2007, 110, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Nejad, L.D.; Biglari, A.; Annese, T.; Ribatti, M. Recombinant fibromodulin and decorin effects on NF-κB and TGFβ1 in the 4T1 breast cancer cell line. Oncol. Lett. 2017, 13, 4475–4480. [Google Scholar] [CrossRef] [PubMed]
- Angelis, C.D.; Molinari, S.; Donne, A.L.; Coletta, M.; Cossu, G. Differential response of embryonic and fetal myoblasts to TGFβ: A possible regulatory mechanism of skeletal muscle histogenesis. Development 1994, 120, 925–933. [Google Scholar]
- Vaidya, T.B.; Rhodes, S.J.; Taparowsky, E.J.; Konieczny, S.F. Fibroblast growth factor and transforming growth factor beta repress transcription of the myogenic regulatory gene MyoD1. Mol. Cell. Boil. 1989, 9, 3576–3579. [Google Scholar] [CrossRef]
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Fmod | ATGGGCAGCACATCTCGATT | TCTTCGGCTTTGCAGGTCAT |
| MyoG | CGGAGGCTGAAGAAGGTGAA | CGGTCCTCTGCCTGGTCAT |
| Mb | CCCTGAGACTTTGGATCGCTT | CTGGGATTTTGTGCTTCGTGG |
| MyoD | GCTACTACACGGAATCACCAAAT | CTGGGCTCCACTGTCACTCA |
| MyHC | CTCCTCACGCTTTGGTAA | TGATAGTCGTATGGGTTGGT |
| Atrogin-1 | TCAACGGGTCGGCAAGTCT | TCCCTCCCATCGCTCAGTC |
| MuRF-1 | GGCAGCAGCATCATCTCGG | CCTCGCAGGTGACGCAGTAG |
| GAPDH | TCCTCCACCTTTGATGCG | GTGCCTGGCTCACTCCTT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, H.; Cui, C.; Han, S.; Chen, Y.; Zhao, J.; He, H.; Li, D.; Zhu, Q. Fibromodulin Modulates Chicken Skeletal Muscle Development via the Transforming Growth Factor-β Signaling Pathway. Animals 2020, 10, 1477. https://doi.org/10.3390/ani10091477
Yin H, Cui C, Han S, Chen Y, Zhao J, He H, Li D, Zhu Q. Fibromodulin Modulates Chicken Skeletal Muscle Development via the Transforming Growth Factor-β Signaling Pathway. Animals. 2020; 10(9):1477. https://doi.org/10.3390/ani10091477
Chicago/Turabian StyleYin, Huadong, Can Cui, Shunshun Han, Yuqi Chen, Jing Zhao, Haorong He, Diyan Li, and Qing Zhu. 2020. "Fibromodulin Modulates Chicken Skeletal Muscle Development via the Transforming Growth Factor-β Signaling Pathway" Animals 10, no. 9: 1477. https://doi.org/10.3390/ani10091477
APA StyleYin, H., Cui, C., Han, S., Chen, Y., Zhao, J., He, H., Li, D., & Zhu, Q. (2020). Fibromodulin Modulates Chicken Skeletal Muscle Development via the Transforming Growth Factor-β Signaling Pathway. Animals, 10(9), 1477. https://doi.org/10.3390/ani10091477





