Comprehensive Transcriptome Analysis Reveals Insights into Phylogeny and Positively Selected Genes of Sillago Species
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
2.2. Sample Collection, RNA Extraction, and Illumina Sequencing
2.3. Transcriptome De Novo Assembly
2.4. Orthology Determination and Phylogenetic Tree Reconstruction
2.5. Prediction of PSGs of Sillago Species
3. Results
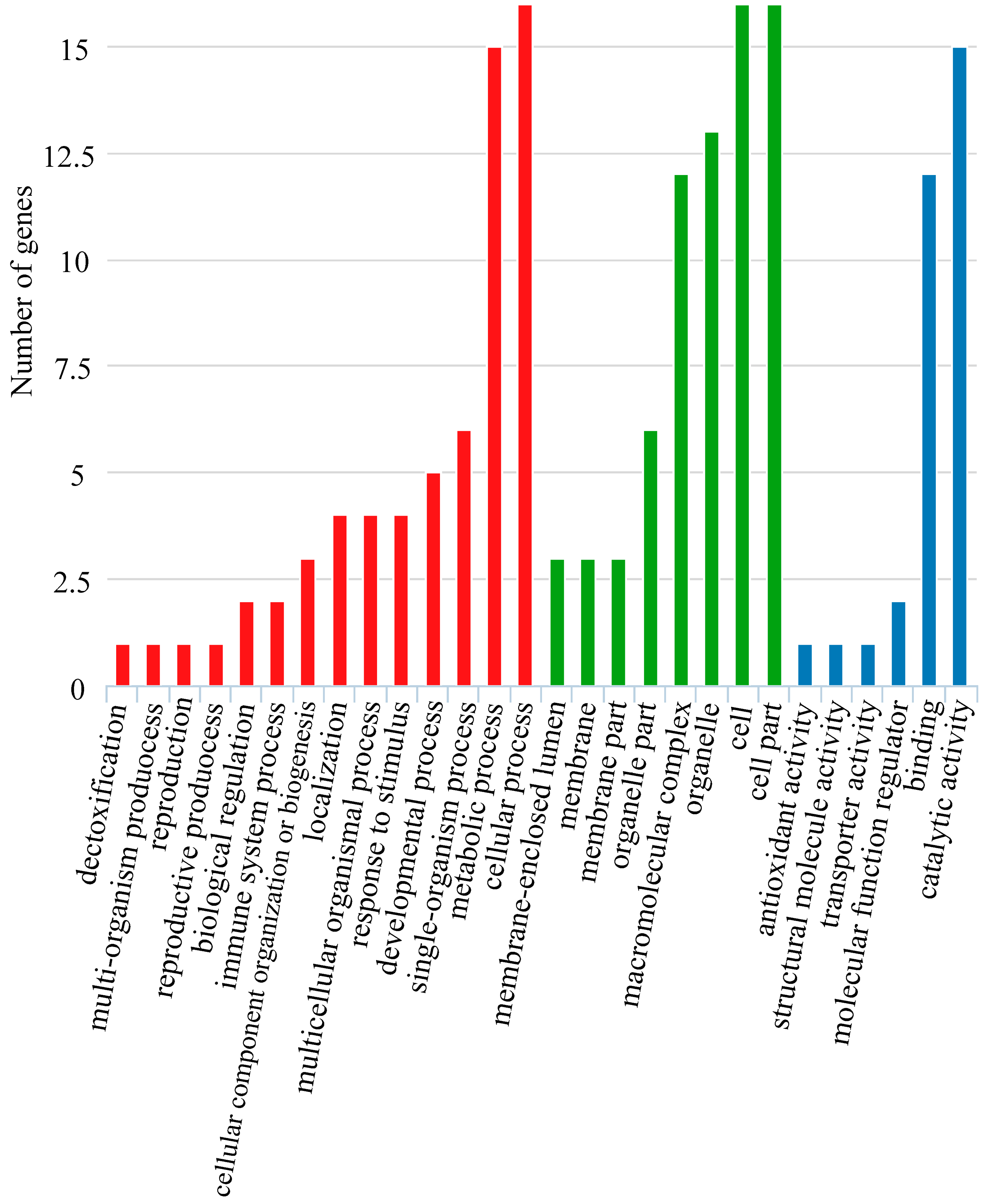
3.1. Illumina Sequencing and the De Novo Assembly of the Seven Sillago Species’ Transcriptomes
3.2. Orthologous Gene Identification and the Phylogenetic Structure of Sillago Species
3.3. PSGs Representative of Sillago Species
4. Discussion
4.1. Transcriptome Data Processing
4.2. More Accurately Determining the Phylogenetic Relationships of Seven Sillago Species
4.3. Positively selected genes Might Contribute to the Ecological Adaptation of Sillago Species
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mckay, R.J. A revision of the fishes of the family Sillaginidae. Mem. Queensl. Mus. 1985, 22, 1–73. [Google Scholar]
- Kaga, T. Phylogenetic systematics of the family Sillaginidae (Percomorpha: Order Perciformes). Zootaxa 2013, 3642, 1–105. [Google Scholar] [CrossRef]
- Mallet, A.L.; Carver, C.E.; Landry, T. Impact of suspended and off-bottom Eastern oyster culture on the benthic environment in eastern Canada. Aquaculture 2006, 255, 362–373. [Google Scholar] [CrossRef]
- Xiao, J.; Song, N.; Han, Z.; Gao, T. Description and DNA barcoding of a new Sillago species, Sillago shaoi (perciformes: Sillaginidae), in the Taiwan Strait. Zool. Stud. 2016, 55, 47. [Google Scholar] [CrossRef]
- Xu, S.; Xiao, S.; Zhu, S.; Zeng, X.; Luo, J.; Gao, T.; Chen, N. A draft genome assembly of the Chinese sillago (Sillago sinica), the first reference genome for Sillaginidae fishes. GigaScience 2018, 7, 108. [Google Scholar] [CrossRef]
- Gao, T.; Ji, D.; Xiao, Y.; Xue, T.; Yanagimoto, T.; Setoguma, T. Description and DNA barcoding of a new Sillago species, Sillago sinica (Perciformes: Sillaginidae), from coastal waters of China. Zool. Stud. 2011, 50, 254–263. [Google Scholar]
- Golani, D.; Fricke, R.; Tikochinski, Y. Sillago suezensis, a new whiting from the northern Red Sea, and status of Sillago erythraea Cuvier (Teleostei: Sillaginidae). J. Nat. Hist. 2014, 48, 413–428. [Google Scholar] [CrossRef]
- Qiu, Y.; Lee, J.; Bernasconiquadroni, F.; Soltis, D.E.; Soltis, P.S.; Zanis, M.; Zimmer, E.A.; Chen, Z.; Savolainen, V.; Chase, M.W. The earliest angiosperms: Evidence from mitochondrial, plastid and nuclear genomes. Nature 1999, 402, 404–407. [Google Scholar] [CrossRef]
- Delsuc, F.; Brinkmann, H.; Philippe, H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005, 6, 361–375. [Google Scholar] [CrossRef]
- Liang, D.; Shen, X.; Zhang, P. One thousand two hundred ninety nuclear genes from a genome-wide survey support Lungfishes as the sister group of Tetrapods. Mol. Biol. Evol. 2013, 30, 1803–1807. [Google Scholar] [CrossRef]
- Faircloth, B.C.; McCormack, J.E.; Crawford, N.G.; Harvey, M.G.; Brumfield, R.T.; Glenn, T.C. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 2012, 61, 717–726. [Google Scholar] [CrossRef]
- Lemmon, A.R.; Emme, S.A.; Lemmon, E.M. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 2012, 61, 727–744. [Google Scholar] [CrossRef]
- Simion, P.; Philippe, H.; Baurain, D.; Jager, M.; Richter, D.J.; Di Franco, A.; Roure, B.; Satoh, N.; Quéinnec, É.; Ereskovsky, A.; et al. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 2017, 27, 958–967. [Google Scholar] [CrossRef]
- Vandepoele, K.; De Vos, W.; Taylor, J.S.; Meyer, A.; Van de Peer, Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. USA 2004, 101, 1638–1643. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. Biol. Sci. 2014, 281, 20132881. [Google Scholar] [CrossRef]
- Kang, J.; Ma, X.; He, S. Evidence of high-altitude adaptation in the glyptosternoid fish, Creteuchiloglanis macropterus from the Nujiang River obtained through transcriptome analysis. BMC Evol. Biol. 2017, 17, 229. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Zhang, Z.; He, S. Comprehensive transcriptome analysis reveals accelerated genic evolution in a Tibet fish, Gymnodiptychus pachycheilus. Genome Biol. Evol. 2014, 7, 251–261. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Froese, R.; Pauly, D. FishBase. World Wide Web electronic publication. 2019. Available online: www.fishbase.org (accessed on 24 February 2020).
- Hughes, L.C.; Ortía, G.; Huang, Y.; Sun, Y.; Baldwin, C.C.; Thompson, A.W.; Arcila, D.; Betancur-R, R.; Li, C.; Becker, L.; et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA 2018, 115, 6249–6254. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Hedges, S.B.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015, 32, 835–845. [Google Scholar] [CrossRef]
- Sanderson, M.J. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 2003, 19, 301–302. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.X.; Zhang, H.R.; Niu, S.F.; Zhai, Y.; Liu, X.F. Development of polymorphic microsatellites for Sillago sihama based on next-generation sequencing and transferability to Sillago japonica. Genet. Mol. Res. 2016, 15, gmr15049046. [Google Scholar] [CrossRef] [PubMed]
- Fitch, W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970, 19, 99–113. [Google Scholar] [CrossRef]
- Near, T.J.; Eytan, R.I.; Dornburg, A.; Kuhn, K.L.; Moore, J.A.; Davis, M.P.; Wainwright, P.C.; Friedman, M.; Smith, W.L. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 2012, 109, 13698–13703. [Google Scholar] [CrossRef]
- Betancur-R, R.; Li, C.; Munroe, T.A.; Ballesteros, J.A.; Ortí, G. Addressing gene tree discordance and non-stationarity to resolve a multi-locus phylogeny of the flatfishes (Teleostei: Pleuronectiformes). Syst. Biol. 2013, 62, 763–785. [Google Scholar] [CrossRef]
- Xiao, J.G. The Taxonomy, Phylogeny and Biogeography of Sillaginidae Species in China; Ocean University of China: Qingdao, China, 2018; (Abstract in English). [Google Scholar]
- Parker, J.; Tsagkogeorga, G.; Cotton, J.A.; Liu, Y.; Provero, P.; Stupka, E.; Rossiter, S.J. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 2013, 502, 228–231. [Google Scholar] [CrossRef]
- Edwards, S.V.; Cloutier, A.; Baker, A.J. Conserved nonexonic elements: A novel class of marker for phylogenomics. Syst. Biol. 2017, 66, 1028–1044. [Google Scholar] [CrossRef]
- Schwarzhans, W.W. Fish Otoliths from the New Zealand Tertiary; New Zealand Geological Survey: Lower Hutt, New Zealand, 1984. [Google Scholar]
- Takahashi, M. On two fish-otoliths of Arctoscopus and Sillago (Teleostei) from the Kaidate Formation (Pliocene), Niigata Prefecture. Earth Sci. 1980, 34, 346–349. [Google Scholar] [CrossRef]
- Voolstra, C.R.; Sunagawa, S.; Matz, M.V.; Bayer, T.; Aranda, M.; Buschiazzo, E.; DeSalvo, M.K.; Lindquist, E.; Szmant, A.M.; Coffroth, M.A.; et al. Rapid evolution of coral proteins responsible for interaction with the environment. PLoS ONE 2011, 6, e20392. [Google Scholar] [CrossRef]
- Loar, J.W.; Seiser, R.M.; Alexandra, E.; Sagerson, H.J.; Ilias, N.; Zobel-Thropp, P.; Craig, E.A.; Lycan, D.E. Genetic and biochemical interactions among Yar1, Ltv1 and RpS3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 2004, 168, 1877–1889. [Google Scholar] [CrossRef] [PubMed]
- Vujcic, S.; Diegelman, P.; Bacchi, C.J.; Kramer, D.L.; Porter, C.W. Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem. J. 2002, 367, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, N.; Venkatraman, P.; Goldberg, A.L. Puromycin-sensitive aminopeptidase is the major peptidase responsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J. 2007, 26, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Menzies, F.M.; Hourez, R.; Imarisio, S.; Raspe, M.; Sadiq, O.; Chandraratna, D.; O’Kane, C.; Rock, K.L.; Reits, E.; Goldberg, A.L.; et al. Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum. Mol. Genet. 2010, 19, 4573–4586. [Google Scholar] [CrossRef] [PubMed]
- Vasiliou, V.; Vasiliou, K.; Nebert, D.W. Human ATP-binding cassette (ABC) transporter family. Hum. Genom. 2008, 3, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, K.; Hirota, Y.; Sawada, N.; Yuge, N.; Watanabe, M.; Uchino, Y.; Okuda, N.; Shimomura, Y.; Suhara, Y.; Okano, T. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature 2010, 468, 117–121. [Google Scholar] [CrossRef]
- Watkin, L.B.; Jessen, B.; Wiszniewski, W.; Vece, T.J.; Jan, M.; Sha, Y.; Thamsen, M.; Santos-Cortez, R.L.; Lee, K.; Gambin, T.; et al. COPA mutations impair ER-Golgi transport and cause hereditary autoimmune-mediated lung disease and arthritis. Nat. Genet. 2015, 47, 654–660. [Google Scholar] [CrossRef]
- Cho, J.G.; Lim, K.H.; Park, S.G. MED28 increases the colony-forming ability of breast cancer cells by stabilizing the ZNF224 protein upon DNA damage. Oncol. Lett. 2018, 15, 3147–3154. [Google Scholar] [CrossRef]
- Harris, C.C. Structure and function of the p53 tumor suppressor gene: Clues for rational cancer therapeutic strategies. J. Natl. Cancer Inst. 1996, 88, 1442–1455. [Google Scholar] [CrossRef]
- He, X.; Zhang, P. Serine/arginine-rich splicing factor 3 (SRSF3) regulates homologous recombination-mediated DNA repair. Mol. Cancer 2015, 14, 158. [Google Scholar] [CrossRef]
- Tannukit, S.; Crabb, T.L.; Hertel, K.J.; Wen, X.; Jans, D.A.; Paine, M.L. Identification of a novel nuclear localization signal and speckletargeting sequence of tuftelin-interacting protein 11, a splicing factor involved in spliceosome disassembly. Biochem. Biophys. Res. Commun. 2009, 390, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Sukhthankar, M.; Choi, C.K.; English, A.; Kim, J.S.; Baek, S.J. A potential proliferative gene, NUDT6, is down-regulated by green tea catechins at the post-transcriptional level. J. Nutr. Biochem. 2010, 21, 98–106. [Google Scholar] [CrossRef] [PubMed]
- García-Díaz, M.; Domínguez, O.; López-Fernández, L.A.; de Lera, L.T.; Saníger, M.L.; Ruiz, J.F.; Párraga, M.; García-Ortiz, M.J.; Kirchhoff, T.; del Mazo, J.; et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2000, 301, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liao, Z.; Lin, D. Review on the research of DEP domain function. Med. Recapitulate 2011, 17, 1772–1774. [Google Scholar] [CrossRef]
- McLennan, A.G. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 2006, 63, 123–143. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Liu, Y.; Wang, H.; Mao, X.; Chen, J.; Liu, J.; Xia, Z.; Zhang, L.; Liu, X.; Yu, T. Ischemic postconditioning influences electron transport chain protein turnover in Langendorff-perfused rat hearts. PeerJ 2016, 4, e1706. [Google Scholar] [CrossRef]
- Quan, X.; Sato-Miyata, Y.; Tsuda, M.; Muramatsu, K.; Asano, T.; Takeo, S.; Aigaki, T. Deficiency of succinyl-CoA synthetase a subunit delays development, impairs locomotor activity and reduces survival under starvation in Drosophila. Biochem. Biophys. Res. Commun. 2017, 483, 566–571. [Google Scholar] [CrossRef]
- Marton, M.J.; Crouch, D.; Hinnebusch, A.G. GCN1, a Translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol. 1993, 13, 3541–3556. [Google Scholar] [CrossRef]
- Varlamov, O.; Wu, F.; Shields, D.; Fricker, L.D. Biosynthesis and packaging of carboxypeptidase D into nascent secretory vesicles in pituitary cell lines. J. Biol. Chem. 1999, 274, 14040–14045. [Google Scholar] [CrossRef]
- Nakatsukasa, K.; Nishimura, T.; Byrne, S.D.; Okamoto, M.; Takahashi-Nakaguchi, A.; Chibana, H.; Okumura, F.; Kamura, T. The ubiquitin ligase SCF (Ucc1) acts as a metabolic switch for the glyoxylate cycle. Mol. Cell 2015, 59, 22–34. [Google Scholar] [CrossRef]
- Jung, Y.; Kim, S.; Lee, S.; Ha, K.S.; Lee, J. Effect of heterologous expression of genes involved in the elongation cycle of fatty acid synthesis on fatty acid production in Saccharomyces cerevisiae. Biotechnol. Bioprocess Eng. 2015, 20, 1–9. [Google Scholar] [CrossRef]
- Accogli, A.; Hamdan, F.F.; Poulin, C.; Nassif, C.; Rouleau, G.A.; Michaud, J.L.; Srour, M. A novel homozygous AP4B1 mutation in two brothers with AP-4 deficiency syndrome and ocular anomalies. Am. J. Med. Genet. 2018, 176, 985–991. [Google Scholar] [CrossRef]
- Luo, H.R.; Moreau, G.A.; Levin, N.; Moore, M.J. The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA 1999, 5, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Pahan, K. Ankyrin repeat and BTB/POZ domain containing protein-2 inhibits the aggregation of alpha-synuclein, Implications for Parkinson’s disease. FEBS Lett. 2013, 587, 3567–3574. [Google Scholar] [CrossRef] [PubMed]
- Contino, G.; Amati, F.; Pucci, S.; Pontieri, E.; Pichiorri, F.; Novelli, A.; Botta, A.; Mango, R.; Nardone, A.M.; Sangiuolo, F.C.; et al. Expression analysis of the gene encoding for the U-box-type ubiquitin ligase UBE4A in human tissues. Gene 2004, 328, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Epis, M.R.; Giles, K.M.; Kalinowski, F.C.; Barker, A.; Cohen, R.J.; Leedman, P.J. Regulation of expression of deoxyhypusine hydroxylase (DOHH), the enzyme that catalyzes the activation of eIF5A, by miR-331-3p and miR-642-5p in prostate cancer cells. J. Biol. Chem. 2012, 42, 35251–35259. [Google Scholar] [CrossRef]
- Yamasaki, S.; Nishida, K.; Yoshida, Y.; Itoh, M.; Hibi, M.; Hirano, T. Gab1 is required for EGF receptor signaling and the transformation by activated ErbB2. Oncogene 2003, 22, 1546–1556. [Google Scholar] [CrossRef]
- Zhang, S.; Shi, M.; Hui, C.C.; Rommens, J.M. Loss of the mouse ortholog of the shwachman-diamond syndrome gene (Sbds) results in early embryonic lethality. Mol. Cell. Biol. 2006, 26, 6656–6663. [Google Scholar] [CrossRef]
- Walker, K.A.; Blackwell, T.K. A broad but restricted requirement for TAF-5 (Human TAFII100) for embryonic transcription in Caenorhabditis elegans. J. Biol. Chem. 2003, 278, 6181–6186. [Google Scholar] [CrossRef]
- Cole, R.A.; Synek, L.; Zarsky, V.; Fowler, J.E. SEC8, a subunit of the putative arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol. 2005, 138, 2005–2018. [Google Scholar] [CrossRef]
- Kantidze, O.L.; Velichko, A.K.; Luzhin, A.V.; Razin, S.V. Heat stress-induced DNA damage. Acta Nat. 2016, 8, 75–78. [Google Scholar] [CrossRef]
- Morales, M.E.; Derbes, R.S.; Ade, C.M.; Ortego, J.C.; Stark, J.; Deininger, P.L.; Roy-Engel, A.M. Heavy metal exposure influences double strand break DNA repair outcomes. PLoS ONE 2016, 11, e0151367. [Google Scholar] [CrossRef] [PubMed]
- Battaglene, S.C.; McBride, S.; Talbot, R.B. Swim bladder inflation in larvae of cultured sand whiting, Sillago ciliata Cuvier (Sillaginidae). Aquaculture 1994, 128, 177–192. [Google Scholar] [CrossRef]
- Fu, S.J.; Cao, Z.D.; Xie, X.J. Feeding metabolism and locomotion metabolism in fishes. Chin. J. Zool. 2008, 43, 150–159. [Google Scholar]
- Bozzano, A. Vision in the rufus snake eel, Ophichthus rufus: Adaptive mechanisms for a burrowing life-style. Mar. Biol. 2003, 143, 167–174. [Google Scholar] [CrossRef]
- Puvanendran, V.; Brown, J.A. Foraging, growth and survival of Atlantic cod larvae reared in different light intensities and photoperiods. Aquaculture 2002, 214, 131–151. [Google Scholar] [CrossRef]
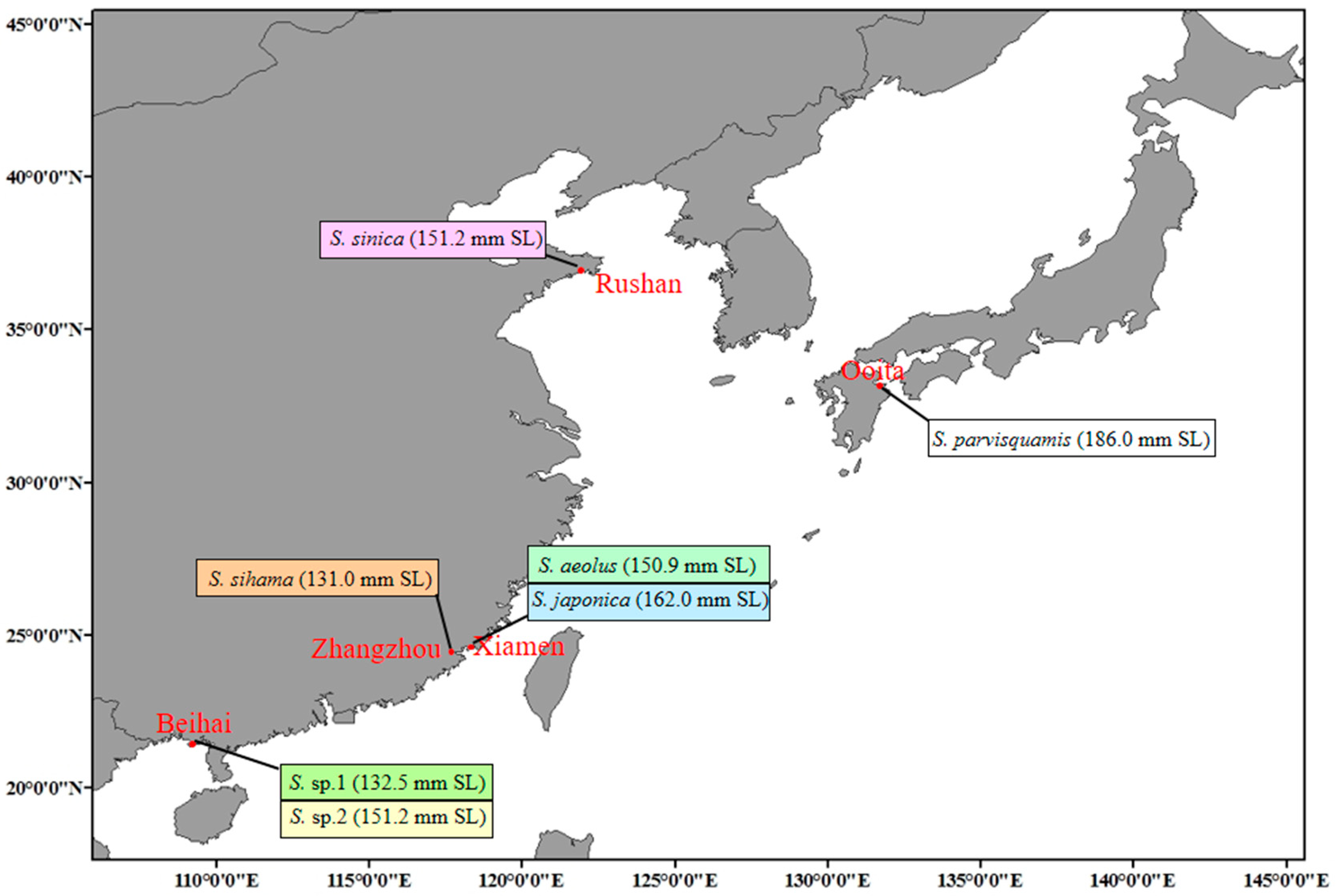
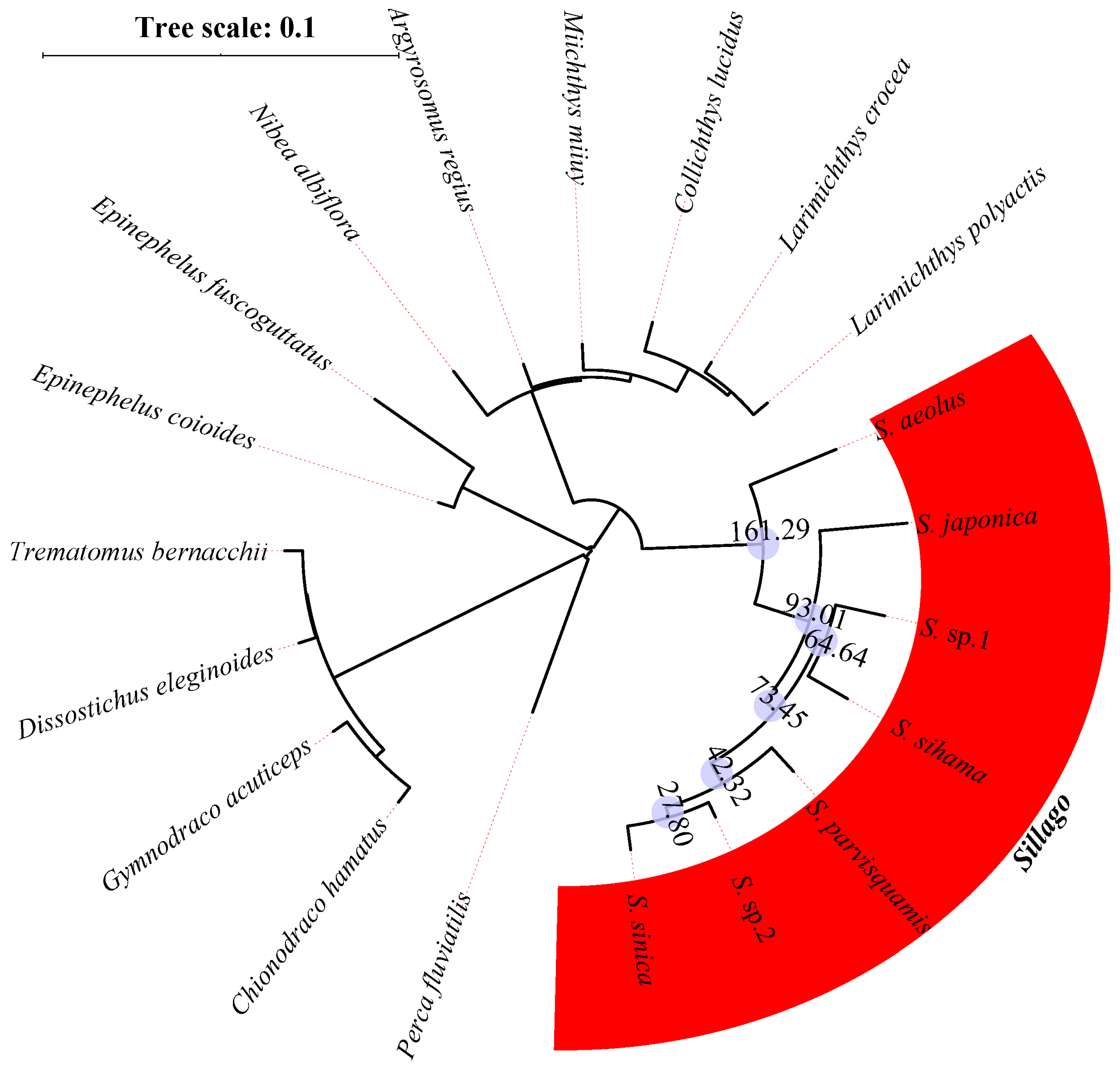
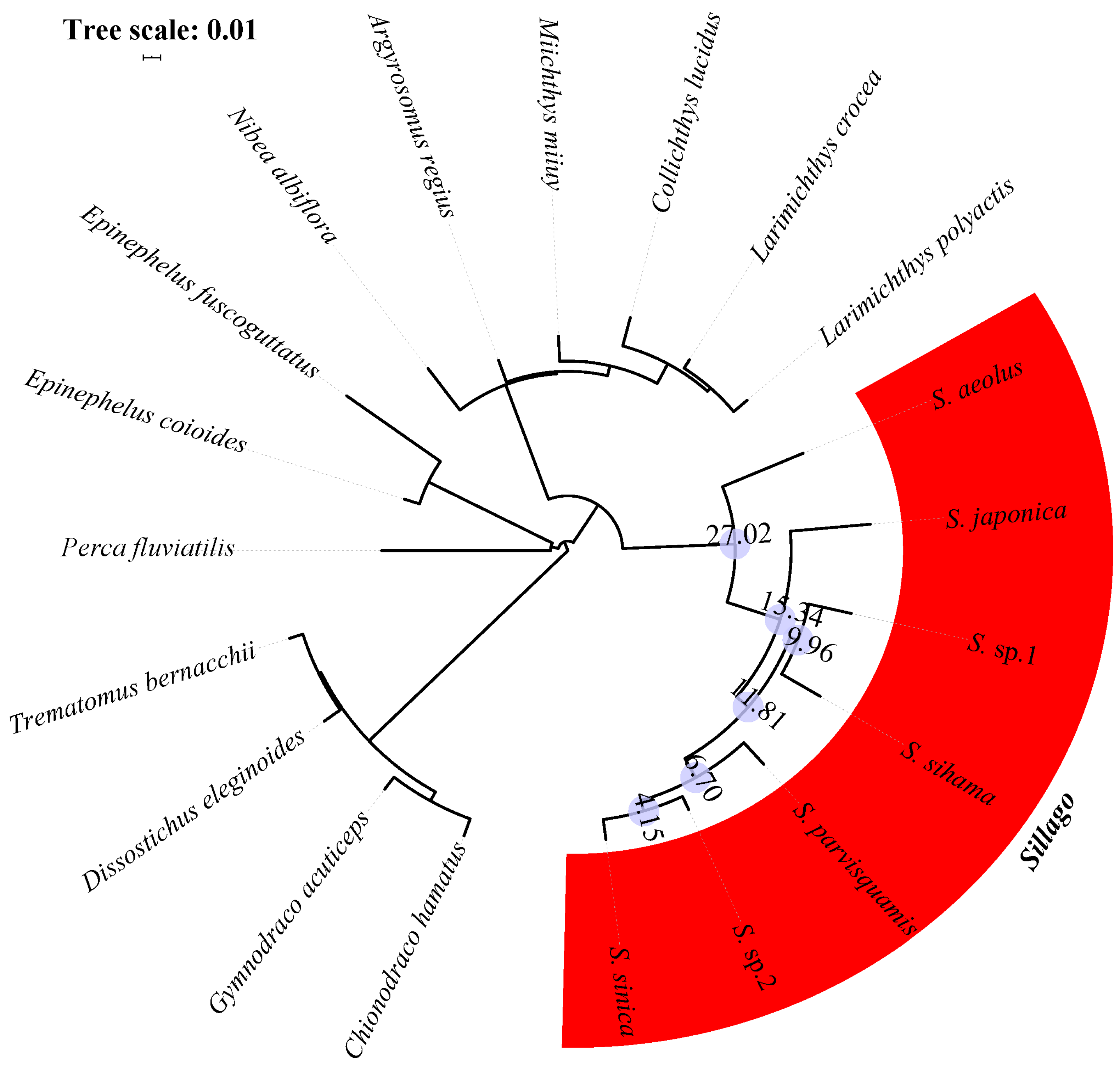
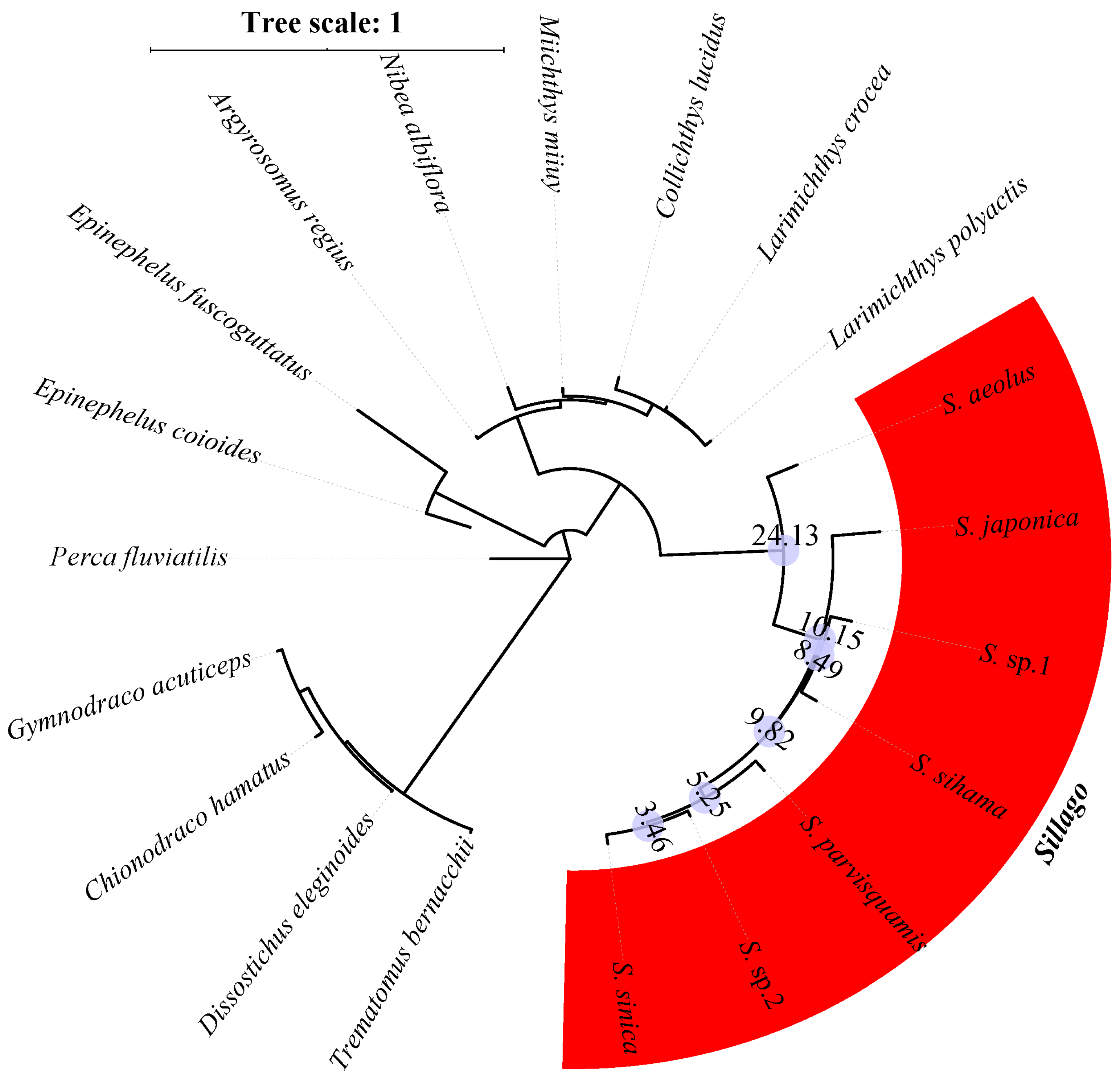
| Species | Classification | Milieu | Climate Zone | Depth Range (M) | Maturity Length (cm) | Feeding Habits | Type of Fish Eggs |
|---|---|---|---|---|---|---|---|
| E. fuscoguttatus | Serranidae | Marine; brackish; reef-associated | Tropical | 1–60 | 50 | Carnivorous | Pelagic |
| E. coioides | Serranidae | Marine; brackish; reef-associated | Subtropical | 1–100 | 25–30 | Carnivorous | Pelagic |
| P. fluviatilis | Percidae | Freshwater; brackish; demersal | Temperate | 1–30 | 11–23.4 | Carnivorous | Adhesive |
| C. hamatus | Channichthyidae | Marine; demersal | Polar | 4–600 | 33–37 | Carnivorous | Pelagic |
| G. acuticeps | Bathydraconidae | Marine; demersal | Polar | 0–550 | - | Carnivorous | Pelagic |
| D. eleginoides | Nototheniidae | Marine; demersal | Temperate | 50–3850 | 38–60 | Carnivorous | Pelagic |
| T. bernacchii | Nototheniidae | Marine; demersal; | Polar | 0–700 | 18 | Carnivorous | Pelagic |
| A. regius | Sciaenidae | Marine; brackish; demersal | Subtropical | 15–300 | 80 | Carnivorous | Pelagic |
| N. albiflora | Sciaenidae | Marine; demersal; coastal waters with mudddy to sanddy-muddy bottoms | Temperate | 25–80 | - | Carnivorous | Pelagic |
| M. miiuy | Sciaenidae | Marine; brackish; demersal; coastal waters with mudddy to sanddy-muddy bottoms | Temperate | 15–100 | - | Carnivorous | Pelagic |
| C. lucidus | Sciaenidae | Marine; demersal; coastal waters with mudddy to sanddy-muddy bottoms | Subtropical | 0–90 | 13 | Carnivorous | Pelagic |
| L. polyactis | Sciaenidae | Marine; demersal; sublittoral zone above 120 m with muddy to sanddy-muddy bottoms | Subtropical | 0–120 | 18.1 | Carnivorous | Pelagic |
| L. crocea | Sciaenidae | Marine; brackish; demersal; coastal waters and estuaries with muddy to muddy-sandy bottoms shallower than 120 m depth | Temperate | 0–120 | 17 | Carnivorous | Pelagic |
| S. aeolus | Sillaginidae | Marine; demersal; nearshore shallow and estuarine waters; burrowing life-style | Tropical | 0–60 | 12 | Carnivorous | Pelagic |
| S. japonica | Sillaginidae | Marine; demersal; nearshore shallow and estuarine waters; burrowing life-style | Subtropical | 0–30 | - | Carnivorous | Pelagic |
| S. parvisquamis | Sillaginidae | Marine; brackish; demersal; nearshore shallow and estuarine waters; burrowing life-style | Subtropical | 0–30 | - | Carnivorous | Pelagic |
| S. sihama | Sillaginidae | Marine; brackish; reef-associated; nearshore shallow and estuarine waters; burrowing life-style | Tropical | 0–60 | 13–19.1 | Carnivorous | Pelagic |
| S. sinica | Sillaginidae | Marine; brackish; demersal; nearshore shallow and estuarine waters; burrowing life-style | Tropical | - | - | Carnivorous | Pelagic |
| S. sp.1 | Sillaginidae | - | - | - | - | - | - |
| S. sp.2 | Sillaginidae | - | - | - | - | - | - |
| Sillago Species | Read Number | GC% | %≥Q30 |
|---|---|---|---|
| S. aeolus | 78,709,246 | 51.14 | 92.37 |
| S. japonica | 50,013,641 | 53.02 | 92.96 |
| S. parvisquamis | 113,351,008 | 52.88 | 93.75 |
| S. sihama | 87,050,702 | 51.34 | 92.63 |
| S. sinica | 97,977,199 | 52.19 | 94.51 |
| S. sp.1 | 51,710,081 | 53.86 | 93.74 |
| S. sp.2 | 70,996,526 | 53.57 | 92.95 |
| Sillago Species | Unigene | |||
|---|---|---|---|---|
| Number | Total Length (bp) | Mean Length (bp) | N50 Length (bp) | |
| S. aeolus | 82,024 | 51,896,226 | 787.32 | 1,403 |
| S. japonica | 58,102 | 23,966,004 | 428.99 | 461 |
| S. parvisquamis | 102,185 | 79,280,211 | 1,019.38 | 1,986 |
| S. sihama | 69,748 | 48,391,713 | 815.81 | 1,369 |
| S. sinica | 102,903 | 78,264,349 | 992.70 | 1,848 |
| S. sp.1 | 63,807 | 34,524,368 | 588.49 | 738 |
| S. sp.2 | 85,990 | 49,751,159 | 652.68 | 902 |
| Gene Name | Description | ×10-Value | FDR-Adjusted p-Value | |
|---|---|---|---|---|
| Stress response | MED27 | mediator of RNA polymerase II transcription subunit 27 | 3.02 × 10−39 | 0.00 |
| MED28 | mediator of RNA polymerase II transcription subunit 28 | 2.33 × 10−29 | 0.00 | |
| LTV1 | protein LTV1 homolog | 1.08 × 10−37 | 7.89 × 10−03 | |
| SMO | Spermine oxidase | 2.21 × 10−22 | 1.27 × 10−14 | |
| PSA | puromycin-sensitive aminopeptidase | 3.70 × 10−41 | 0.00 | |
| ABCB7 | ATP-binding cassette sub-family B member 7, mitochondrial | 5.54 × 10−31 | 0.00 | |
| COPA | coatomer subunit alpha | 4.99 × 10−45 | 0.00 | |
| SF3A1 | splicing factor 3A subunit 1 | 2.31 × 10−44 | 0.00 | |
| SF3B5 | splicing factor 3B subunit 5 | 8.30 × 10−60 | 0.00 | |
| DEPDC5 | GATOR complex protein DEPDC5 isoform X3 | 1.72 × 10−46 | 0.00 | |
| POLλ | DNA polymerase lambda | 1.48 × 10−80 | 0.00 | |
| TFIP11 | tuftelin-interacting protein 11 | 2.10 × 10−72 | 0.00 | |
| NUDT6 | Nucleoside diphosphate-linked moiety X motif 6 | 1.32 × 10−83 | 0.00 | |
| UBIAD1 | UbiA prenyltransferase domain-containing protein 1 | 7.59 × 10−96 | 0.00 | |
| Energy metabolism | APF | bis(5′-nucleosyl)-tetraphosphatase [asymmetrical] | 5.30 × 10−64 | 0.00 |
| IMMT | MICOS complex subunit MIC60 isoform X2 | 1.66 × 10−126 | 1.38 × 10−04 | |
| Carbohydrate metabolism | SUCLG1 | succinate-CoA ligase [ADP/GDP−forming] subunit alpha, mitochondrial | 2.37 × 10−23 | 0.00 |
| Amino acid metabolism | GCN1 | eIF-2-alpha kinase activator GCN1 | 1.82 × 10−41 | 0.00 |
| CPD | Carboxypeptidase D | 6.74 × 10−46 | 0.00 | |
| Lipid metabolism | HUWE1 | E3 ubiquitin-protein ligase HUWE1 isoform X1 | 3.95 × 10−35 | 1.15 × 10−03 |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 isoform X1 | 4.55 × 10−24 | 0.00 | |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 isoform X1 | 8.49 × 10−54 | 7.85 × 10−03 | |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 isoform X1 | 1.31 × 10−38 | 0.00 | |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 isoform X1 | 1.48 × 10−40 | 0.00 | |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 isoform X1 | 1.40 × 10−41 | 0.00 | |
| FABZ | hydroxyacyl-thioester dehydratase type 2, mitochondrial | 7.36 × 10−80 | 0.00 | |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 isoform X1 | 2.11 × 10−117 | 5.07 × 10−03 | |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 isoform X1 | 1.57 × 10−117 | 0.00 | |
| Visual sense | AP4B1 | AP-4 complex subunit beta-1 | 5.00 × 10−45 | 0.00 |
| PRF8 | Pre-mRNA-processing-splicing factor 8 | 2.12 × 10−52 | 0.00 | |
| Growth and differentiation | ABTB1 | ankyrin repeat and BTB/POZ domain-containing protein 1 | 1.96 × 10−36 | 0.00 |
| UBE4A | ubiquitin conjugation factor E4 B isoform X2 | 1.31 × 10−29 | 1.11 × 10−12 | |
| GAB1 | GRB2-associated-binding protein 1 isoform X1 | 3.65 × 10−61 | 0.00 | |
| DOHH | deoxyhypusine hydroxylase | 2.66 × 10−64 | 0.00 | |
| Embryogenesis | SBDS | ribosome maturation protein SBDS | 2.55 × 10−39 | 0.00 |
| SEC8 | exocyst complex component 8 | 1.38 × 10−90 | 0.00 | |
| TAF5L | TAF5-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 5L | 0.00 | 0.00 | |
| Others | CCDC25 | Coiled-coil domain-containing protein 25 | 9.63 × 10−17 | 0.00 |
| PIGY | phosphatidylinositol N-acetylglucosaminyltransferase subunit Y | 1.12 × 10−40 | 0.00 | |
| - | fumarylacetoacetate hydrolase domain-containing protein 2-like isoform X2 | 6.37 × 10−43 | 0.00 | |
| USP24 | ubiquitin carboxyl-terminal hydrolase 24 isoform X2 | 5.15 × 10−31 | 0.00 | |
| RPN2 | 26S proteasome non-ATPase regulatory subunit 1 | 2.38 × 10−44 | 0.00 | |
| CXORF56 | UPF0428 protein CXorf56 homolog | 1.53 × 10−136 | 0.00 | |
| TALIN | talin rod domain-containing protein 1 | 0.00 | 3.81 × 10−06 |
| Pathway | Pathway_ID | Key Enzyme | Gene Name |
|---|---|---|---|
| Nicotinate and nicotinamide metabolism | map00760 | diphosphatase | APF |
| Carbon fixation pathways in prokaryotes | map00720 | ligase (ADP-forming) | SUCLG1 |
| Purine metabolism | map00230 | adenylpyrophosphatase; diphosphatase; phosphatase | ABCB7, APF, ABCB7 |
| C5-Branched dibasic acid metabolism | map00660 | ligase (ADP-forming) | SUCLG1 |
| Starch and sucrose metabolism | map00500 | diphosphatase | APF |
| Arginine biosynthesis | map00220 | synthase (NADPH) | UBIAD1 |
| Riboflavin metabolism | map00740 | diphosphatase | APF |
| Pantothenate and CoA biosynthesis | map00770 | diphosphatase | APF |
| Pyrimidine metabolism | map00240 | diphosphatase | APF |
| Biosynthesis of antibiotics | map01130 | synthase (NADPH); ligase (ADP-forming); ligase (GDP-forming) | UBIAD1, SUCLG1, SUCLG1 |
| Citrate cycle (TCA cycle) | map00020 | ligase (ADP-forming); ligase (GDP-forming) | SUCLG1, SUCLG1 |
| Propanoate metabolism | map00640 | ligase (ADP-forming); ligase (GDP-forming) | SUCLG1, SUCLG1 |
| Thiamine metabolism | map00730 | Phosphatase | ABCB7 |
| Arginine and proline metabolism | map00330 | synthase (NADPH) | UBIAD1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, F.; Zhang, Y.; Song, N.; Ji, D.; Gao, T. Comprehensive Transcriptome Analysis Reveals Insights into Phylogeny and Positively Selected Genes of Sillago Species. Animals 2020, 10, 633. https://doi.org/10.3390/ani10040633
Lou F, Zhang Y, Song N, Ji D, Gao T. Comprehensive Transcriptome Analysis Reveals Insights into Phylogeny and Positively Selected Genes of Sillago Species. Animals. 2020; 10(4):633. https://doi.org/10.3390/ani10040633
Chicago/Turabian StyleLou, Fangrui, Yuan Zhang, Na Song, Dongping Ji, and Tianxiang Gao. 2020. "Comprehensive Transcriptome Analysis Reveals Insights into Phylogeny and Positively Selected Genes of Sillago Species" Animals 10, no. 4: 633. https://doi.org/10.3390/ani10040633
APA StyleLou, F., Zhang, Y., Song, N., Ji, D., & Gao, T. (2020). Comprehensive Transcriptome Analysis Reveals Insights into Phylogeny and Positively Selected Genes of Sillago Species. Animals, 10(4), 633. https://doi.org/10.3390/ani10040633





