Ashwin Gene Expression Profiles in Oocytes, Preimplantation Embryos, and Fetal and Adult Bovine Tissues
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Organ Samples
2.2. Total RNA Isolation and Polymerase Chain Reactions
2.3. Oocyte Maturation, In Vitro Fertilization, and Embryo Culture
2.4. Immunohistochemical Analysis
2.5. Statistical Analysis
3. Results
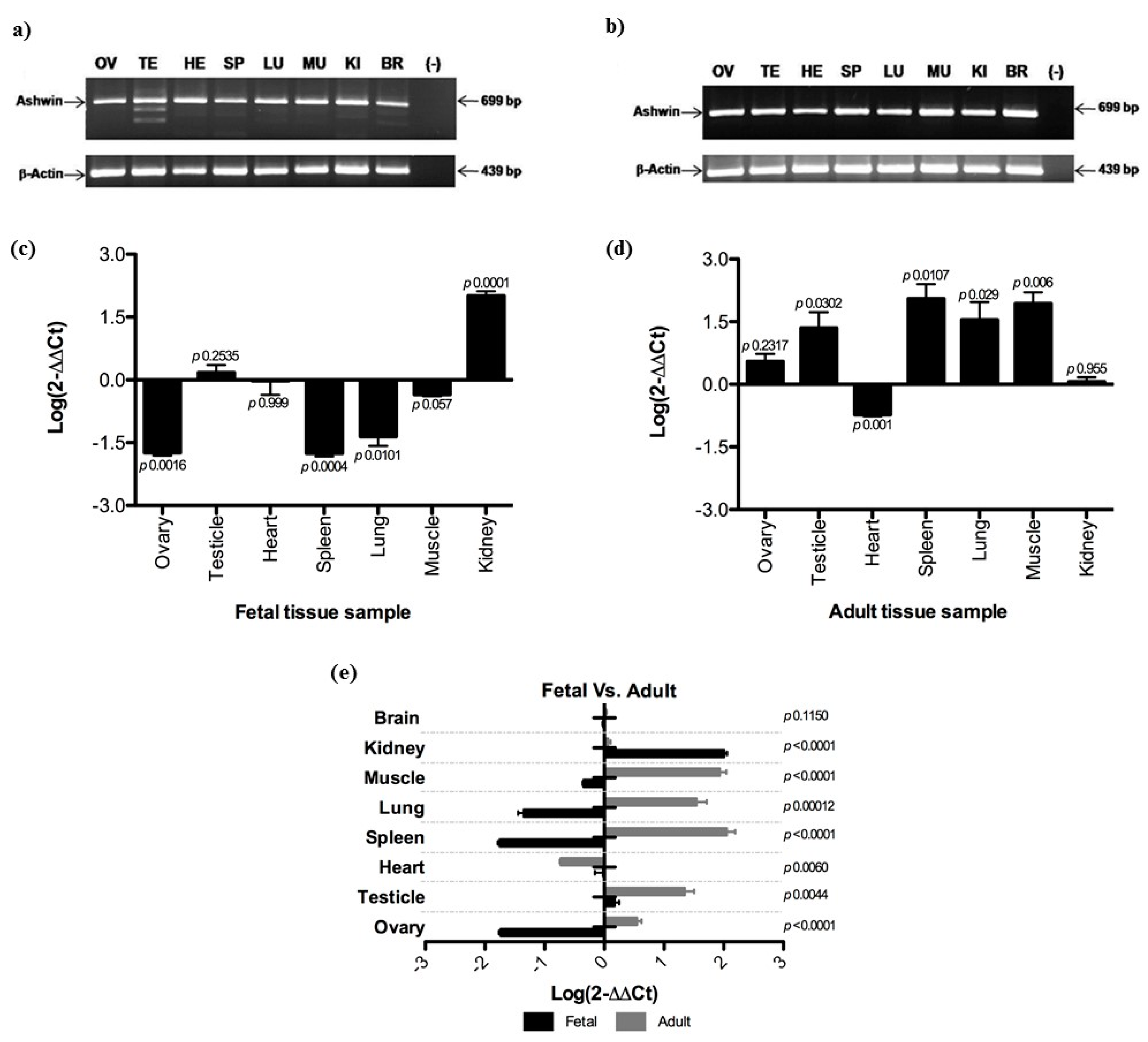
3.1. Ashwin mRNA Expression in Fetal and Adult Tissues
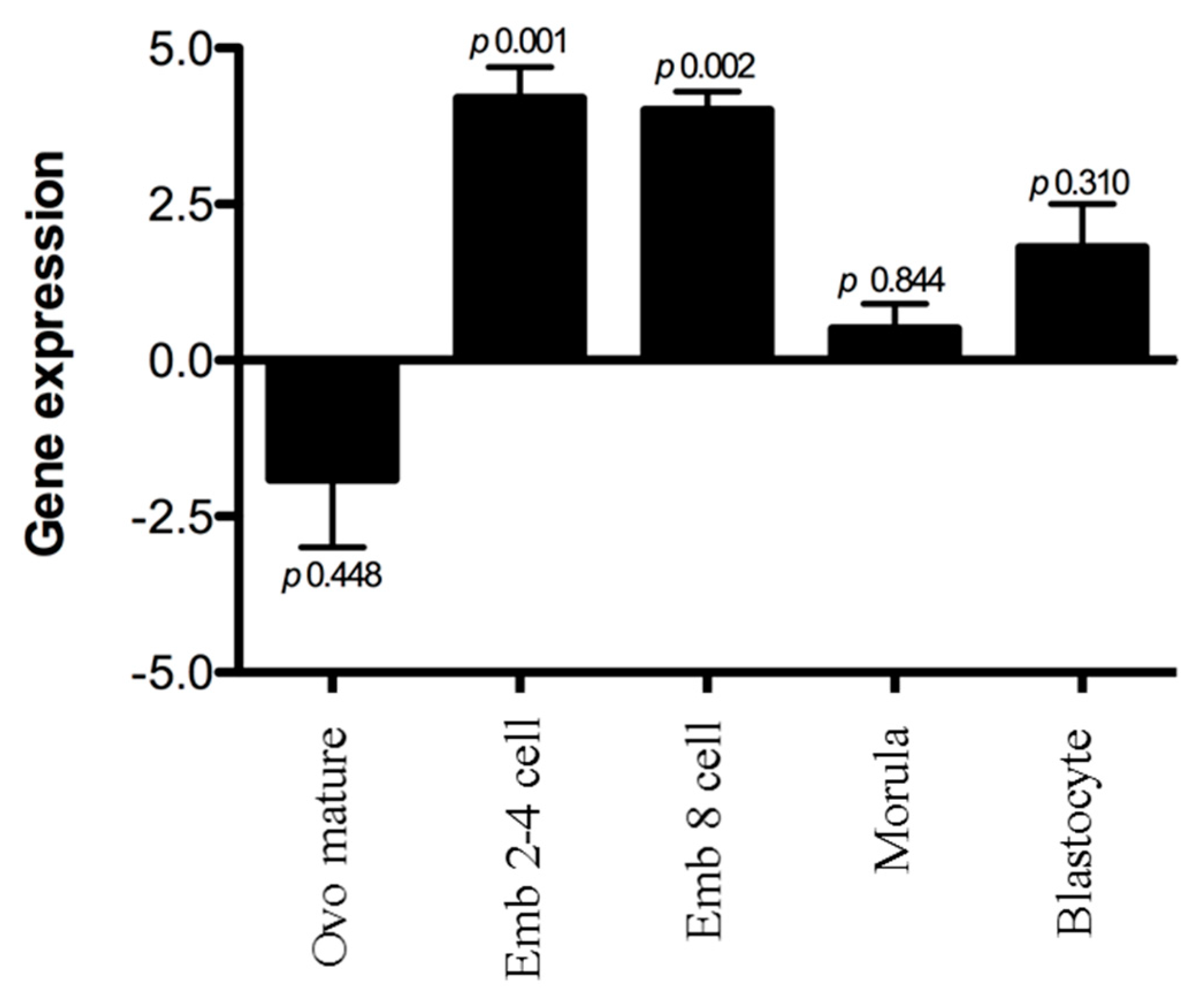
3.2. Expression of Ashwin mRNA in Early Embryonic Development
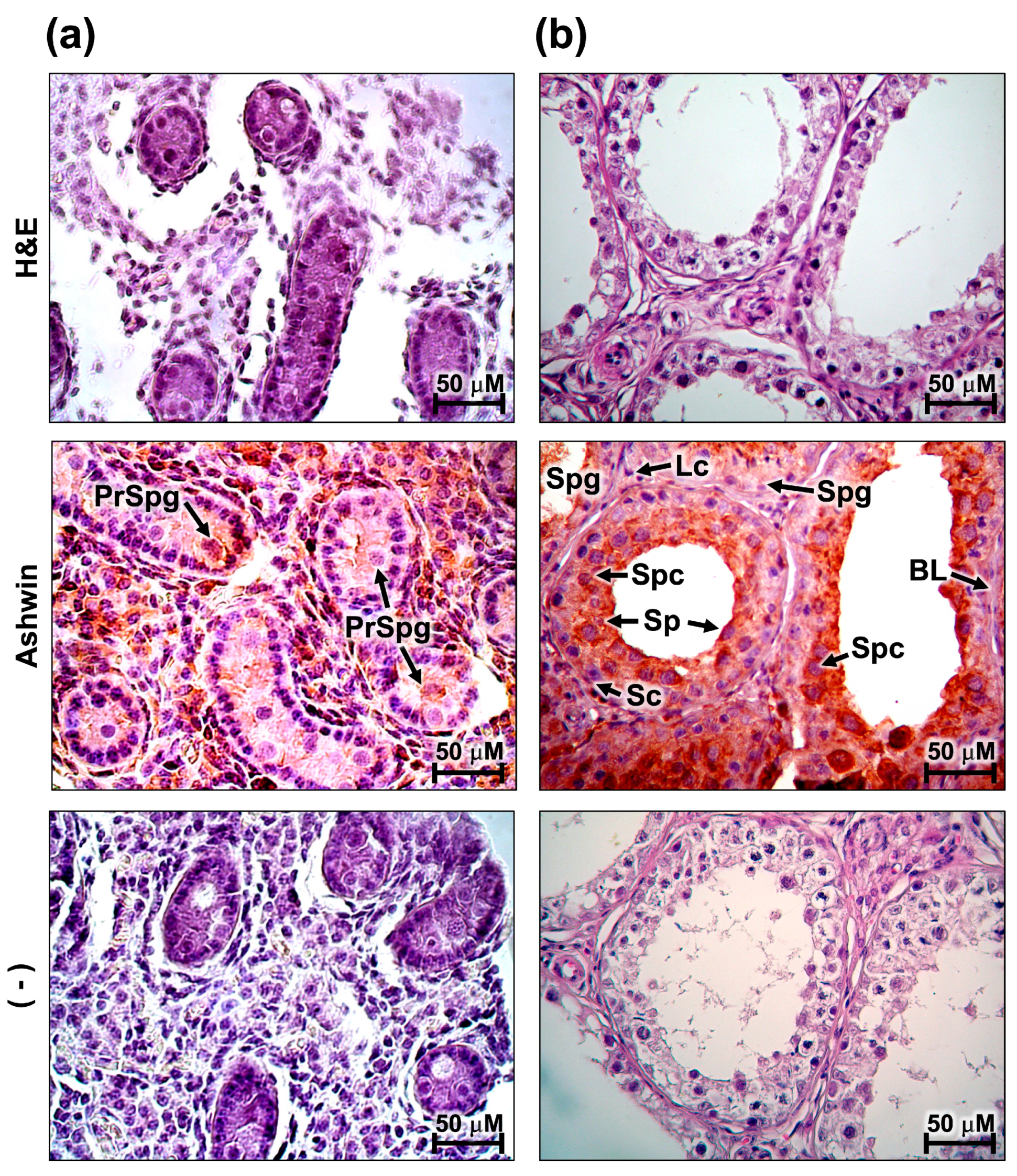
3.3. Localization of the Ashwin Protein in the Ovary and Testicle
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Patil, S.S.; Alexander, T.B.; Uzman, J.A.; Lou, C.H.; Gohil, H.; Sater, A.K. Novel gene ashwin functions in Xenopus cell survival and anteroposterior patterning. Dev. Dyn. 2006, 235, 1895–1907. [Google Scholar] [CrossRef] [PubMed]
- Hurst, H.C. Transcription factors 1: bZIP proteins. Protein Profile 1995, 2, 101–168. [Google Scholar] [PubMed]
- Krieg, P.A.; Sakaguchi, D.S.; Kintner, C.R. Primary structure and developmental expression of a large cytoptasmic domain form of Xenopus laevis neural cell adhesion molecule (NCAM). Nucleic Acids Res. 1989, 17, 10321–10335. [Google Scholar] [CrossRef] [PubMed]
- Houtmeyers, R.; Souopgui, J.; Tejpar, S.; Arkell, R. The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell. Mol. Life Sci. 2013, 70, 3791–3811. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.C.; Knecht, A.K.; Wu, M.; Harland, R.M. Secreted noggin protein mimics the Spemann organizer in dorsalizing Xenopus mesoderm. Nature 1993, 361, 547–549. [Google Scholar] [CrossRef] [PubMed]
- Krause, C.; Guzman, A.; Knaus, P. Noggin. Int. J. Biochem. Cell Biol. 2011, 43, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Shimasaki, S.; Moore, R.K.; Otsuka, F.; Erickson, G.F. The Bone Morphogenetic Protein System in Mammalian Reproduction. Endocr Rev. 2004, 25, 72–101. [Google Scholar] [CrossRef]
- Pereda-Espinoza, B.; Burrola-Barraza, M.E.; Rodríguez-Almeida, F.; Antillon-Ruiz, J.; Anchondo-Garay, A. Reduced mineral oil ratio improves blastocyst yield in well-of-the-well (WOW) and polyester mesh (PM) single-embryo cultures. Veterinarski Arhiv. 2016, 83, 467–474. [Google Scholar]
- Burrola-Barraza, M.E.; Hernández-Seáñez, R.; Barceló-Fimbres, M.; Rodríguez-Almeida, F.A.; González-Rodríguez, E.; García-Quiñónez, S.; Grado-Ahuir, J.A.; Moreno-Brito, V. Dicer gene expression during early bovine embryo development. Mol. Reprod. Dev. 2011, 78, 622. [Google Scholar] [CrossRef] [PubMed]
- Braw-Tal, R.; Yossefi, S. Studies in vivo and in vitro on the initiation of follicle growth in the bovine ovary. J. Reprod. Fertil. 1997, 109, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Hamatani, T.; Carter, M.G.; Sharov, A.A.; Ko, M.S.H. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004, 6, 117–131. [Google Scholar] [CrossRef]
- Hamatani, T.; Ko, M.S.; Yamada, M.; Kuji, N.; Mizusawa, Y.; Shoji, M.; Hada, T.; Asada, H.; Maruyama, T.; Yoshimura, Y. Global gene expression profiling of preimplantation embryos. Hum. Cell. 2006, 19, 98–117. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, P.; Dean, J. Maternal control of early mouse development. Development 2010, 137, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lu, X.; Dean, J. The maternal to zygotic transition in mammals. Mol. Aspects. Med. 2013, 34, 919–938. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, I.; Camargo, L.S.A.; Pereira, M.M.; Fernandez-Martin, R.; Paz, D.A.; Salamone, D.F. Effects of bone morphogenic protein 4 (BMP4) and its inhibitor, Noggin, on in vitro maturation and culture of bovine preimplantation embryos. Reprod. Biol. Endocrinol. 2011, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bebbere, D.; Masala, L.; Albertini, D.F.; Ledda, S. The subcortical maternal complex: multiple functions for one biological structure? J. Assist. Reprod. Genet. 2016, 33, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Gao, Z.; Qin, D.; Li, L. A Maternal Functional Module in the Mammalian Oocyte-To-Embryo Transition. Trends Mol. Med. 2017, 23, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.L.; Ealy, A.D.; Monterroso, V.H.; Hansen, P.J. Ontogeny of temperature-regulated heat shock protein 70 synthesis in preimplantation bovine embryos. Mol. Reprod. Dev. 1997, 48, 23–33. [Google Scholar] [CrossRef]
- Wang, G.; Li, Y.; Yang, Q.; Xu, S.; Ma, S.; Yan, R.; Zhang, R.; Jia, G.; Ai, D.; Yang, Q. Gene expression dynamics during the gonocyte to spermatogonia transition and spermatogenesis in the domestic yak. J. Anim. Sci. Biotechnol. 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.P.; Tian, L.; O’ Neill, C.; King, N.J.C. Regulation of cellular adhesion molecule expression in murine oocytes, peri-implantation and post-implantation embryos. Cell Res. 2002, 12, 373–383. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Brito, V.; Morales-Adame, D.; Soto-Orduño, E.; González-Chávez, S.A.; Pacheco-Tena, C.; Espino-Solis, G.P.; Leal-Berumen, I.; González-Rodríguez, E. Ashwin Gene Expression Profiles in Oocytes, Preimplantation Embryos, and Fetal and Adult Bovine Tissues. Animals 2020, 10, 276. https://doi.org/10.3390/ani10020276
Moreno-Brito V, Morales-Adame D, Soto-Orduño E, González-Chávez SA, Pacheco-Tena C, Espino-Solis GP, Leal-Berumen I, González-Rodríguez E. Ashwin Gene Expression Profiles in Oocytes, Preimplantation Embryos, and Fetal and Adult Bovine Tissues. Animals. 2020; 10(2):276. https://doi.org/10.3390/ani10020276
Chicago/Turabian StyleMoreno-Brito, Verónica, Daniel Morales-Adame, Elier Soto-Orduño, Susana Aideé González-Chávez, César Pacheco-Tena, Gerardo Pavel Espino-Solis, Irene Leal-Berumen, and Everardo González-Rodríguez. 2020. "Ashwin Gene Expression Profiles in Oocytes, Preimplantation Embryos, and Fetal and Adult Bovine Tissues" Animals 10, no. 2: 276. https://doi.org/10.3390/ani10020276
APA StyleMoreno-Brito, V., Morales-Adame, D., Soto-Orduño, E., González-Chávez, S. A., Pacheco-Tena, C., Espino-Solis, G. P., Leal-Berumen, I., & González-Rodríguez, E. (2020). Ashwin Gene Expression Profiles in Oocytes, Preimplantation Embryos, and Fetal and Adult Bovine Tissues. Animals, 10(2), 276. https://doi.org/10.3390/ani10020276





