Nutrigenomic Effects of Long-Term Grape Pomace Supplementation in Dairy Cows
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Study Design
2.2. Extraction and Determination of GP Total Polyphenols Content (TPC)
2.3. Sampling
2.4. Milk and Feed Composition Analysis
2.5. Library Preparation and RNA-Seq Analysis
2.6. Functional Analysis and Statistics
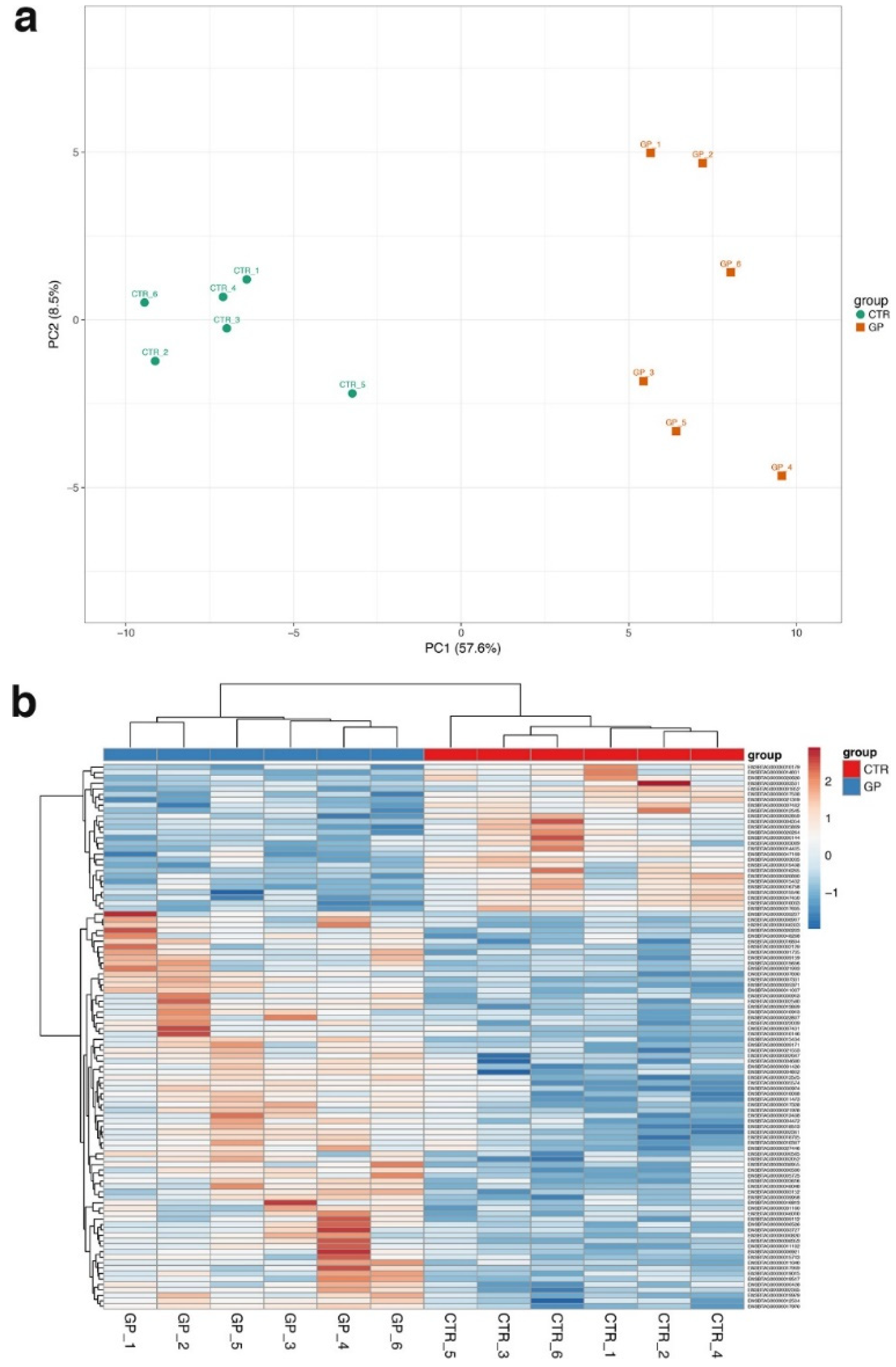
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Van Kernebeek, H.R.J.; Oosting, S.J.; Van Ittersum, M.K.; Bikker, P.; De Boer, I.J.M. Saving land to feed a growing population: Consequences for consumption of crop and livestock products. Int. J. Life Cycle Assess. 2016, 21, 677–687. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.S.; Feugang, J.M.; Liao, S.F. A Nutrigenomics Approach Using RNA Sequencing Technology to Study Nutrient-Gene Interactions in Agricultural Animals. Curr. Dev. Nutr. 2019, 3, nzz082. [Google Scholar] [CrossRef] [Green Version]
- EU. A General Union Environment Action Programme to 2020 “Living well, within the limits of our planet”. Off. J. Eur. Union 2013, L354, 171–200. [Google Scholar]
- Leiva-Candia, D.E.; Pinzi, S.; Redel-Macías, M.D.; Koutinas, A.; Webb, C.; Dorado, M.P. The potential for agro-industrial waste utilization using oleaginous yeast for the production of biodiesel. Fuel 2014, 123, 33–42. [Google Scholar] [CrossRef]
- Schieber, A.; Stintzing, F.C.; Carle, R. By-products of plant food processing as a source of functional compounds—Recent developments. Trends Food Sci. Technol. 2001, 12, 401–413. [Google Scholar] [CrossRef]
- Vauzour, D.; Rodriguez-Mateos, A.; Corona, G.; Oruna-Concha, M.J.; Spencer, J.P.E. Polyphenols and human health: Prevention of disease and mechanisms of action. Nutrients 2010, 2, 1106–1131. [Google Scholar] [CrossRef] [Green Version]
- Federici, F.; Fava, F.; Kalogerakis, N.; Mantzavinos, D. Valorisation of agro-industrial by-products, effluents and waste: Concept, opportunities and the case of olive mill waste waters. J. Chem. Technol. Biotechnol. 2009, 84, 895–900. [Google Scholar] [CrossRef]
- Mirzaei-Aghsaghali, A.; Maheri-Sis, N. Nutritive value of some agro-industrial by-products for ruminants—A review. World J. Zool. 2008, 3, 40–46. [Google Scholar]
- Makris, D.P.; Boskou, G.; Andrikopoulos, N.K. Polyphenolic content and in vitro antioxidant characteristics of wine industry and other agri-food solid waste extracts. J. Food Compos. Anal. 2007, 20, 125–132. [Google Scholar] [CrossRef]
- Santos, N.W.; Santos, G.T.D.; Silva-Kazama, D.C.; Grande, P.A.; Pintro, P.M.; de Marchi, F.E.; Jobim, C.C.; Petit, H.V. Production, composition and antioxidants in milk of dairy cows fed diets containing soybean oil and grape residue silage. Livest. Sci. 2014, 159, 37–45. [Google Scholar] [CrossRef]
- Llobera, A.; Cañellas, J. Antioxidant activity and dietary fibre of Prensal Blanc white grape (Vitis vinifera) by-products. Int. J. Food Sci. Technol. 2008, 43, 1953–1959. [Google Scholar] [CrossRef]
- Negro, C.; Tommasi, L.; Miceli, A. Phenolic compounds and antioxidant activity from red grape marc extracts. Bioresour. Technol. 2003, 87, 41–44. [Google Scholar] [CrossRef]
- Viveros, A.; Chamorro, S.; Pizarro, M.; Arija, I.; Centeno, C.; Brenes, A. Effects of dietary polyphenol-rich grape products on intestinal microflora and gut morphology in broiler chicks. Poult. Sci. 2011, 90, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Gessner, D.K.; Fiesel, A.; Most, E.; Dinges, J.; Wen, G.; Ringseis, R.; Eder, K. Supplementation of a grape seed and grape marc meal extract decreases activities of the oxidative stress-responsive transcription factors NF-κB and Nrf2 in the duodenal mucosa of pigs. Acta Vet. Scand. 2013, 55, 18. [Google Scholar] [CrossRef] [Green Version]
- Fiesel, A.; Gessner, D.K.; Most, E.; Eder, K. Effects of dietary polyphenol-rich plant products from grape or hop on pro-inflammatory gene expression in the intestine, nutrient digestibility and faecal microbiota of weaned pigs. BMC Vet. Res. 2014, 10, 196. [Google Scholar] [CrossRef] [Green Version]
- Ishida, K.; Kishi, Y.; Oishi, K.; Hirooka, H.; Kumagai, H. Effects of feeding polyphenol-rich winery wastes on digestibility, nitrogen utilization, ruminal fermentation, antioxidant status and oxidative stress in wethers. Anim. Sci. J. 2015, 86, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, F.; Palazzo, F.; Castellani, F.; Masetti, G.; Grotta, L.; Cichelli, A.; Martino, G. Rumen microbiome in dairy calves fed copper and grape-pomace dietary supplementations: Composition and predicted functional profile. PLoS ONE 2018, 13, e0205670. [Google Scholar] [CrossRef]
- Kafantaris, I.; Kotsampasi, B.; Christodoulou, V.; Kokka, E.; Kouka, P.; Terzopoulou, Z.; Gerasopoulos, K.; Stagos, D.; Mitsagga, C.; Giavasis, I.; et al. Grape pomace improves antioxidant capacity and faecal microflora of lambs. J. Anim. Physiol. Anim. Nutr. 2017, 101, e108–e121. [Google Scholar] [CrossRef] [PubMed]
- Nudda, A.; Correddu, F.; Marzano, A.; Battacone, G.; Nicolussi, P.; Bonelli, P.; Pulina, G. Effects of diets containing grape seed, linseed, or both on milk production traits, liver and kidney activities, and immunity of lactating dairy ewes. J. Dairy Sci. 2015, 98, 1157–1166. [Google Scholar] [CrossRef] [Green Version]
- Gessner, D.K.; Koch, C.; Romberg, F.J.; Winkler, A.; Dusel, G.; Herzog, E.; Most, E.; Eder, K. The effect of grape seed and grape marc meal extract on milk performance and the expression of genes of endoplasmic reticulum stress and inflammation in the liver of dairy cows in early lactation. J. Dairy Sci. 2015, 98, 8856–8868. [Google Scholar] [CrossRef] [Green Version]
- Manso, T.; Gallardo, B.; Salvá, A.; Guerra-Rivas, C.; Mantecón, A.R.; Lavín, P.; de la Fuente, M.A. Influence of dietary grape pomace combined with linseed oil on fatty acid profile and milk composition. J. Dairy Sci. 2016, 99, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Chedea, V.S.; Pelmus, R.S.; Lazar, C.; Pistol, G.C.; Calin, L.G.; Toma, S.M.; Dragomir, C.; Taranu, I. Effects of a diet containing dried grape pomace on blood metabolites and milk composition of dairy cows. J. Sci. Food Agric. 2017, 97, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Ianni, A.; Martino, G. Dietary Grape Pomace Supplementation in Dairy Cows: Effect on Nutritional Quality of Milk and Its Derived Dairy Products. Foods 2020, 9, 168. [Google Scholar] [CrossRef] [Green Version]
- Iannaccone, M.; Elgendy, R.; Giantin, M.; Martino, C.; Giansante, D.; Ianni, A.; Dacasto, M.; Martino, G. RNA sequencing-based whole-transcriptome analysis of friesian cattle fed with grape pomace-supplemented diet. Animals 2018, 8, 188. [Google Scholar] [CrossRef] [Green Version]
- European Union. Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Off. J. Eur. Union 2010, 53, 33–79. [Google Scholar]
- Ianni, A.; Di Luca, A.; Martino, C.; Bennato, F.; Marone, E.; Grotta, L.; Cichelli, A.; Martino, G. Dietary supplementation of dried grape pomace increases the amount of linoleic acid in beef, reduces the lipid oxidation and modifies the volatile profile. Animals 2019, 9, 578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singleton, V.L.; Rossi, J. Colorimetry of total phenolics with phosphomolybdic. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Helrick, K. Official Methods of Analysis (AOAC); Association of Official Analytical Chemist: Washington, DC, USA, 1990; ISBN 8589439488. [Google Scholar]
- Goering, H.K.; Van Soest, P.J. Forage Fiber Analyses (Apparatus, Reagent, Procedures and Some Applications): Agriculture Handbook No. 379; Agricultural Research Service, US Department of Agriculture: Washington, DC, USA, 1970. [Google Scholar]
- Andrews, S.; Krueger, F.; Seconds-Pichon, A.; Biggins, F.; Wingett, S. FastQC. A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 April 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Averilla, J.N.; Oh, J.; Kim, H.J.; Kim, J.S.; Kim, J.S. Potential health benefits of phenolic compounds in grape processing by-products. Food Sci. Biotechnol. 2019, 28, 1607–1615. [Google Scholar] [CrossRef]
- Ianni, A.; Di Maio, G.; Pittia, P.; Grotta, L.; Perpetuini, G.; Tofalo, R.; Cichelli, A.; Martino, G. Chemical–nutritional quality and oxidative stability of milk and dairy products obtained from Friesian cows fed with a dietary supplementation of dried grape pomace. J. Sci. Food Agric. 2019, 99, 3635–3643. [Google Scholar] [CrossRef] [PubMed]
- Ianni, A.; Innosa, D.; Martino, C.; Bennato, F.; Martino, G. Short communication: Compositional characteristics and aromatic profile of caciotta cheese obtained from Friesian cows fed with a dietary supplementation of dried grape pomace. J. Dairy Sci. 2019, 102, 1025–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scuderi, R.A.; Ebenstein, D.B.; Lam, Y.W.; Kraft, J.; Greenwood, S.L. Inclusion of grape marc in dairy cattle rations alters the bovine milk proteome. J. Dairy Res. 2019, 86, 154–161. [Google Scholar] [CrossRef] [Green Version]
- Cotoras, M.; Vivanco, H.; Melo, R.; Aguirre, M.; Silva, E.; Mendoza, L. In vitro and in vivo evaluation of the antioxidant and prooxidant activity of phenolic compounds obtained from grape (Vitis vinifera) pomace. Molecules 2014, 19, 21154–21167. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Cabral, C.; Kumar, R.; Ganguly, R.; Rana, H.K.; Gupta, A.; Lauro, M.R.; Carbone, C.; Reis, F.; Pandey, A.K. Beneficial effects of dietary polyphenols on gut microbiota and strategies to improve delivery efficiency. Nutrients 2019, 11, 2216. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Tsao, R. Dietary polyphenols, oxidative stress and antioxidant and anti-inflammatory effects. Curr. Opin. Food Sci. 2016, 8, 33–42. [Google Scholar] [CrossRef]
- Yahfoufi, N.; Alsadi, N.; Jambi, M.; Matar, C. The immunomodulatory and anti-inflammatory role of polyphenols. Nutrients 2018, 10, 1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Focaccetti, C.; Izzi, V.; Benvenuto, M.; Fazi, S.; Ciuffa, S.; Giganti, M.G.; Potenza, V.; Manzari, V.; Modesti, A.; Bei, R. Polyphenols as immunomodulatory compounds in the tumor microenvironment: Friends or foes? Int. J. Mol. Sci. 2019, 20, 1714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, H.; Sureda, A.; Belwal, T.; Çetinkaya, S.; Süntar, İ.; Tejada, S.; Devkota, H.P.; Ullah, H.; Aschner, M. Polyphenols in the treatment of autoimmune diseases. Autoimmun. Rev. 2019, 18, 647–657. [Google Scholar] [CrossRef]
- Jin, Q.; O’Hair, J.; Stewart, A.C.; O’Keefe, S.F.; Neilson, A.P.; Kim, Y.T.; McGuire, M.; Lee, A.; Wilder, G.; Huang, H. Compositional characterization of different industrial white and red grape pomaces in Virginia and the potential valorization of the major components. Foods 2019, 8, 667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St. Laurent, G.; Shtokalo, D.; Tackett, M.R.; Yang, Z.; Vyatkin, Y.; Milos, P.M.; Seilheimer, B.; McCaffrey, T.A.; Kapranov, P. On the importance of small changes in RNA expression. Methods 2013, 63, 18–24. [Google Scholar]
- Garosi, P.; De Filippo, C.; van Erk, M.; Rocca-Serra, P.; Sansone, S.-A.; Elliott, R. Defining best practice for microarray analyses in nutrigenomic studies. Br. J. Nutr. 2005, 93, 425–432. [Google Scholar] [CrossRef]
- Pagmantidis, V.; Méplan, C.; Van Schothorst, E.M.; Keijer, J.; Hesketh, J.E. Supplementation of healthy volunteers with nutritionally relevant amounts of selenium increases the expression of lymphocyte protein biosynthesis genes. Am. J. Clin. Nutr. 2008, 87, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Elgendy, R.; Giantin, M.; Castellani, F.; Grotta, L.; Palazzo, F.; Dacasto, M.; Martino, G. Transcriptomic signature of high dietary organic selenium supplementation in sheep: A nutrigenomic insight using a custom microarray platform and gene set enrichment analysis. J. Anim. Sci. 2016, 94, 3169–3184. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Liu, Q.; Liang, C.; Zhou, L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 2020, 29, 378–390. [Google Scholar] [CrossRef]
- Elia, G.; Santoro, M.G. Regulation of heat shock protein synthesis by quercetin in human erythroleukaemia cells. Biochem. J. 1994, 300, 201–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, N.; Nakai, A.; Nagata, K. Quercetin suppresses heat shock response by down regulation of HSF1. Biochem. Biophys. Res. Commun. 1995, 208, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- Aalinkeel, R.; Bindukumar, B.; Reynolds, J.L.; Sykes, D.E.; Mahajan, S.D.; Chadha, K.C.; Schwartz, S.A. The dietary bioflavonoid, quercetin, selectively induces apoptosis of prostate cancer cells by down-regulating the expression of heat shock protein 90. Prostate 2008, 68, 1773–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youle, R.J.; Van Der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilkerson, R. A disturbance in the force: Cellular stress sensing by the mitochondrial network. Antioxidants 2018, 7, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue-Yamauchi, A.; Oda, H. Depletion of mitochondrial fission factor DRP1 causes increased apoptosis in human colon cancer cells. Biochem. Biophys. Res. Commun. 2012, 421, 81–85. [Google Scholar] [CrossRef]
- Tak, H.; Eun, J.W.; Kim, J.; Park, S.J.; Kim, C.; Ji, E.; Lee, H.; Kang, H.; Cho, D.H.; Lee, K.; et al. T-cell-restricted intracellular antigen 1 facilitates mitochondrial fragmentation by enhancing the expression of mitochondrial fission factor. Cell Death Differ. 2017, 24, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Prieto, J.; León, M.; Ponsoda, X.; García-García, F.; Bort, R.; Serna, E.; Barneo-Muñoz, M.; Palau, F.; Dopazo, J.; López-García, C.; et al. Dysfunctional mitochondrial fission impairs cell reprogramming. Cell Cycle 2016, 15, 3240–3250. [Google Scholar] [CrossRef] [Green Version]
- Toyama, E.Q.; Herzig, S.; Courchet, J.; Lewis, T.L., Jr.; Losón, O.C.; Hellberg, K.; Young, N.P.; Chen, H.; Polleux, F.; Chan, D.C.; et al. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 2016, 351, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.W.A.; Chen, J.; Han, Q.; Xu, Y.; Ishfaq, M.; Teng, X. Ammonia inhalation impaired immune function and mitochondrial integrity in the broilers bursa of fabricius: Implication of oxidative stress and apoptosis. Ecotoxicol. Environ. Saf. 2020, 190, 110078. [Google Scholar] [CrossRef]
- Yang, J.; Guo, W.; Wang, J.; Yang, X.; Zhang, Z.; Zhao, Z. T-2 toxin-induced oxidative stress leads to imbalance of mitochondrial fission and fusion to activate cellular apoptosis in the human liver 7702 cell line. Toxins 2020, 12, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, J.; Zhang, C.; Sun, Y.C.; Zhang, Q.; Lv, M.W.; Guo, K.; Li, J.L. Cadmium exposure triggers mitochondrial dysfunction and oxidative stress in chicken (Gallus gallus) kidney via mitochondrial UPR inhibition and Nrf2-mediated antioxidant defense activation. Sci. Total Environ. 2019, 689, 1160–1171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, J.; Zhu, J.; Chen, Y.; Han, X. Microcystin-leucine-arginine induced neurotoxicity by initiating mitochondrial fission in hippocampal neurons. Sci. Total Environ. 2020, 703, 134702. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Huang, J.; Shen, J.; Bai, Q. Quercetin improves endothelial insulin sensitivity in obese mice by inhibiting Drp1 phosphorylation at serine 616 and mitochondrial fragmentation. Acta Biochim. Biophys. Sin. 2019, 51, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, S.; Li, J.; Liu, K.; Huang, F.; Liu, B. Metformin and resveratrol inhibit Drp1-mediated mitochondrial fission and prevent ER stress-associated NLRP3 inflammasome activation in the adipose tissue of diabetic mice. Mol. Cell. Endocrinol. 2016, 434, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Naoi, M.; Wu, Y.; Shamoto-Nagai, M.; Maruyama, W. Mitochondria in Neuroprotection by Phytochemicals: Bioactive Polyphenols Modulate Mitochondrial Apoptosis System, Function and Structure. Int. J. Mol. Sci. 2019, 20, 2451. [Google Scholar] [CrossRef] [Green Version]
- Ferraz, R.C.; Camara, H.; De-Souza, E.A.; Pinto, S.; Pinca, A.P.F.; Silva, R.C.; Sato, V.N.; Castilho, B.A.; Mori, M.A. IMPACT is a GCN2 inhibitor that limits lifespan in Caenorhabditis elegans. BMC Biol. 2016, 14, 87. [Google Scholar] [CrossRef] [Green Version]
- Cambiaghi, T.D.; Pereira, C.M.; Shanmugam, R.; Bolech, M.; Wek, R.C.; Sattlegger, E.; Castilho, B.A. Evolutionarily conserved IMPACT impairs various stress responses that require GCN1 for activating the eIF2 kinase GCN2. Biochem. Biophys. Res. Commun. 2014, 443, 592–597. [Google Scholar] [CrossRef]
- Pereira, C.M.; Filev, R.; Dubiela, F.P.; Brandão, B.B.; Queiroz, C.M.; Ludwig, R.G.; Hipolide, D.; Longo, B.M.; Mello, L.E.; Mori, M.A.; et al. The GCN2 inhibitor IMPACT contributes to diet-induced obesity and body temperature control. PLoS ONE 2019, 14, e0217287. [Google Scholar] [CrossRef] [Green Version]
- Castilho, B.A.; Shanmugam, R.; Silva, R.C.; Ramesh, R.; Himme, B.M.; Sattlegger, E. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 1948–1968. [Google Scholar] [CrossRef] [Green Version]
- Ravishankara, B.; Liua, H.; Shindea, R.; Chaudharya, K.; Xiaoa, W.; Bradleya, J.; Koritzinsky, M.; Madaio, M.P.; McGaha, T.L. The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity. Proc. Natl. Acad. Sci. USA 2015, 112, 10774–10779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murguía, J.R.; Serrano, R. New functions of protein kinase Gcn2 in yeast and mammals. IUBMB Life 2012, 64, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Hachimura, S.; Totsuka, M.; Hosono, A. Immunomodulation by food: Impact on gut immunity and immune cell function. Biosci. Biotechnol. Biochem. 2018, 82, 584–599. [Google Scholar] [CrossRef] [PubMed]
- Revelo, X.S.; Winer, S.; Winer, D.A. Starving Intestinal Inflammation with the Amino Acid Sensor GCN2. Cell Metab. 2016, 23, 763–765. [Google Scholar] [CrossRef] [Green Version]
- Alqinyah, M.; Hooks, S.B. Regulating the regulators: Epigenetic, transcriptional, and post-translational regulation of RGS proteins. Cell. Signal. 2018, 42, 77–87. [Google Scholar] [CrossRef]
- Ota, A.; Sawai, M.; Sakurai, H. Stress-induced transcription of regulator of G protein signaling 2 (RGS2) by heat shock transcription factor HSF1. Biochimie 2013, 95, 1432–1436. [Google Scholar] [CrossRef]
- Zmijewski, J.W.; Song, L.; Jope, R.S.; Harkins, L.; Cobbs, C.S. Oxidative stress and heat shock stimulate RGS2 expression in 1321N1 astrocytoma cells. Arch. Biochem. Biophys. 2001, 392, 192–196. [Google Scholar] [CrossRef]
- Salim, S.; Asghar, M.; Taneja, M.; Hovatta, I.; Wu, Y.L.; Saha, K.; Sarraj, N.; Hite, B. Novel role of RGS2 in regulation of antioxidant homeostasis in neuronal cells. FEBS Lett. 2011, 585, 1375–1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salaga, M.; Storr, M.; Martemyanov, K.A.; Fichna, J. RGS proteins as targets in the treatment of intestinal inflammation and visceral pain: New insights and future perspectives. BioEssays 2016, 38, 344–354. [Google Scholar] [CrossRef] [Green Version]
- Wang, D. The essential role of G protein-coupled receptor (GPCR) signaling in regulating T cell immunity. Immunopharmacol. Immunotoxicol. 2018, 40, 187–192. [Google Scholar] [CrossRef]
- Vasiliou, V.; Vasiliou, K.; Nebert, D.W. Human ATP-binding cassette (ABC) transporter family. Hum. Genomics 2009, 3, 281. [Google Scholar] [CrossRef]
- Chaiworapongsa, T.; Romero, R.; Whitten, A.; Tarca, A.L.; Bhatti, G.; Draghici, S.; Chaemsaithong, P.; Miranda, J.; Hassan, S.S. Differences and similarities in the transcriptional profile of peripheral whole blood in early and lateonset preeclampsia: Insights into the molecular basis of the phenotype of preeclampsia. J. Perinat. Med. 2013, 41, 485–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dréan, A.; Rosenberg, S.; Lejeune, F.X.; Goli, L.; Nadaradjane, A.A.; Guehennec, J.; Schmitt, C.; Verreault, M.; Bielle, F.; Mokhtari, K.; et al. ATP binding cassette (ABC) transporters: Expression and clinical value in glioblastoma. J. Neurooncol. 2018, 138, 479–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Tellez, G.; Escobar, J.; Vazquez-Anon, M. Impact of Trace Minerals on Wound Healing of Footpad Dermatitis in Broilers. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gessner, D.K.; Ringseis, R.; Eder, K. Potential of plant polyphenols to combat oxidative stress and inflammatory processes in farm animals. J. Anim. Physiol. Anim. Nutr. 2017, 101, 605–628. [Google Scholar] [CrossRef]
- Ding, S.; Jiang, H.; Fang, J. Regulation of immune function by polyphenols. J. Immunol. Res. 2018. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, E.R.; Brogi, S.; Lucon-Júnior, J.F.; Campiani, G.; Gemma, S.; Maquiaveli, C.D.C. Dietary polyphenols rutin, taxifolin and quercetin related compounds target: Leishmania amazonensis arginase. Food Funct. 2019, 10, 3172–3180. [Google Scholar] [CrossRef]
- Anthony, J.P.; Fyfe, L.; Stewart, D.; McDougall, G.J. Differential effectiveness of berry polyphenols as anti-giardial agents. Parasitology 2011, 138, 1110–1116. [Google Scholar] [CrossRef] [Green Version]
- Bottari, N.B.; Schetinger, M.R.C.; Pillat, M.M.; Palma, T.V.; Ulrich, H.; Alves, M.S.; Morsch, V.M.; Melazzo, C.; de Barros, L.D.; Garcia, J.L.; et al. Resveratrol as a Therapy to Restore Neurogliogenesis of Neural Progenitor Cells Infected by Toxoplasma gondii. Mol. Neurobiol. 2019, 56, 2328–2338. [Google Scholar] [CrossRef]
- Soliman, R.H.; Ismail, O.A.; Badr, M.S.; Nasr, S.M. Resveratrol ameliorates oxidative stress and organ dysfunction in Schistosoma mansoni infected mice. Exp. Parasitol. 2017, 174, 52–58. [Google Scholar] [CrossRef]
- Bahramsoltani, R.; Sodagari, H.R.; Farzaei, M.H.; Abdolghaffari, A.H.; Gooshe, M.; Rezaei, N. The preventive and therapeutic potential of natural polyphenols on influenza. Expert Rev. Anti. Infect. Ther. 2016, 14, 57–80. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, P.; Sahuc, M.E.; Rouillé, Y.; Rivière, C.; Bonneau, N.; Vandeputte, A.; Brodin, P.; Goswami, M.; Bandyopadhyay, T.; Dubuisson, J.; et al. Theaflavins, polyphenols of black tea, inhibit entry of hepatitis C virus in cell culture. PLoS ONE 2018, 13, e0198226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Wu, Q.; Chen, Y.; Duan, M.; Tian, G.; Deng, X.; Sun, Y.; Zhou, T.; Zhang, G.; Chen, W.; et al. Inhibition of proanthocyanidin A2 on porcine reproductive and respiratory syndrome virus replication in vitro. PLoS ONE 2018, 13, e0193309. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Cui, Q.; Fu, Q.; Song, X.; Jia, R.; Yang, Y.; Zou, Y.; Li, L.; He, C.; Liang, X.; et al. Antiviral properties of resveratrol against pseudorabies virus are associated with the inhibition of IκB kinase activation. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Chedea, V.S.; Palade, L.M.; Pelmus, R.S.; Dragomir, C.; Taranu, I. Red grape pomace rich in polyphenols diet increases the antioxidant status in key organs—Kidneys, liver, and spleen of piglets. Animals 2019, 9, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herosimczyk, A.; Lepczyński, A.; Ożgo, M.; Tuśnio, A.; Taciak, M.; Barszcz, M. Effect of dietary inclusion of 1% or 3% of native chicory inulin on the large intestinal mucosa proteome of growing pigs. Animal 2020. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Matzapetakis, M.; Horvatić, A.; Terré, M.; Bach, A.; Kuleš, J.; Yeste, N.; Gómez, N.; Arroyo, L.; Rodríguez-Tomàs, E.; et al. Metabolome and proteome changes in skeletal muscle and blood of pre-weaning calves fed leucine and threonine supplemented diets. J. Proteomics 2020, 216, 103677. [Google Scholar] [CrossRef] [PubMed]


| Chemical Composition of GP | |
|---|---|
| Dry Matter, % | 92.20 |
| Crude protein 1, % | 12.40 |
| Ether extract 1, % | 4.20 |
| Crude fibre 1, % | 31.80 |
| Neutral-detergent fibre 1, % | 42.20 |
| Acid detergent fibre 1, % | 41.30 |
| Acid-detergent lignin 1, % | 28.80 |
| Starch 1, % | - |
| Ash 1, % | 8.50 |
| Total Polyphenols Content (TCP) in GP | |
| TPC (GAE 2 mg/g) | 16.07 ± 1.41 |
| Diets | ||
|---|---|---|
| Ingredients of the diets | CTR/basal | GP |
| Corn silage, % | 45.70 | 45.50 |
| Second-cut alfalfa hay, % | 20.30 | 20.20 |
| Wheat straw, % | 3.80 | 1.30 |
| Corn grain meal, % | 13.50 | 13.10 |
| Soybean meal, % | 8.40 | 7.60 |
| Barley meal, % | 2.50 | 2.50 |
| Cotton seed, % | 3.80 | 2.80 |
| Dried grape pomace, % | - | 5.00 |
| Vitamins and minerals, % | 2.00 | 2.00 |
| Nutrient composition | ||
| Dry Matter, % | 61.50 | 61.70 |
| Crude protein 1, % | 15.30 | 15.90 |
| Ether extract 1, % | 2.30 | 2.20 |
| Crude fibre 1, % | 17.80 | 18.90 |
| Neutral-detergent fibre 1, % | 34.40 | 36.30 |
| Acid detergent fibre 1, % | 20.30 | 22.50 |
| Acid-detergent lignin 1, % | 4.60 | 5.90 |
| Starch 1, % | 25.20 | 24.80 |
| Ash 1, % | 4.50 | 5.10 |
| Net energy (Milk UF/kg DM) | 0.89 | 0.91 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pauletto, M.; Elgendy, R.; Ianni, A.; Marone, E.; Giantin, M.; Grotta, L.; Ramazzotti, S.; Bennato, F.; Dacasto, M.; Martino, G. Nutrigenomic Effects of Long-Term Grape Pomace Supplementation in Dairy Cows. Animals 2020, 10, 714. https://doi.org/10.3390/ani10040714
Pauletto M, Elgendy R, Ianni A, Marone E, Giantin M, Grotta L, Ramazzotti S, Bennato F, Dacasto M, Martino G. Nutrigenomic Effects of Long-Term Grape Pomace Supplementation in Dairy Cows. Animals. 2020; 10(4):714. https://doi.org/10.3390/ani10040714
Chicago/Turabian StylePauletto, Marianna, Ramy Elgendy, Andrea Ianni, Elettra Marone, Mery Giantin, Lisa Grotta, Solange Ramazzotti, Francesca Bennato, Mauro Dacasto, and Giuseppe Martino. 2020. "Nutrigenomic Effects of Long-Term Grape Pomace Supplementation in Dairy Cows" Animals 10, no. 4: 714. https://doi.org/10.3390/ani10040714
APA StylePauletto, M., Elgendy, R., Ianni, A., Marone, E., Giantin, M., Grotta, L., Ramazzotti, S., Bennato, F., Dacasto, M., & Martino, G. (2020). Nutrigenomic Effects of Long-Term Grape Pomace Supplementation in Dairy Cows. Animals, 10(4), 714. https://doi.org/10.3390/ani10040714






