Isolation and Polyphasic Characterization of Desulfuromonas versatilis sp. Nov., an Electrogenic Bacteria Capable of Versatile Metabolism Isolated from a Graphene Oxide-Reducing Enrichment Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Growth Conditions
2.2. Morphological, Physiological, and Biochemical Analyses
2.3. Chemotaxonomic Analysis
2.4. Genetic Characterization
3. Results
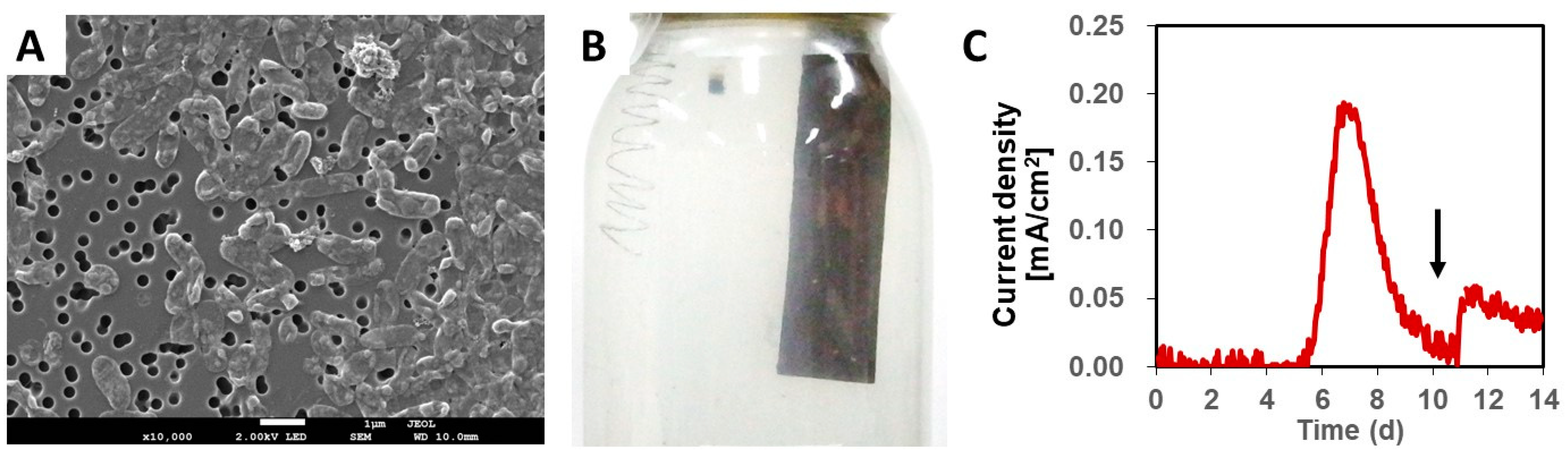
3.1. Isolation of NIT-T3
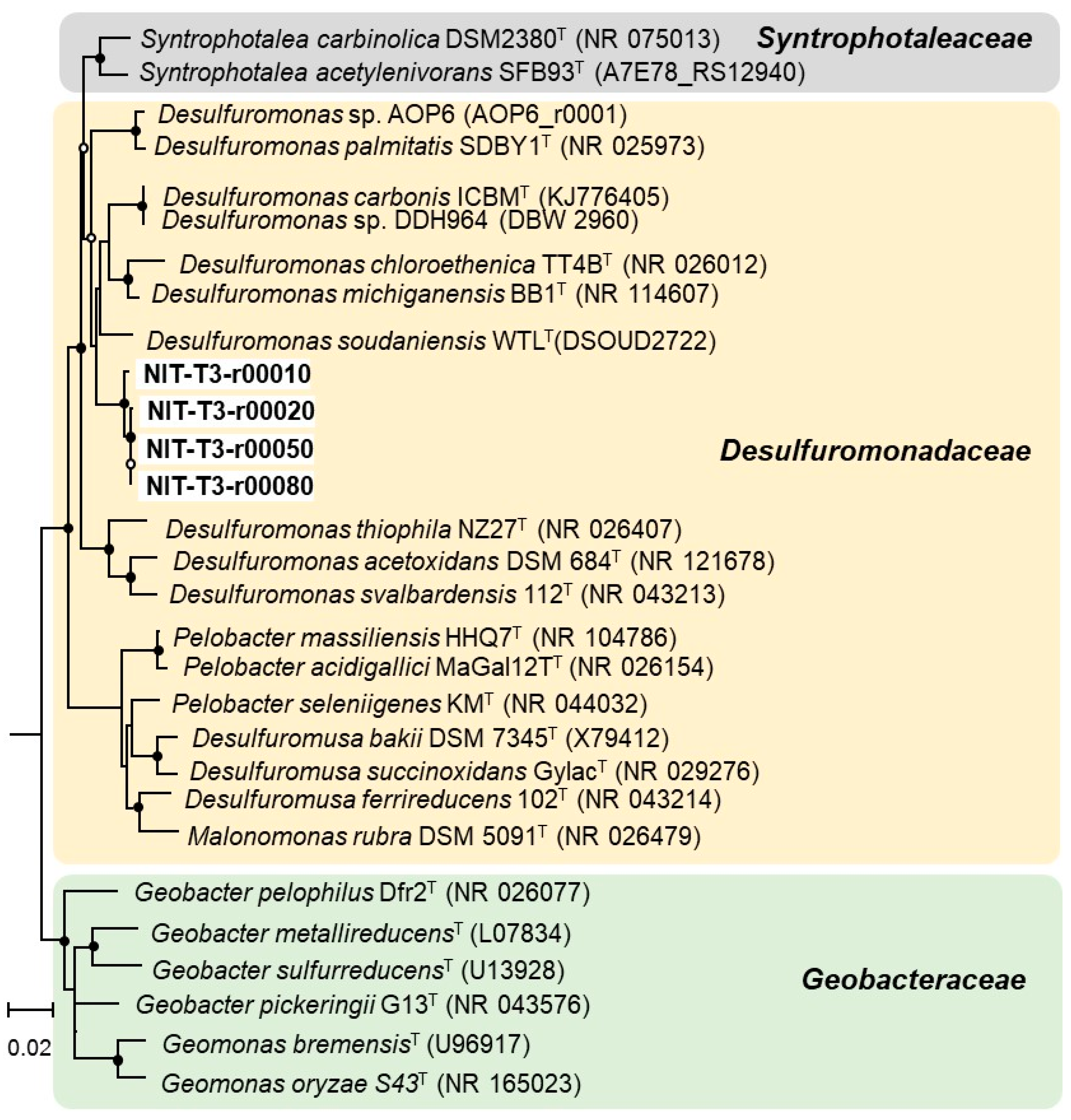
3.2. Phylogenetic Identification Based on 16S rRNA Sequencing
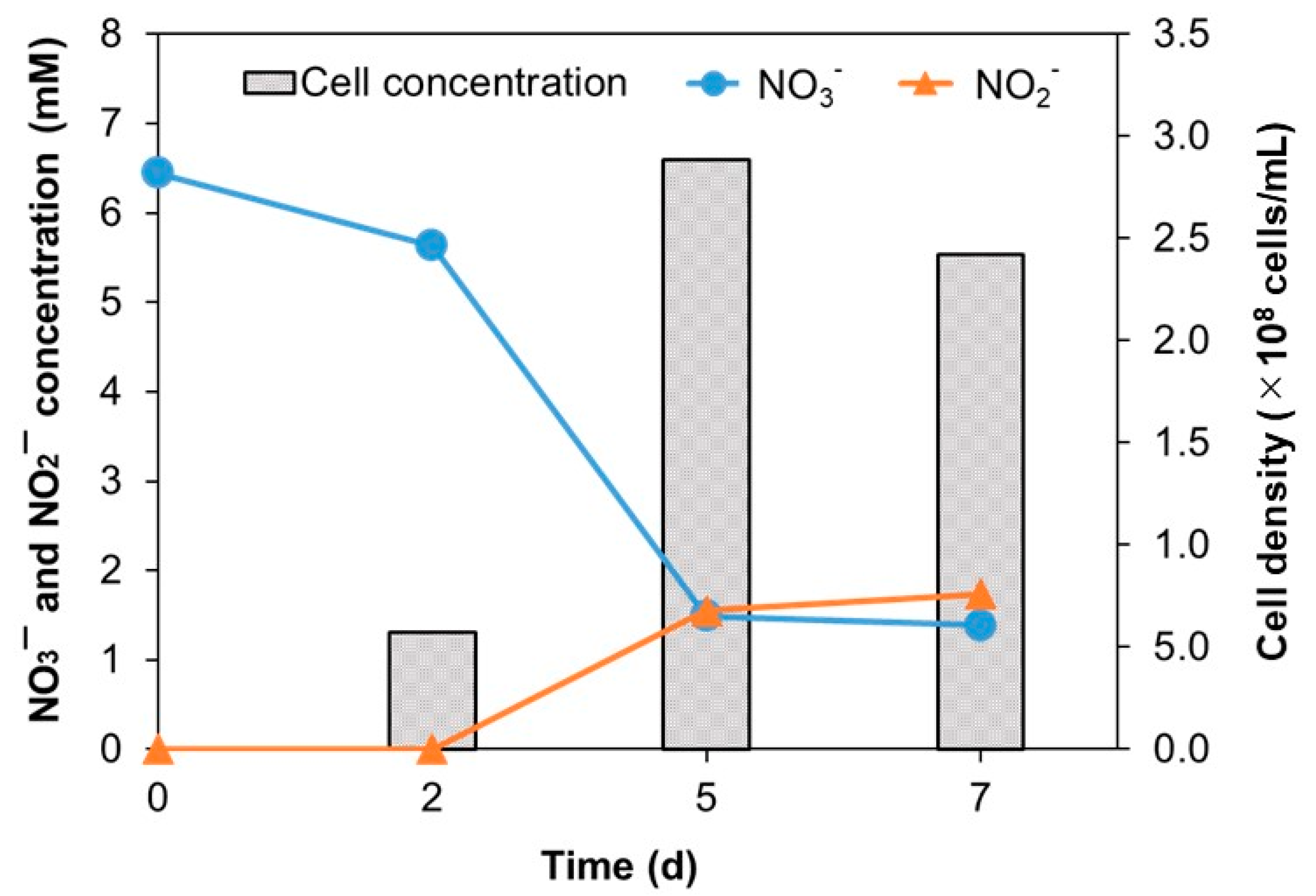
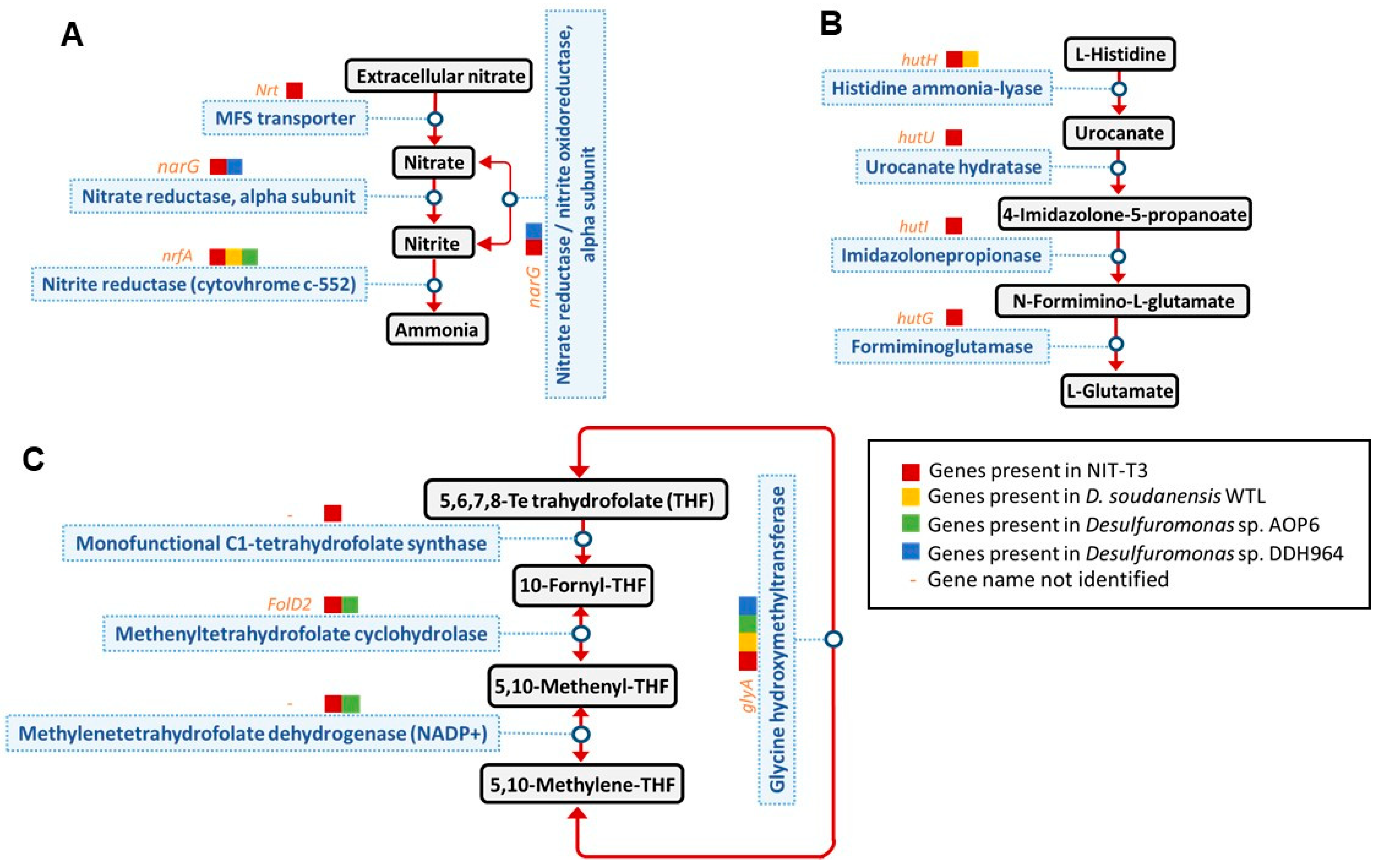
3.3. Physiological and Biochemical Characterization
3.4. Chemotaxonomic Characterization
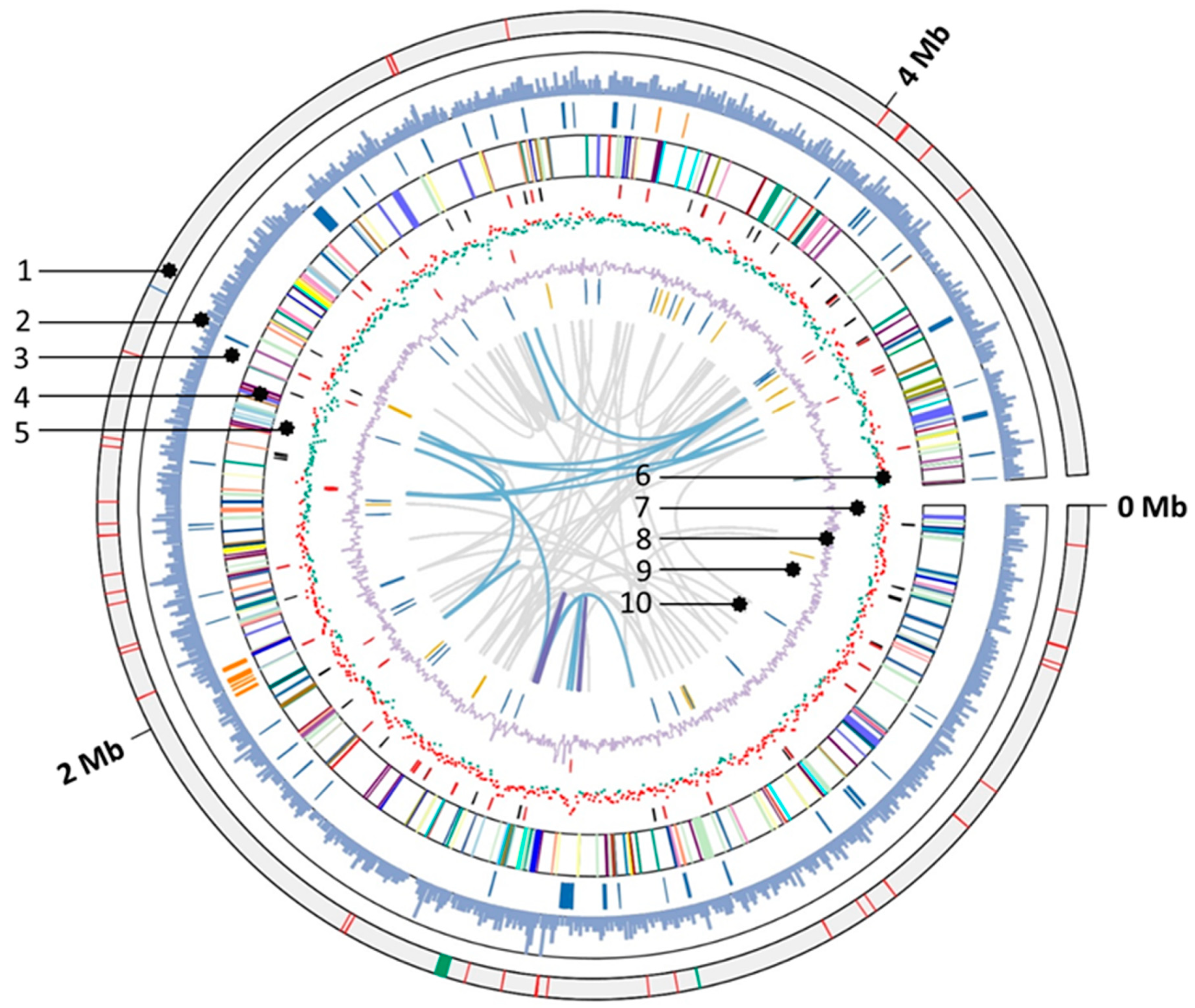
3.5. General Genomic Features
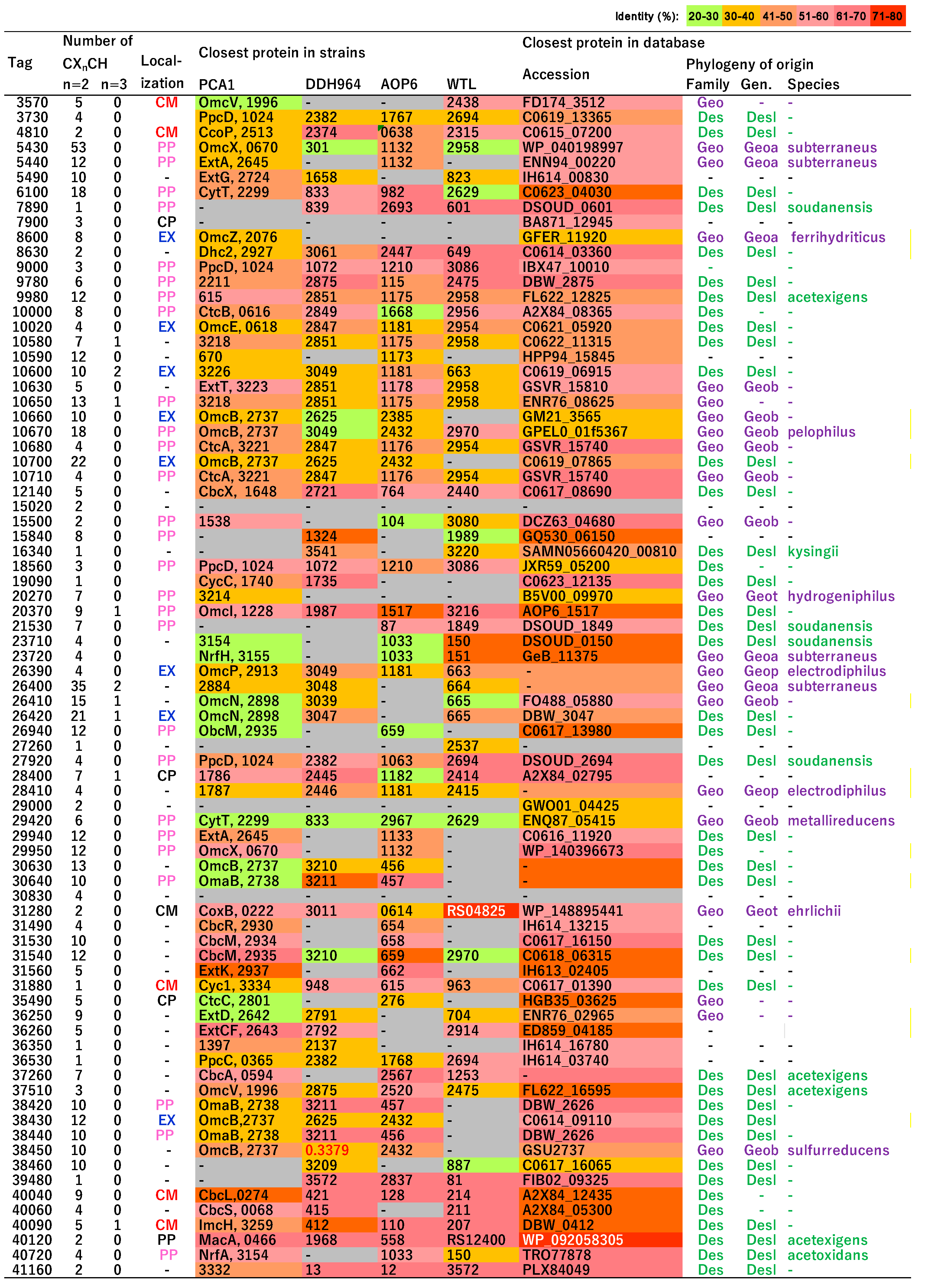
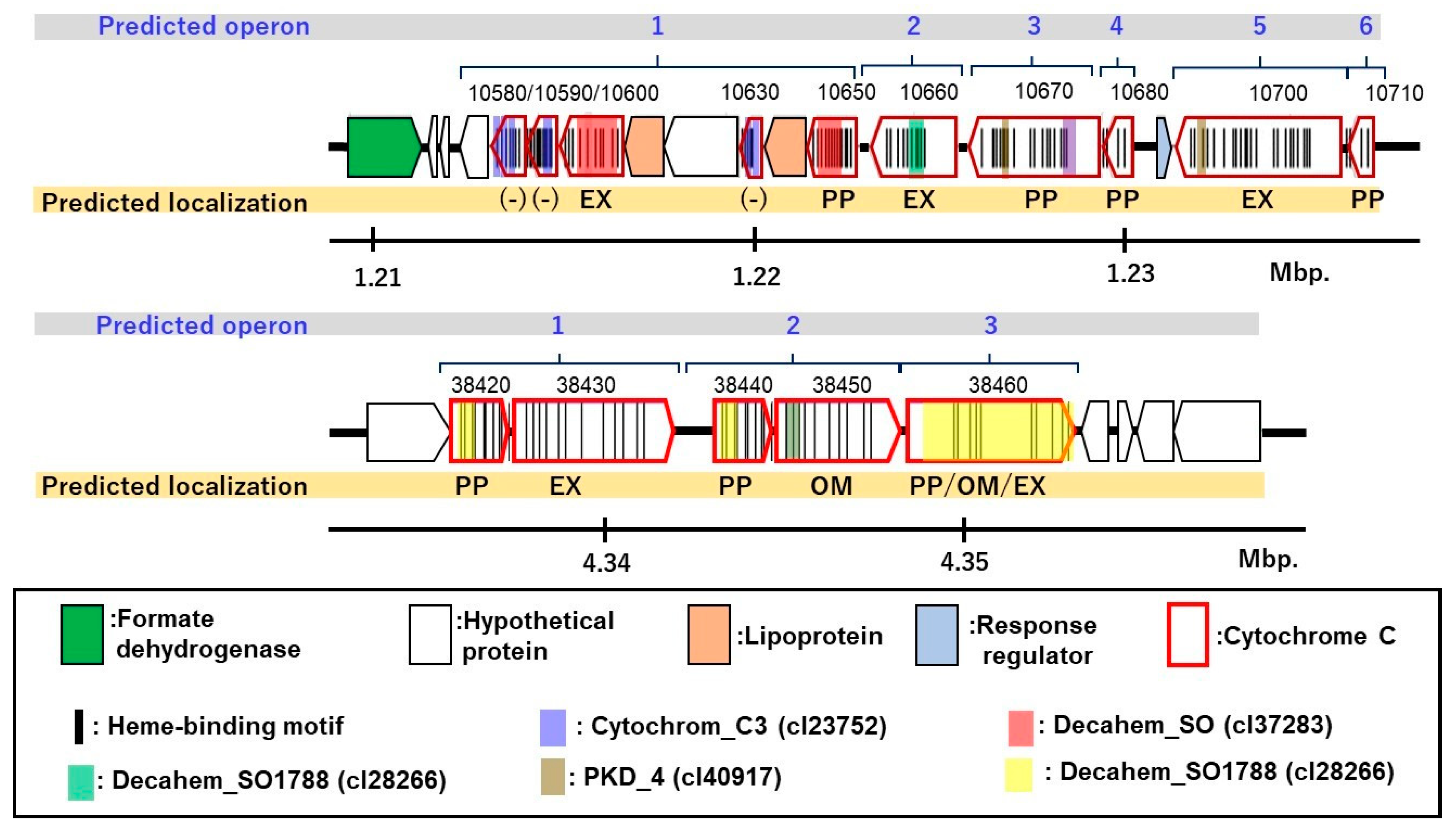
3.6. Putative c-Type Cytochromes
3.7. Homologs of c-Type Cytochromes to Those in G. sulfurreducens PCA
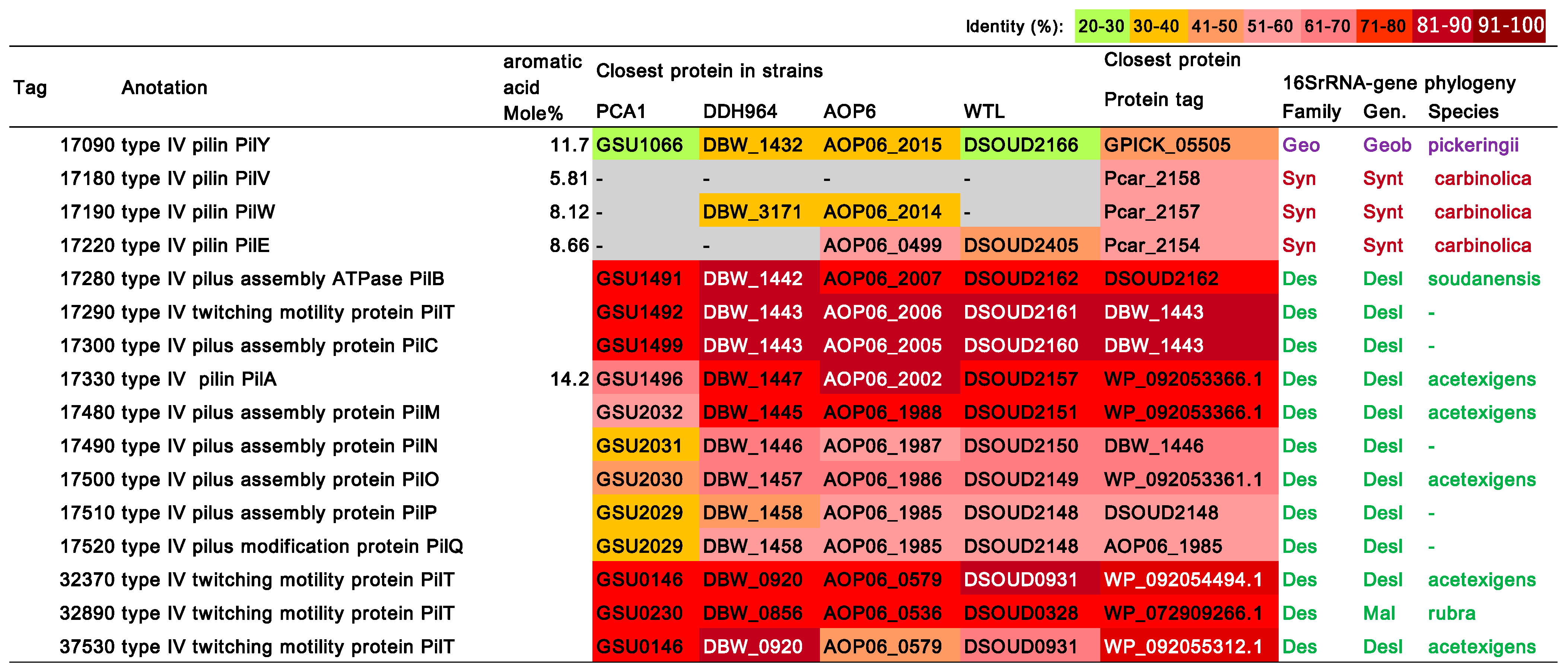
3.8. Type IV Pilus (T4P)-Related Genes
4. Discussion
5. Conclusions
Description of Desulfuromonas versatilis sp. Nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Logan, B.E.; Hamelers, B.; Rozendal, R.; Schröder, U.; Keller, J.; Freguia, S.; Aelterman, P.; Verstraete, W.; Rabaey, K. Microbial fuel cells: Methodology and technology. Environ. Sci. Technol. 2006, 40, 5181–5192. [Google Scholar] [CrossRef]
- Zou, S.; He, Z. Efficiently “pumping out” value-added resources from wastewater by bioelectrochemical systems: A review from energy perspectives. Water Res. 2018, 131, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Aulenta, F.; Puig, S.; Esteve-Núñez, A.; He, Y.; Mu, Y.; Rabaey, K. Microbial electrochemistry for bioremediation. Environ. Sci. Ecotechnol. 2020, 1, 100013. [Google Scholar] [CrossRef]
- Zhi, W.; Ge, Z.; He, Z.; Zhang, H. Methods for understanding microbial community structures and functions in microbial fuel cells: A review. Bioresour. Technol. 2014, 171, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Saheb-Alam, S.; Persson, F.; Wilén, B.M.; Hermansson, M.; Modin, O. Response to starvation and microbial community composition in microbial fuel cells enriched on different electron donors. Microb. Biotechnol. 2019, 12, 962–975. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, M.; Tsui, T.-H.; Yin, Z.; Yin, C. Comparative evaluation on the utilization of applied electrical potential in a conductive granule packed biotrickling filter for continuous abatement of xylene: Performance, limitation, and microbial community. J. Environ. Manag. 2020, 274, 111145. [Google Scholar] [CrossRef] [PubMed]
- Katuri, K.P.; Kamireddy, S.; Kavanagh, P.; Mohammad, A.; Conghaile, P.; Kumar, A.; Saikaly, P.E.; Leech, D. Electroactive biofilms on surface functionalized anodes: The anode respiring behavior of a novel electroactive bacterium, Desulfuromonas acetexigens. bioRxiv 2020. [Google Scholar] [CrossRef]
- Marone, A.; Carmona-Martínez, A.A.; Sire, Y.; Meudec, E.; Steyer, J.P.; Bernet, N.; Trably, E. Bioelectrochemical treatment of table olive brine processing wastewater for biogas production and phenolic compounds removal. Water Res. 2016, 100, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Zubchenko, L.; Kuzminskiy, Y. Characteristics of biofilm formation process in the bioelectrochemical systems, working in batch-mode of cultivation. Chem. Chem. Technol. 2017, 11, 105–110. [Google Scholar] [CrossRef]
- Kouzuma, A.; Ishii, S.; Watanabe, K. Metagenomic insights into the ecology and physiology of microbes in bioelectrochemical systems. Bioresour. Technol. 2018, 255, 302–307. [Google Scholar] [CrossRef]
- Nijenhuis, I.; Nikolausz, M.; Köth, A.; Felföldi, T.; Weiss, H.; Drangmeister, J.; Großmann, J.; Kästner, M.; Richnow, H.H. Assessment of the natural attenuation of chlorinated ethenes in an anaerobic contaminated aquifer in the Bitterfeld/Wolfen area using stable isotope techniques, microcosm studies and molecular biomarkers. Chemosphere 2007, 67, 300–311. [Google Scholar] [CrossRef]
- Dowideit, K.; Scholz-Muramatsu, H.; Miethling-Graff, R.; Vigelahn, L.; Freygang, M.; Dohrmann, A.B.; Tebbe, C.C. Spatial heterogeneity of dechlorinating bacteria and limiting factors for in situ trichloroethene dechlorination revealed by analyses of sediment cores from a polluted field site. FEMS Microbiol. Ecol. 2010, 71, 444–459. [Google Scholar] [CrossRef][Green Version]
- Chang, Y.H.; Cheng, T.W.; Lai, W.J.; Tsai, W.Y.; Sun, C.H.; Lin, L.H.; Wang, P.L. Microbial methane cycling in a terrestrial mud volcano in eastern Taiwan. Environ. Microbiol. 2012, 14, 895–908. [Google Scholar] [CrossRef]
- Pfennig, N.; Biebl, H. Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch. Microbiol. 1976. [Google Scholar] [CrossRef] [PubMed]
- Finster, K.; Bak, F.; Pfennig, N. Desulfuromonas acetexigens sp. nov., a dissimilatory sulfur-reducing eubacterium from anoxic freshwater sediments. Arch. Microbiol. 1994. [Google Scholar] [CrossRef]
- Coates, J.D.; Lonergan, D.J.; Philips, E.J.P.; Jenter, H.; Lovley, D.R.; Lovley, D.R. Desulfuromonas palmitatis sp. nov., a marine dissimilatory Fe(ill) reducer that can oxidize long-chain fatty acids. Arch. Microbiol. 1995, 164, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Finster, K.; Coates, J.D.; Liesack, W.; Pfennig, N. Desulfuromonas thiophila sp. nov., a new obligately sulfur-reducing bacterium from anoxic freshwater sediment. Int. J. Syst. Bacteriol. 1997, 47, 754–758. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krumholz, L.R. Desulfuromonas chloroethenica sp. nov. uses tetrachloroethylene and trichloroethylene as electron acceptors. Int. J. Syst. Bacteriol. 1997, 47, 1262–1263. [Google Scholar] [CrossRef]
- Sung, Y.; Ritalahti, K.M.; Sanford, R.A.; Urbance, J.W.; Flynn, S.J.; Tiedje, J.M.; Löffler, F.E. Characterization of two tetrachloroethene-reducing, acetate-oxidizing anaerobic bacteria and their description as Desulfuromonas michiganensis sp. nov. Appl. Environ. Microbiol. 2003, 69, 2964–2974. [Google Scholar] [CrossRef] [PubMed]
- Vandieken, V.; Mußmann, M.; Niemann, H.; Jørgensen, B.B. Desulfuromonas svalbardensis sp. nov. and Desulfuromusa ferrireducens sp. nov., psychrophilic, Fe(III)-reducing bacteria isolated from Arctic sediments, Svalbard. Int. J. Syst. Evol. Microbiol. 2006, 56, 1133–1139. [Google Scholar] [CrossRef]
- An, T.T.; Picardal, F.W. Desulfuromonas carbonis sp. nov., an Fe(III)-, S0- and Mn(IV)-reducing bacterium isolated from an active coalbed methane gas well. Int. J. Syst. Evol. Microbiol. 2015, 65, 1686–1693. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.; Cha, I.; Min, D.; Kim, J.; Chung, W.; Chae, J.; Jeon, C.O.; Rhee, S. Metabolic versatility of toluene-degrading, iron-reducing bacteria in tidal flat sediment, characterized by stable isotope probing-based metagenomic analysis. Environ. Microbiol. 2014, 16, 189–204. [Google Scholar] [CrossRef]
- Guo, Y.; Aoyagi, T.; Inaba, T.; Sato, Y.; Habe, T.H.H. Complete Genome Sequence of Desulfuromonas sp. Strain AOP6, an Iron (III) Reducer Isolated from Subseafloor Sediment. Microbiol. Resour. Announc. 2020, 9, e01325-19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Bain, T.S.; Barlett, M.A.; Dar, S.A.; Snoeyenbos-West, O.L.; Nevin, K.P.; Lovley, D.R. Sulfur oxidation to sulfate coupled with electron transfer to electrodes by Desulfuromonas strain TZ1. Microbiology 2014, 160, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Badalamenti, J.P.; Summers, Z.M.; Chan, C.H.; Gralnick, J.A.; Bond, D.R. Isolation and genomic characterization of “Desulfuromonas soudanensis WTL”, a metal- and electrode-respiring bacterium from anoxic deep subsurface brine. Front. Microbiol. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, T.; Bardiaux, B.; Francetic, O.; Izadi-Pruneyre, N.; Nilges, M. Structure and function of minor pilins of type IV pili. Med. Microbiol. Immunol. 2020, 209, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Goto, Y.; Miyata, Y.; Thakur, V.K. Selective growth of and electricity production by marine exoelectrogenic bacteria in self-aggregated hydrogel of microbially reduced graphene oxide. C. J. Carbon Res. 2016, 2016, 15. [Google Scholar] [CrossRef]
- Yoshida, N.; Miyata, Y.; Doi, K.; Goto, Y.; Nagao, Y.; Tero, R.; Hiraishi, A. Graphene oxide-dependent growth and self-Aggregation into a hydrogel complex of exoelectrogenic bacteria. Sci. Rep. 2016, 6, 15. [Google Scholar] [CrossRef]
- Goto, Y.; Yoshida, N.; Umeyama, Y.; Yamada, T.; Tero, R.; Hiraishi, A. Enhancement of electricity production by graphene oxide in soil microbial fuel cells and plant microbial fuel cells. Front. Bioeng. Biotechnol. 2015, 3, 42. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Yoshida, N. Scaling up microbial fuel cells for treating. Water 2019, 11, 1803. [Google Scholar] [CrossRef]
- Goto, Y.; Yoshida, N. Microbially reduced graphene oxide shows efficient electricity recovery from artificial dialysis wastewater. J. Gen. Appl. Microbiol. 2017, 63, 165–171. [Google Scholar] [CrossRef]
- Yoshida, N.; Miyata, Y.; Mugita, A.; Iida, K. Electricity recovery from municipal sewage wastewater using a hydrogel complex composed of microbially reduced graphene oxide and sludge. Materials 2016, 9, 742. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Asahi, K.; Sakakibara, Y.; Miyake, K.; Katayama, A. Isolation and quantitative detection of tetrachloroethene (PCE)-dechlorinating bacteria in unsaturated subsurface soils contaminated with chloroethenes. J. Biosci. Bioeng. 2007, 104, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Ismaeil, M.; Yoshida, N.; Katayama, A. Bacteroides sedimenti sp. nov., isolated from a chloroethenes-dechlorinating consortium enriched from river sediment. J. Microbiol. 2018, 56, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Nagahashi, W.; Yoshida, N. Comparative evaluation of fibrous artificial carbons and bamboo charcoal in terms of recovery of current from sewage wastewater. J. Gen. Appl. Microbiol. 2021, in press. [Google Scholar] [CrossRef]
- Tamaoka, J.; Katayama-Fujimura, Y.; Kuraishi, H. Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J. Appl. Bacteriol. 1983, 54, 31–36. [Google Scholar] [CrossRef]
- Ismaeil, M.; Yoshida, N.; Katayama, A. Identification of multiple dehalogenase genes involved in tetrachloroethene-to-ethene dechlorination in a dehalococcoides-dominated enrichment culture. Biomed. Res. Int. 2017, 2017, 9191086. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the rapid annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, 206–214. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wang, A.; Cheng, S.; Yates, M.; Logan, B.E. Geobacter anodireducens sp. nov., an exoelectrogenic microbe in bioelectrochemical systems. Int. J. Syst. Evol. Microbiol. 2014, 64, 3485–3491. [Google Scholar] [CrossRef] [PubMed]
- Viulu, S.; Nakamura, K.; Okada, Y.; Saitou, S.; Takamizawa, K. Geobacter luticola sp. nov., an Fe(III)-reducing bacterium isolated from lotus field mud. Int. J. Syst. Evol. Microbiol. 2013, 63, 442–448. [Google Scholar] [CrossRef]
- Shelobolina, E.S.; Vrionis, H.A.; Findlay, R.H.; Lovley, D.R. Geobacter uraniireducens sp. nov., isolated from subsurface sediment undergoing uranium bioremediation. Int. J. Syst. Evol. Microbiol. 2008, 58, 1075–1078. [Google Scholar] [CrossRef] [PubMed]
- Shelobolina, E.S.; Nevin, K.P.; Blakeney-Hayward, J.D.; Johnsen, C.V.; Plaia, T.W.; Krader, P.; Woodard, T.; Holmes, D.E.; VanPraagh, C.G.; Lovley, D.R. Geobacter pickeringii sp. nov., Geobacter argillaceus sp. nov. and Pelosinus fermentans gen. nov., sp. nov., isolated from subsurface kaolin lenses. Int. J. Syst. Evol. Microbiol. 2007, 57, 126–135. [Google Scholar] [CrossRef]
- Hedrick, D.B.; Peacock, A.D.; Lovley, D.R.; Woodard, T.L.; Nevin, K.P.; Long, P.E.; White, D.C. Polar lipid fatty acids, LPS-hydroxy fatty acids, and respiratory quinones of three Geobacter strains, and variation with electron acceptor. J. Ind. Microbiol. Biotechnol. 2009, 36, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Kunapuli, U.; Jahn, M.K.; Lueders, T.; Geyer, R.; Heipieper, H.J.; Meckenstock, R.U. Desulfitobacterium aromaticivorans sp. nov. and Geobacter toluenoxydans sp. nov., iron-reducing bacteria capable of anaerobic degradation of monoaromatic hydrocarbons. Int. J. Syst. Evol. Microbiol. 2010, 60, 686–695. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; Cerutti, L.; Hulo, N.; Gattiker, A.; Falquet, L.; Pagni, M.; Bairoch, A.; Bucher, P. PROSITE: A documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 2002, 3, 265–274. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, M.; Shi, L. Molecular underpinnings for microbial extracellular electron transfer during biogeochemical cycling of earth elements. Sci. China Life Sci. 2019, 62, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Levar, C.E.; Chan, C.H.; Mehta-Kolte, M.G.; Bond, D.R. An inner membrane cytochrome required only for reduction of high redox potential extracellular electron acceptors. MBio 2014, 5, e02034-14. [Google Scholar] [CrossRef]
- Levar, C.E.; Hoffman, C.L.; Dunshee, A.J.; Toner, B.M.; Bond, D.R. Redox potential as a master variable controlling pathways of metal reduction by Geobacter sulfurreducens. ISME J. 2017, 11, 741–752. [Google Scholar] [CrossRef]
- Morgado, L.; Brulx, M.; Pessanha, M.; Londer, Y.Y.; Salgueiro, C.A. Thermodynamic characterization of a triheme cytochrome family from geobacter sulfurreducens reveals mechanistic and functional diversity. Biophys. J. 2010, 99, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fredrickson, J.K.; Zachara, J.M.; Shi, L. Direct involvement of ombB, omaB, and omcB genes in extracellular reduction of Fe (III) by Geobacter sulfurreducens PCA. Front. Microbiol. 2015, 6, 1075. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Z.; Liu, J.; Levar, C.; Edwards, M.J.; Babauta, J.T.; Kennedy, D.W.; Shi, Z.; Beyenal, H.; Bond, D.R.; et al. A trans-outer membrane porin-cytochrome protein complex for extracellular electron transfer by Geobacter sulfurreducens PCA. Environ. Microbiol. Rep. 2014, 6, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Otero, F.J.; Chan, C.H.; Bond, D.R. Identification of different putative outer membrane electron conduits necessary for Fe(III) citrate, Fe(III) oxide, Mn(IV) oxide, or electrode reduction by Geobacter sulfurreducens. J. Bacteriol. 2018, 200, e00347-18. [Google Scholar] [CrossRef]
- Trumpower, B. The protonmotive Q cycle. J. Biol. Chem. 1990, 265, 11409–11412. [Google Scholar] [CrossRef]
- Mollaei, M.; Timmers, P.H.A.; Suarez-Diez, M.; Boeren, S.; van Gelder, A.H.; Stams, A.J.M.; Plugge, C.M. Comparative proteomics of Geobacter sulfurreducens PCAT in response to acetate, formate and/or hydrogen as electron donor. Environ. Microbiol. 2021, 23, 299–315. [Google Scholar] [CrossRef]
- Seidel, J.; Hoffmann, M.; Ellis, K.E.; Seidel, A.; Spatzal, T.; Gerhardt, S.; Elliott, S.J.; Einsle, O. MacA is a second cytochrome c peroxidase of Geobacter sulfurreducens. Biochemistry 2012, 51, 2747–2756. [Google Scholar] [CrossRef]
- Aklujkar, M.; Coppi, M.V.; Leang, C.; Kim, B.C.; Chavan, M.A.; Perpetua, L.A.; Giloteaux, L.; Liu, A.; Holmes, D.E. Proteins involved in electron transfer to Fe (III) and Mn (IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 2013, 159, 515–535. [Google Scholar] [CrossRef]
- Butler, J.E.; Young, N.D.; Lovley, D.R. Evolution of electron transfer out of the cell: Comparative genomics of six Geobacter genomes. BMC Genom. 2010, 11, 40. [Google Scholar] [CrossRef]
- Zheng, S.; Liu, F.; Li, M.; Xiao, L.; Wang, O. Comparative transcriptomic insights into the mechanisms of electron transfer in Geobacter co-cultures with activated carbon and magnetite. Sci. China Life Sci. 2018, 61, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.; Forest, K.T.; Maier, B. Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 2019, 17, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R.; Walker, D.J.F. Geobacter protein nanowires. Front. Microbiol. 2019, 10, 2078. [Google Scholar] [CrossRef]
- Lovley, D.R. Electrically conductive pili: Biological function and potential applications in electronics. Curr. Opin. Electrochem. 2017, 4, 190–198. [Google Scholar] [CrossRef]
- Walker, D.J.; Adhikari, R.Y.; Holmes, D.E.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Lovley, D.R. Electrically conductive pili from pilin genes of phylogenetically diverse microorganisms. ISME J. 2018, 12, 48–58. [Google Scholar] [CrossRef]
- Herzberg, M.; Kaye, I.K.; Peti, W.; Wood, T.K. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 2006, 188, 587–598. [Google Scholar] [CrossRef]
- Button, J.E.; Silhavy, T.J.; Ruiz, N. A suppressor of cell death caused by the loss of σE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J. Bacteriol. 2007, 189, 1523–1530. [Google Scholar] [CrossRef][Green Version]
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Motility | NM | ND | NM | Motile | Motile | NM | Motile | NM | Motile | Motile |
| G + C content (%) | 63.1 | 61.2 | 61.2 | 50.1 | ND | ND | 61.6 | 54.7 | 62.3 | 62.3 |
| Temp. range (°C) (Optimum) | 10–35 (25) | 24 | 10–37 (25–37) | 2–20 (14) | 10–35 (25) | 21–31 | 10–39 (26–30) | 40 | (30–35) | (30–35) |
| pH range (Optimum) | 6.4–8.4 (6.8–7.1) * | 6.8 | 6.5–8.0 (7.0) | 6.5–7.5 (7.3) | 6.8–8.0 (7.0–7.5) | 6.5–7.4 (7.4) | 6.5–8.2 (6.9–7.9) | ND | 6.4–8.5 (7.6–7.8) | 6.4–8.5 (7.6–7.8) |
| Electron donors | ||||||||||
| Hydrogen | + | + | (+) | - | - | - | - | (+) | - | - |
| Lactate | + | + | + | - | + | - | - | + | - | - |
| Fumarate | + | - | + | - | + | - | + | + | - | - |
| Succinate | + | - | + | - | + | - | + | + | - | - |
| Malate | + | ND | + | - | + | - | - | ND | - | - |
| Acetate | + | + | + | + | + | + | + | + | + | + |
| Pyruvate | + | + | + | + | + | + | + | - | ND | - |
| Glucose | - | - | - | - | - | ND | - | - | - | - |
| Butyrate | - | - | - | - | - | ND | - | - | - | - |
| Glycerol | - | - | ND | - | - | ND | - | ND | - | - |
| Peptone | + | ND | ND | ND | ND | ND | - | ND | ND | - |
| Isopropanol | + | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Ethanol | - | + | ND | + | - | - | - | - | - | + |
| Benzoate | - | - | - | ND | ND | - | - | - | - | - |
| Methanol | - | - | ND | ND | - | ND | - | - | - | - |
| Phenol | - | ND | - | ND | ND | ND | ND | - | ND | ND |
| Fructose | - | ND | - | - | - | ND | - | - | - | - |
| Isobutyrate | - | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Caproate | - | ND | ND | ND | ND | ND | - | ND | - | ND |
| Butanol | - | ND | ND | + | ND | ND | - | ND | - | + |
| Fermentation of | Fumarate | ND | Fumarate | ND | Fumarate malate | None | ND | ND | ND | ND |
| Electron acceptors | ||||||||||
| Nitrate | + | ND | - | - | - | - | - | - | - | - |
| Sulfur | + | ND | + | + | + | ND | + | + | + | + |
| Sulfate | - | ND | - | - | - | - | - | - | - | - |
| Thiosulfate | - | ND | - | - | - | - | - | - | - | - |
| Ferric iron (Fe(III)) | + | ND | + | + | + | + | (+) | + | ND | ND |
| Malate | + | ND | - | - | + | - | - | - | + | + |
| Fumarate | ND | ND | - | + | + | + | - | + | + | + |
| AQDS | + | ND | - | ND | ND | ND | ND | ND | ND | ND |
| GO | + | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Major fatty acids (>10%) | C16: 1ω7c (26.2%) C16: 0 (18.3%) C15: 0 (13.2) | ND | C16: 0 (39.3%) C16:1ω7c and/or iso-C15:0 2-OH (36.6%) | C16: 0 (43%) C16:1ω7c (35%) C15: 0 (10%) | ND | ND | ND | ND | ND | ND |
| Major respiratory quinones | MK-8 (93%) MK-9 (5.3%) MK-7 (1.9%) | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Strains: 1, NIT-T3 (this study); 2, Desulfuromonas soudanensis WTLT [21]; 3, Desulfuromonas carbonis ICBMT [18]; 4, Desulfuromonas svalbardensis 112T [17]; 5, Desulfuromonas michiganensis BB1T [16]; 6, Desulfuromonas chloroethenica TT4BT [15]; 7, Desulfuromonas thiophila NZ27T [14]; 8, Desulfuromonas palmitatis SDBY1T [13]; 9, Desulfuromonas acetexigens 2873T [12]; 10, Desulfuromonas acetoxidans DSM 684T [11]. The data for NIT-T3 was obtained in this study and others are brought from references [14,15,16,17,18,19,20,21]. | ||||||||||
| +, good growth; (+), hydrogen was oxidized but no growth; -, no growth; [motile], only a small population was motile; ND, not determined; NM, not motile; temp., temperature; AQDS, Anthraquinone-2,6-disulfonate; GO, graphene oxide. | ||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, L.; Yoshida, N.; Ishii, S.; Meng, L. Isolation and Polyphasic Characterization of Desulfuromonas versatilis sp. Nov., an Electrogenic Bacteria Capable of Versatile Metabolism Isolated from a Graphene Oxide-Reducing Enrichment Culture. Microorganisms 2021, 9, 1953. https://doi.org/10.3390/microorganisms9091953
Xie L, Yoshida N, Ishii S, Meng L. Isolation and Polyphasic Characterization of Desulfuromonas versatilis sp. Nov., an Electrogenic Bacteria Capable of Versatile Metabolism Isolated from a Graphene Oxide-Reducing Enrichment Culture. Microorganisms. 2021; 9(9):1953. https://doi.org/10.3390/microorganisms9091953
Chicago/Turabian StyleXie, Li, Naoko Yoshida, Shun’ichi Ishii, and Lingyu Meng. 2021. "Isolation and Polyphasic Characterization of Desulfuromonas versatilis sp. Nov., an Electrogenic Bacteria Capable of Versatile Metabolism Isolated from a Graphene Oxide-Reducing Enrichment Culture" Microorganisms 9, no. 9: 1953. https://doi.org/10.3390/microorganisms9091953
APA StyleXie, L., Yoshida, N., Ishii, S., & Meng, L. (2021). Isolation and Polyphasic Characterization of Desulfuromonas versatilis sp. Nov., an Electrogenic Bacteria Capable of Versatile Metabolism Isolated from a Graphene Oxide-Reducing Enrichment Culture. Microorganisms, 9(9), 1953. https://doi.org/10.3390/microorganisms9091953






