1. Introduction
On a global scale, cattle meat and milk production has more than doubled between 1961 and 2014, increasing from 28 million to 68 million tonnes per year for meat products, and 344 million to 792 million tonnes for milk products [
1]. To support this increasing demand, the use of artificial insemination (AI) has become widespread in the cattle breeding industry. In many breeding programmes, the emphasis on genomic selection is on genotype and pedigree analysis, with relatively little attention being paid to the underlying fertility of the animal. However, the extensive use of artificial insemination using a small pool of high genetic-merit bulls, and the rising use of in vitro-produced embryos, means that the importance of selecting parents that also have optimal fertility is vital. Impaired fertility reduces genetic gain, increases veterinary costs, and reduces milk and meat production, all of which result in heavy financial and environmental losses for the breeding company; costs that is ultimately passed on to the end consumer.
Semen analysis is commonly used as a fertility indicator in livestock breeding programmes where volume, morphology, motility and concentration are routinely measured [
2]. However, this type of analysis is thought to be an unreliable indicator of fertility, and does not allow for the detection of underlying subfertility on a chromosomal level [
3]. Instead, the most widely used parameter for the detection of subfertility in cattle is the ‘nonreturn rate’ (the number of females returning to the oestrus cycle, being therefore indicative of a failure to conceive) [
4].
Chromosomal rearrangements, including Robertsonian (centromeric end fusion) and reciprocal (nonhomologous exchange) translocations, can have a significant detrimental effect on the fertility of cattle. Where these rearrangements occur, the process of meiotic pairing and chromosome segregation during gametogenesis is disturbed, leading to gametes that can be genetically unbalanced [
5]. These unbalanced gametes inevitably result in early embryonic loss due to reduced viability. In recent decades, efforts have been made to diagnose fertility issues in domestic breeding animals using chromosome analysis. In 1964, Ingemar Gustavsson first reported the presence of the 1;29 centromeric fusion (Robertsonian translocation) in a population of Swedish Red and White cattle [
6]. Since then, the 1;29 translocation has been the most commonly seen rearrangement of the 44 that have been identified in cattle so far [
7], with cases found in all breeds, except Holstein-Fresian [
8]. In one 15-year study of the Italian breeding population, 7.1% animals were identified as carrying a Roberstonian translocation [
9]. These heterozygous 1;29 carriers are phenotypically normal, but suffer a reduction in fertility of 3–5% [
10]. Homozygous carriers are rare, but have been reported by several groups, although incidence varies between breeds. Reported examples of the homozygous 1;29 state include the presence in 8.5% of Blonde d’Aquitaine bulls [
10], along with cases found in five of the eight Portuguese cattle breeds [
11]. All cattle (
Bos taurus and
Bos indicus) have the same chromosome complement with 2n = 60, and thus, any novel approach must be applicable to all commercial breeds.
Reciprocal translocations have been identified in cattle, albeit much less frequently, with the aforementioned Italian study reporting a rate of 0.03% [
9]. To date, only 19 reciprocal translocations involving different chromosomes in cattle have been reported [
12]. In fact, De Lorenzi and colleagues suggested that the frequency of reciprocal translocations is grossly underreported, largely due to the inherent difficulties in detecting these rearrangements using routine cytogenetics [
12]. The cattle karyotype is notoriously difficult to analyse reliably because of a diploid number of 60, largely made up of similar-sized acrocentric chromosomes, and is therefore problematic for the detection of anything other than Robertsonian translocations. A molecular approach that will detect reciprocal and Robertsonian translocations is, therefore, essential.
Recently, we developed an approach for the detection of cryptic and overt translocations in boars [
13]. This method uses a panel of subtelomeric fluorescence
in-situ hybridisation (FISH) probes on a multihybridisation device as a means of highlighting the ends of each chromosome, thereby facilitating the identification of rearrangements between chromosomes. The purpose of this study was to use similar technology to isolate and visualise each end of every chromosome in cattle. Here, as proof of principle, we use a small sample size; however, all cattle have the same chromosomes, thereby allowing for the detection of translocations, particularly the challenging reciprocal ones.
2. Materials and Methods
Heparinised whole blood samples from 39 Holstein bulls were obtained from local suppliers. Samples were collected as part of standard procedures used for commercial evaluation by in house trained veterinarians via standard phlebotomy in heparin tubes. Whole blood samples were cultured for 72 h in PB MAX Karyotyping medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C, 5% CO2. Cell division was arrested by the addition of colcemid at a concentration of 10.0 μg/mL (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) for 35 min prior to hypotonic treatment with 75 M KCl and fixation to glass slides using 3:1 methanol:acetic acid.
2.1. Selection and Preparation of Fluorescence In-Situ Hybridisation (FISH) Probes
Bacterial artificial chromosome (BAC) clones of approximately 150 kb in size were selected from the Btau 4.6.1 NCBI database (
www.ncbi.nlm.nih.gov) and ordered from the CHORI-240 Bovine BAC library for each autosome and the X chromosome (see
Table 1). A lack of available BACs for the Y chromosome meant that this chromosome was excluded from the study. BAC DNA was isolated using the Qiagen Miniprep Kit (Qiagen, Hilden, Germany), the products of which were then amplified and directly labelled by nick translation with FITC-Fluroescein-12-UTP (Roche, Basel, Switzerland) for subcentromeric probes and Texas Red-12-dUTP (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) for distal q-arm probes prior to purification. A list of BACs is given in
Table 1.
2.2. Development of Multiprobe Device
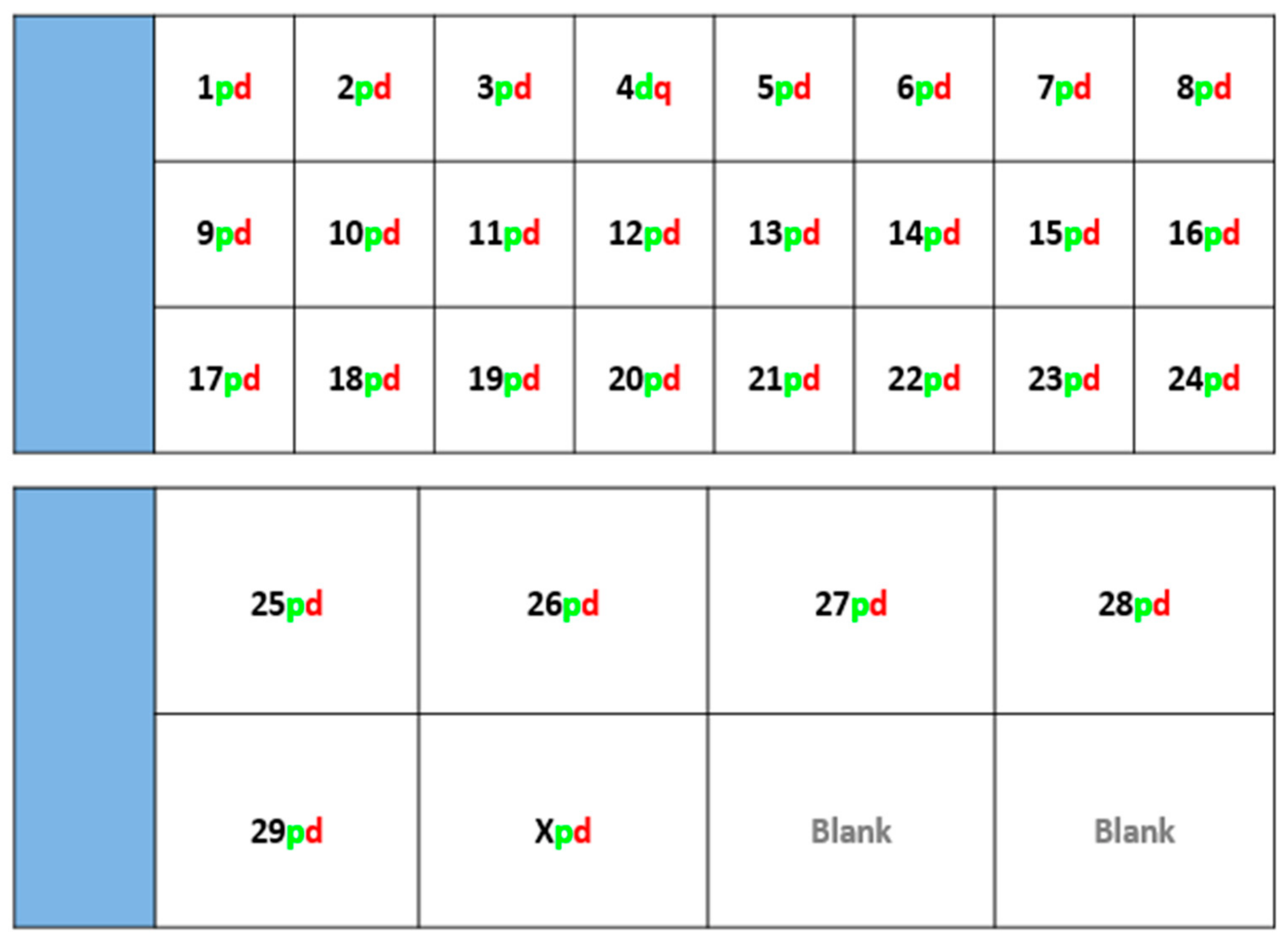
Fluorescently-labelled probes were diluted to a concentration of 10 ng/μL in sterile distilled water along with competitor DNA (Bovine Hybloc, Applied Genetics Laboratories, Melbourne, FL, USA). Each probe combination contained a probe isolated from each end of the chromosome, and was individually assigned with the appropriate chromosome number followed by the letter p (proximal) or d (distal). Using a proprietary Chromoprobe Multiprobe System device manufactured by Cytocell Ltd., Cambridge, UK, each probe combination (e.g., 1pd) for chromosomes 1 to 24 was air dried onto a square of the device. The corresponding glass slide was subdivided into 24 squares, designed to align to the 24 squares on the device upon which chromosome suspensions were fixed. A second 8-square device was used to facilitate the larger number of chromosomes in the cattle karyotype. The precise orientation of the clones and development of the bespoke device is given in the results section (
Figure 1).
2.3. Fluorescence In-Situ Hybridisation (FISH)
Slides were dropped with fixed metaphase preparations and dehydrated through an ethanol series (2 min each in 2× sodium saline citrate (SSC), 70%, 85% and 100% ethanol at room temperature). Formamide-based hybridisation buffer (Cytocell Hyb I, Cambridge, UK) was pipetted onto each square of the device in order to resuspend the probes. The glass slide and the device were sandwiched together and warmed on a 37 °C hotplate for 10 min. The probe and target DNA were subsequently denatured on a 75 °C hotplate for 5 min prior to overnight hybridisation in a dry hybridisation chamber in a 37 °C water bath. Slides were washed post hybridisation for 2 min in 0.4× SSC at 72 °C and 30 s in 2× SSC/0.05% Tween 20 at room temperature, then counterstained using DAPI in VECTASHIELD. Metaphases for karyotyping were stained with DAPI in VECTASHIELD antifade medium (Vector Laboratories, Peterborough, UK). Image capturing was performed using an Olympus BX61 epifluorescence microscope (Olympus, Tokyo, Japan) with a cooled CCD camera and SmartCapture (Digital Scientific UK, Cambridge, UK) system. The SmartType software (Digital Scientific UK, London, UK) was used for karyotyping purposes.
4. Discussion
The consequences of using a subfertile bull in an AI breeding programme are many. First of all, while using a subfertile bull may result in a small number of pregnancies, the pregnancy rates will inevitably be significantly lower than expected. This then leads to extended calving intervals and an increased likelihood that a higher proportion of cows will be culled for presumed sterility. Both of these factors result not just in reduced financial returns, but also in a large degree of wastage, raising ethical and environmental concerns. This is particularly important when the very large bull to cow ratios employed in AI programmes are taken into account. With average calving rates of only 50–60% in UK domestic dairy cattle, it is vital to prevent any further potential loss of fertility in order to maximise the opportunities for each cow to conceive and to improve productivity [
14].
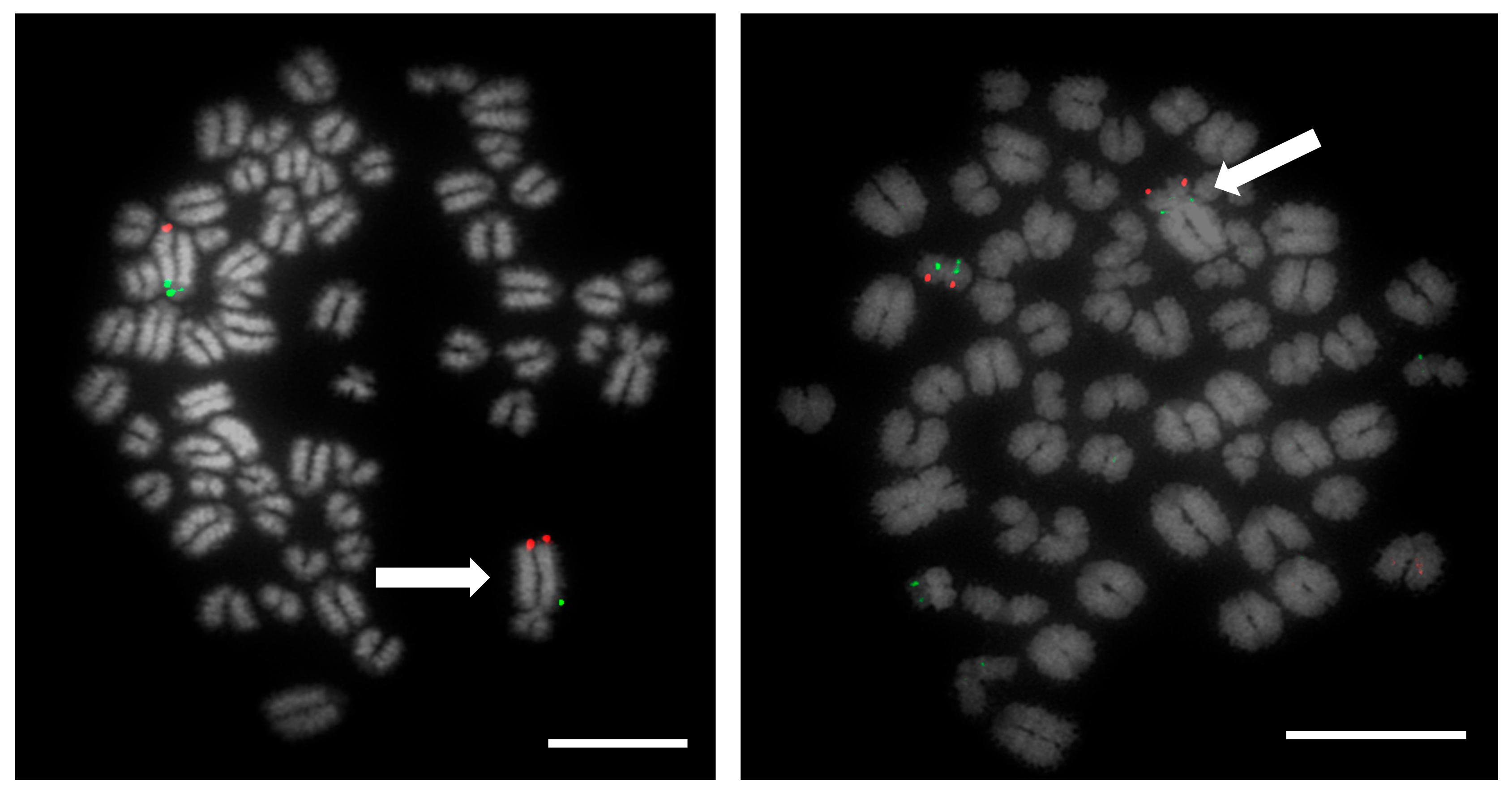
Prior to the results reported here, standard karyotype analysis, a time-consuming and error prone method, was the only means of diagnosing chromosomal rearrangements in cattle. De Lorenzi and colleagues calculated that for a translocation to be observable through karyotyping alone, an abnormal chromosome derivative must be either at least 15% (185 Mb) longer than chromosome 1, or 40% (26.4 Mb) shorter than chromosome 25 [
12]. Even with optimum G-banding preparations, it is likely that most reciprocal translocations involving chromosomes 2–24 would be indistinguishable from other autosomes. It is vital, therefore, that efficient and accurate methods are implemented for the detection of chromosome translocations as part of a routine screening programme for cattle destined for AI. The results generated in this study demonstrate the validity of a FISH-based screening device for the detection of reciprocal translocations, two of which would remain undiagnosed if standard karyotyping alone had been used. Efforts to eradicate chromosomal translocations from the cattle breeding herd are ongoing; however, the results presented here demonstrate that both Robertsonian and reciprocal translocations are present in the breeding population. Many of these may be de novo rearrangements, but it is highly likely that the reciprocal translocation identified here is one that has been carried through multiple generations but which had not been identified due to the inherent difficulties in screening using traditional methods. Screening for chromosomal translocations that result in economic loss is, therefore, more important than ever, and this study demonstrates, in only a small group of animals, a means by which it could be achieved in the future.
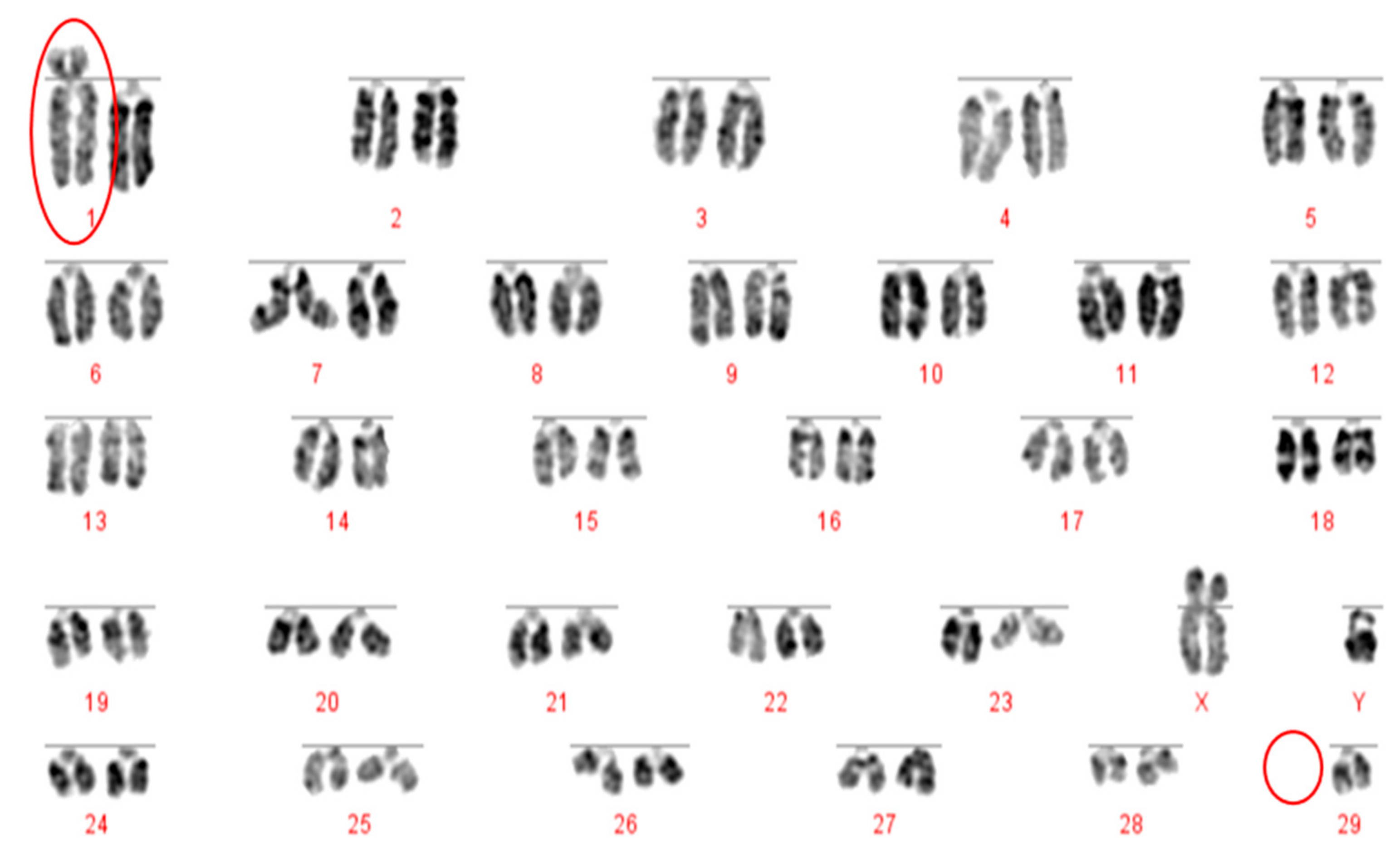
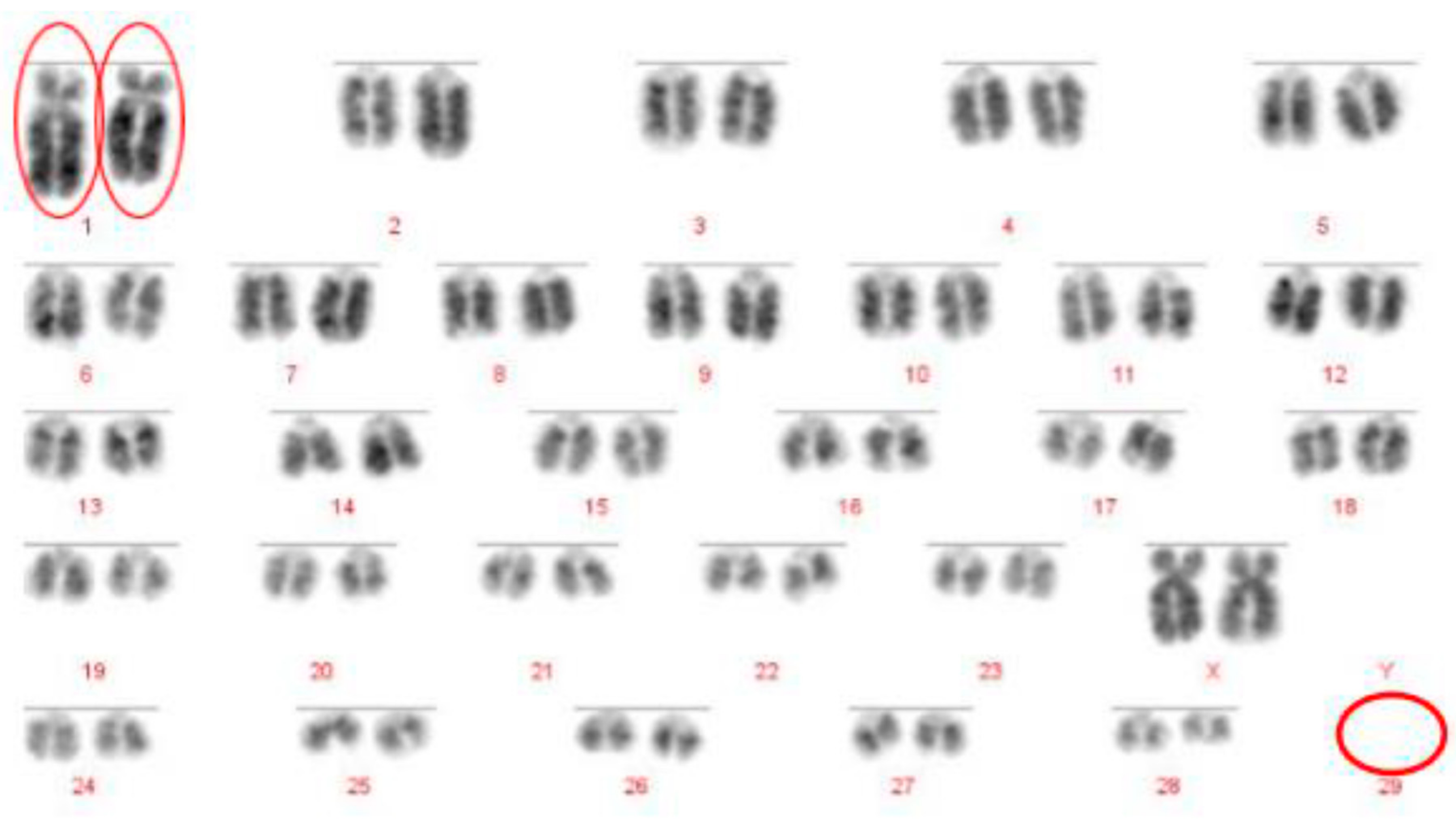
The Robertsonian translocations identified in this study, while detectable with basic karyotyping, can also easily and accurately be identified using FISH. Interestingly, despite great efforts in many breeding programmes to eliminate the 1;29 translocation, our results suggest that either these efforts have not been wholly successful, or that this rearrangement continues to recur de novo. Without historical data and ongoing routine screening of all animals entering the breeding population, it is difficult to ascertain what proportion of these rearrangements fall into this category.
This paper provides a small proof of principle for an approach that could potentially have wide applicability. The development and implementation of this FISH-based assay has, however, already markedly improved the efficiency and accuracy of translocation screening, allowing multiple hybridisation experiments in a single assay. Whilst chromosomal translocations have been demonstrated to significantly affect fertility in all species tested, in cattle, many of these rearrangements have remained undetected due to the inherent difficulties in finding them using previous technologies. The method presented here resolves this issue, allowing for the rapid identification of an abnormality, and corresponding rapid removal of affected animals from the herd. Not only will this lead to a reduction in the economic losses associated with using a subfertile bull, it will also reduce the need for the unnecessary culling of cows and bulls that are suspected (but unproven) to be sterile, thereby reducing economic and environmental loss. Moreover, the karyotype of both Bos Taurus and Bos indicus is identical (aside from the morphology of the Y chromosome), and therefore, although only established on a small number of individuals, this approach is universally applicable to all commercial breeding bulls.
In addition, an increasing emphasis on the use of in vitro production (IVP) methods to improve cattle breeding means that the requirement for high genetic merit gametes is not just limited to the analysis of bulls, but that the need to screen cows, and ultimately oocytes or embryos, for chromosome abnormalities will also become increasingly important [
15]. This screening approach allows both donor parents to be screened for underlying chromosomal abnormalities prior to their use in IVP programmes, thereby improving the genetic quality of the embryos generated using these methods. Other efforts in our laboratory allow for the screening of oocytes and embryos [
15].
Finally, with the success of this screening method in place and the success of our previously developed method in screening for chromosome abnormalities in pigs, it is plausible to suggest that this technique could be applied to any animal of interest, with the horse being an ideal future candidate. The domestic horse (2n = 64) is of significant interest to many different groups worldwide; the thoroughbred breeding industry, for example, could benefit from a similar screening service. Previous cytogenetic studies have identified chromosomal translocations that affect fertility in thoroughbred mares [
16], another group for which breeders place a significant importance on high genetic merit in the breeding population. Having the tools to examine and diagnose chromosomal abnormalities in a similarly fast and efficient manner would, therefore, be beneficial to this industry.