RNA-Seq Provides New Insights into the Gene Expression Changes in Azoarcus olearius BH72 under Nitrogen-Deficient and Replete Conditions beyond the Nitrogen Fixation Process
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth and Harvesting of Bacterial Cultures
2.2. Extraction of RNA from Bacterial Cells
2.3. Preparation and Sequencing of Directional RNA-Seq Library
2.4. Mapping of Reads to the Genome Sequence and Analysis of Differential Expression
2.5. Verification of RNA-Seq Data by Quantitative RT-PCR
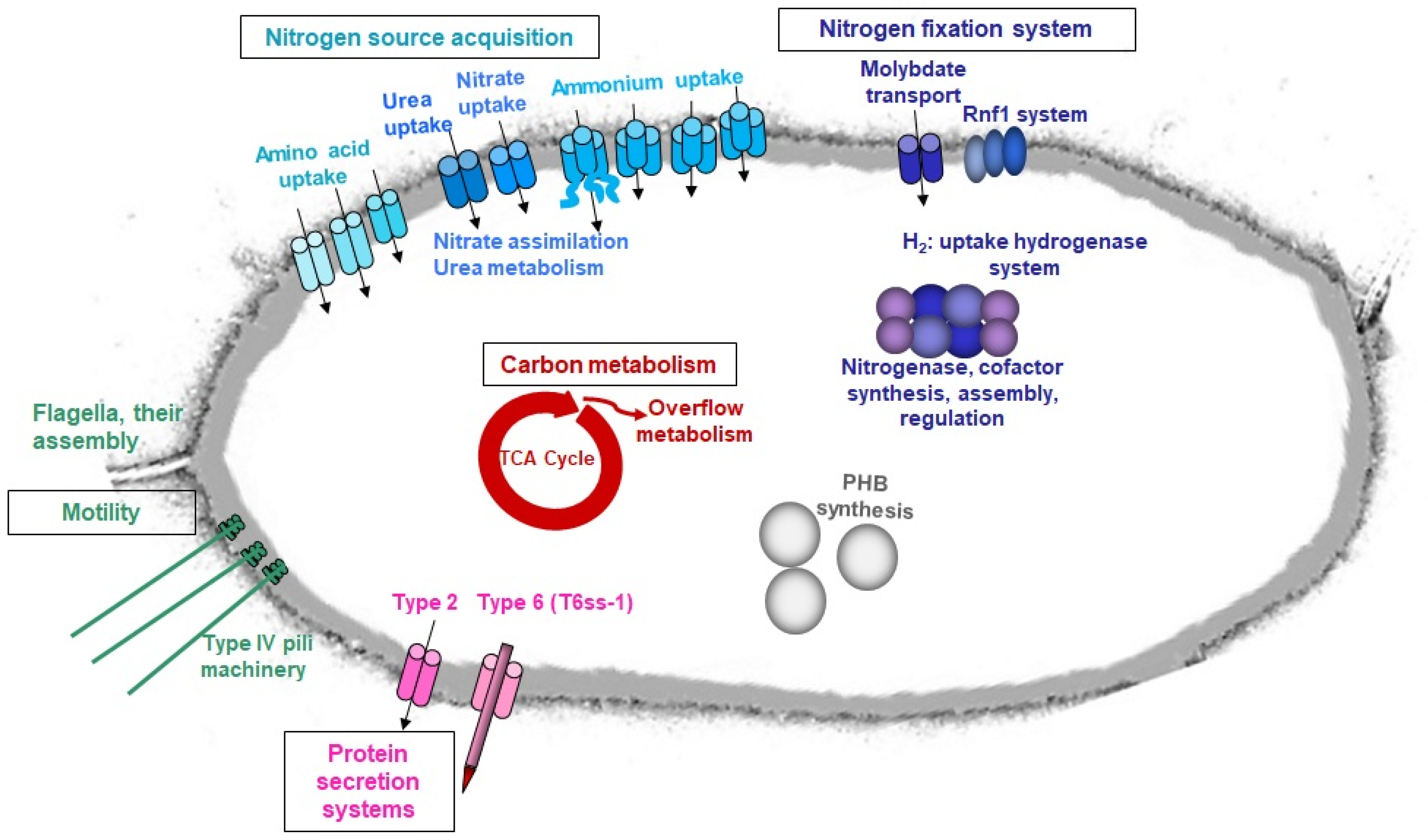
3. Results and Discussion
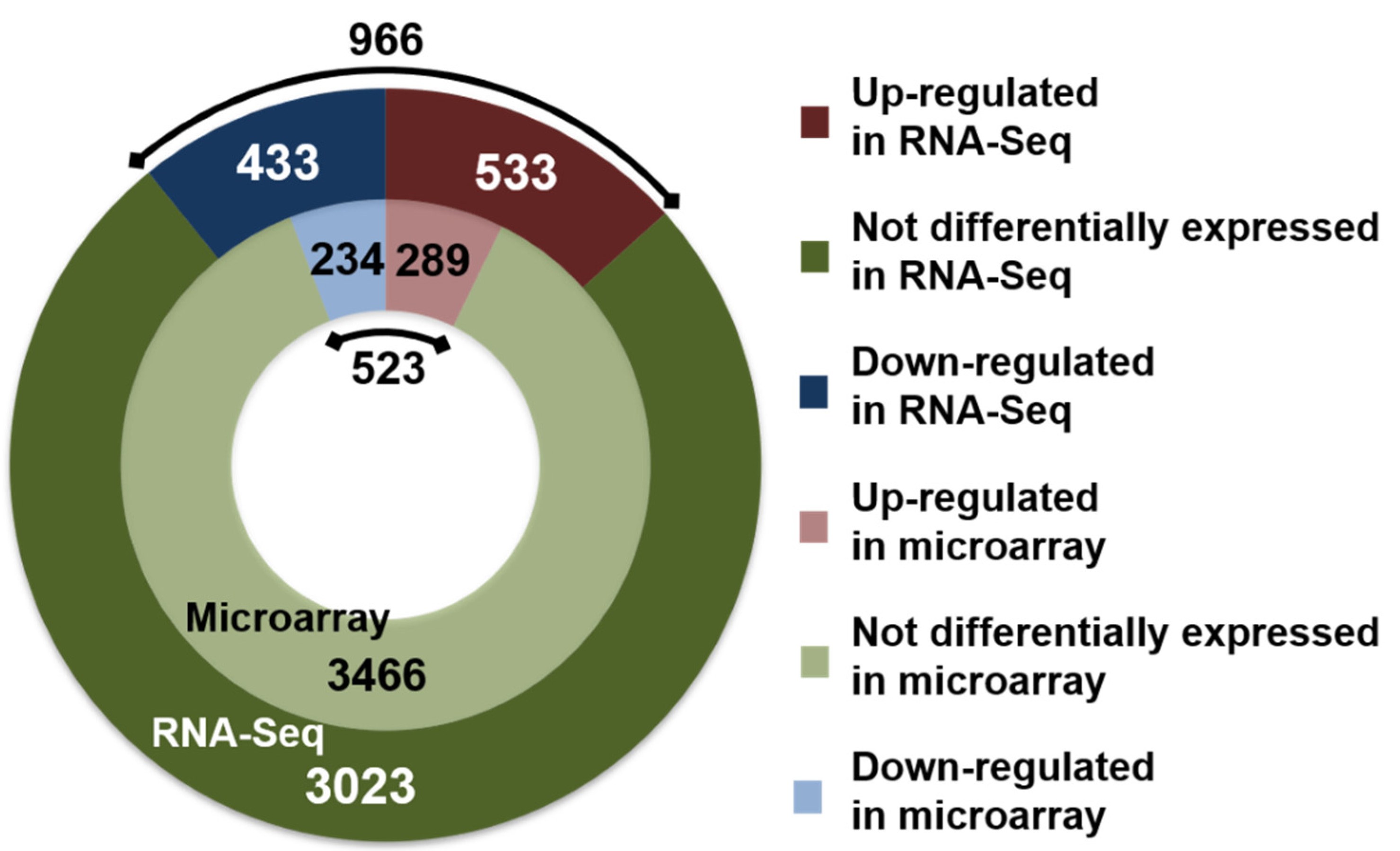
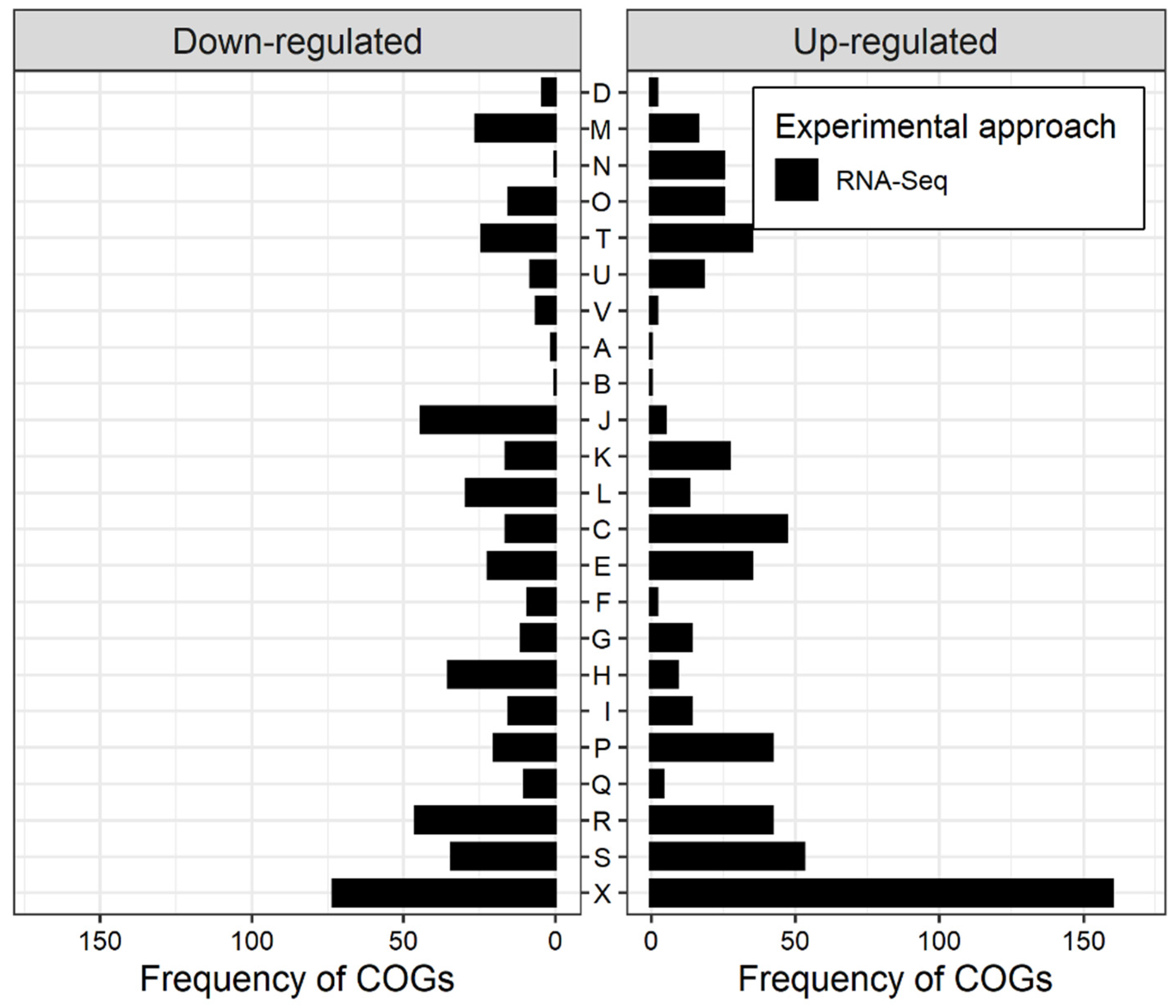
3.1. New Information on the Transcriptome under N2-Fixing Conditions Provided by RNA-Seq Compared to Previous Microarray-Based Transcriptome Analyses
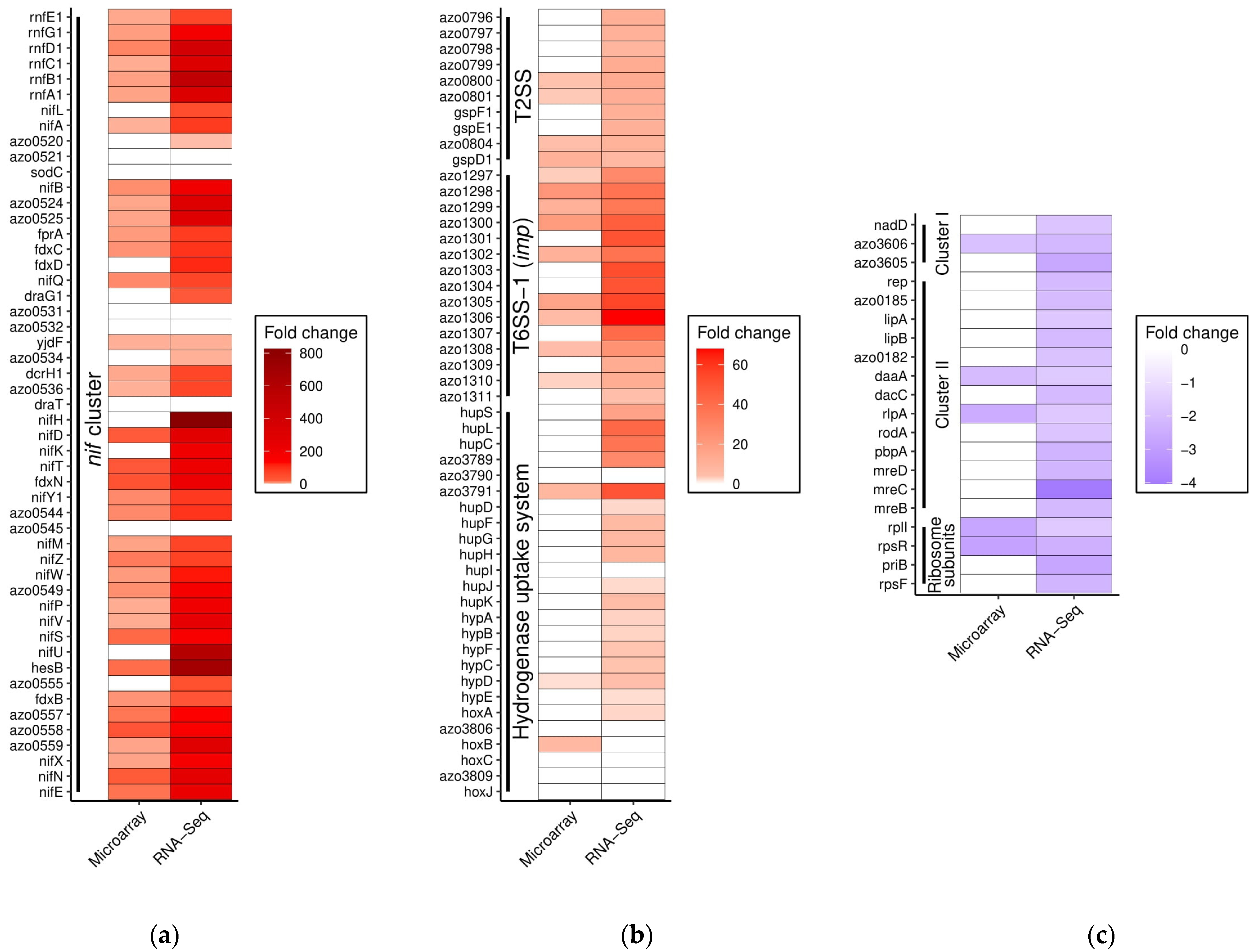
3.2. The Higher Sensitivity of RNA-Seq Confirms Known Regulation Patterns
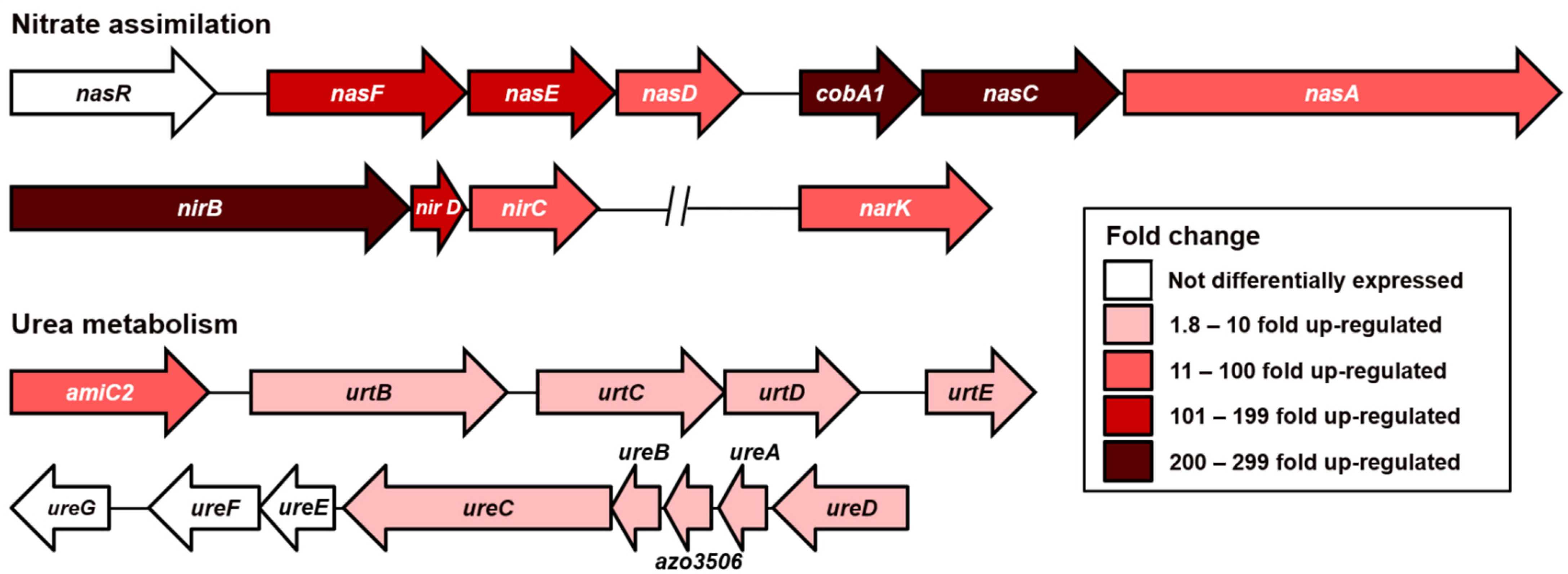
3.3. The Quest for Alternative Nitrogen Sources during Nitrogen Fixation
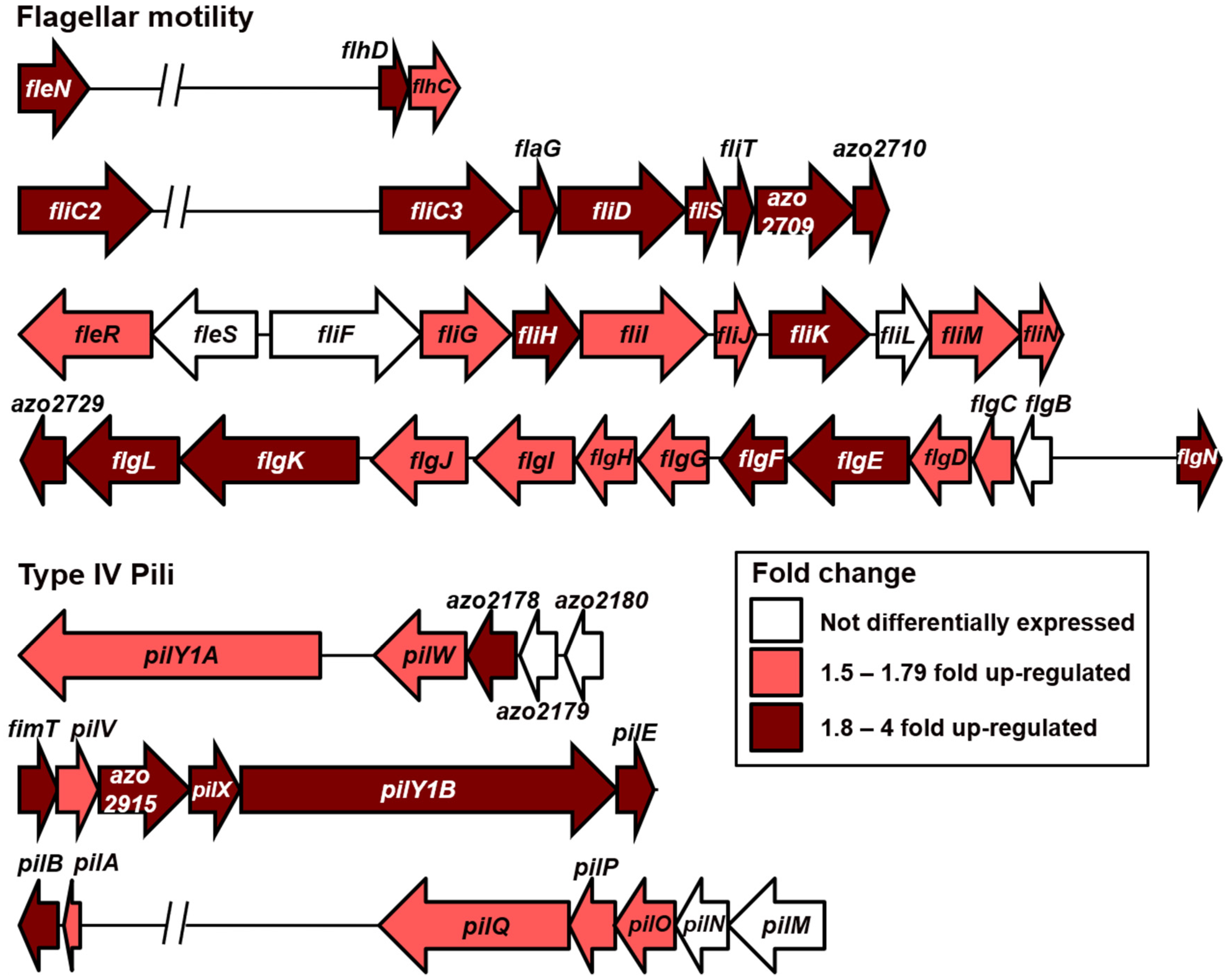
3.4. Motility Fostered by N2-Deplete Environment–Search for More Favourable Niches?
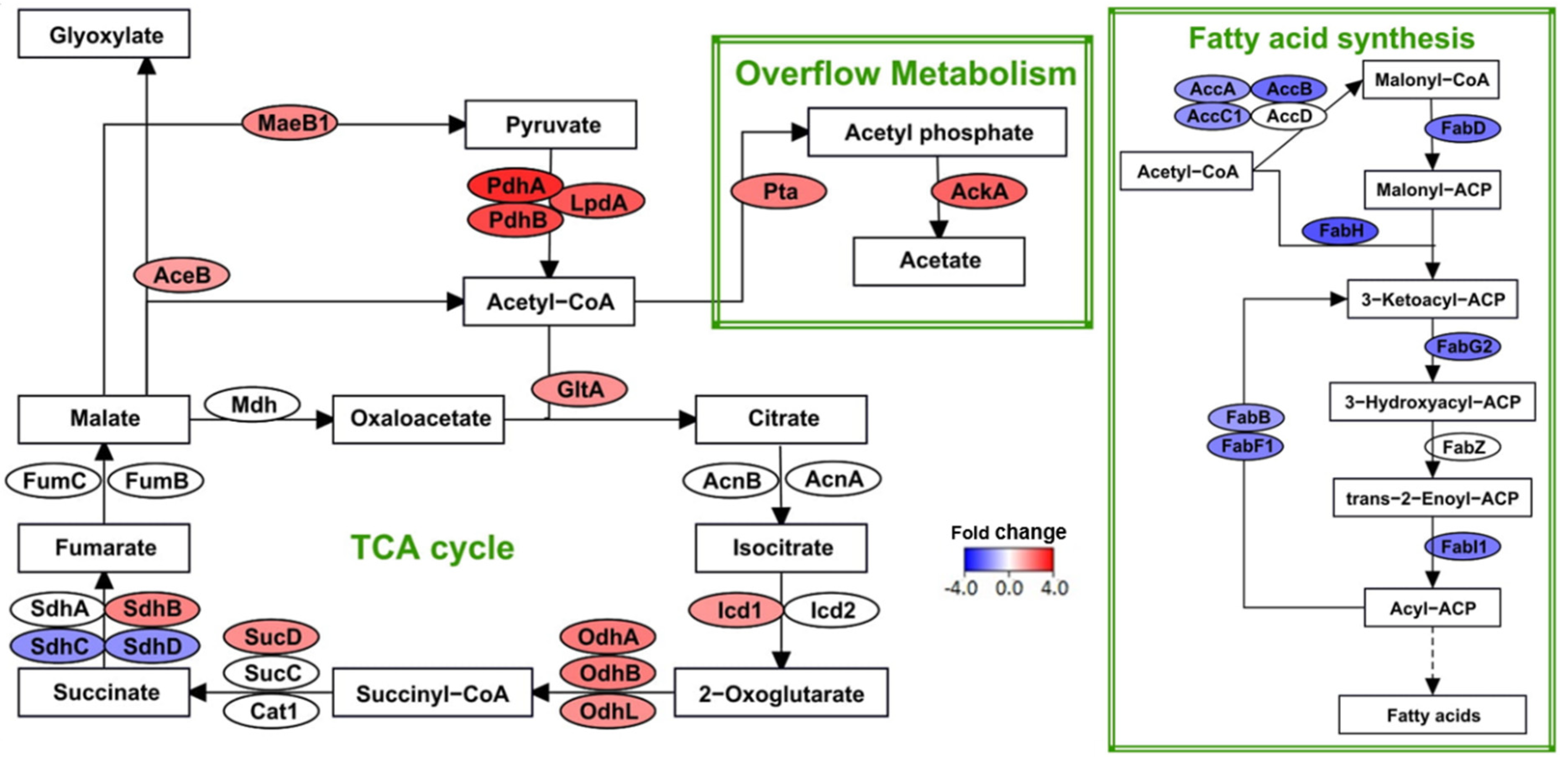
3.5. Increased Energy Requirements under Diazotrophic Reflected in Transcriptome
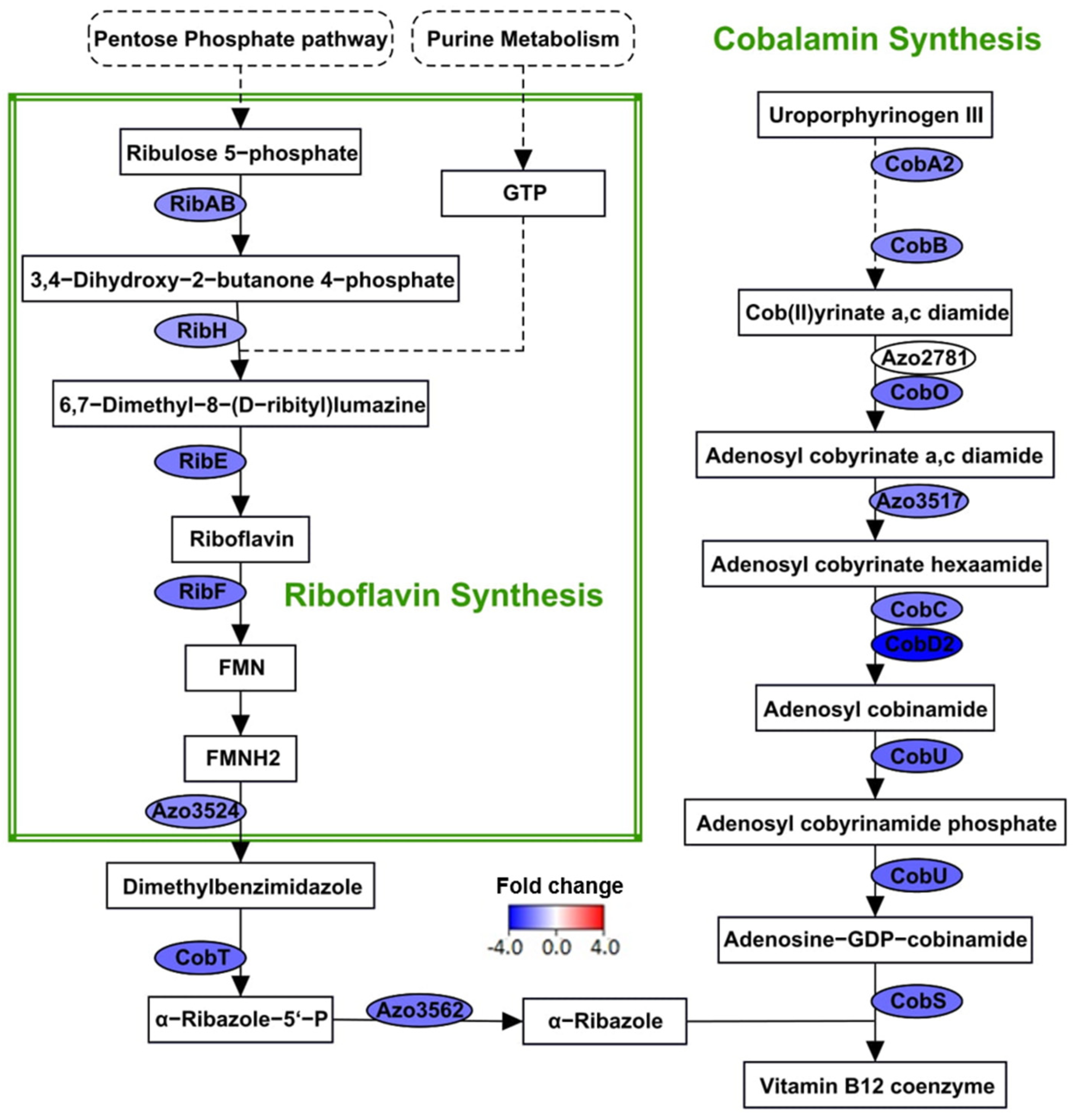
3.6. Suppression of Vitamin Biosynthesis during N2-Fixation
3.7. Down-Regulation of Ribosomal Genes Corresponding to Decreased Growth Rates
3.8. Differential Expression of Transcriptional Regulators
3.9. Preparing for Sustained N Starvation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| BNF | Biological nitrogen fixation |
| COGs | Cluster of orthologous groups |
| DEGs | Differentially expressed genes |
| EMP | Embden–Meyerhof–Parnas pathway |
| GTP | Guanosine triphosphate |
| nif-cluster | Nitrogen fixation gene cluster |
| ORFs | Open reading frames |
| padj | Benjamini–Hochberg adjusted p-value |
| PHB | poly-ß-hydroxybutyrate |
| PPP | Pentose Phosphate Pathway |
| PRPP | 5-phosphoribosyl 1-pyrophosphate |
| RPKM | Reads Per Kilobase per Million mapped reads |
| SM | Synthetic malate |
| T2SS | Type 2 protein secretion system |
| T6SS | Type 6 protein secretion system |
| TCA | Tricarboxylic acid cycle |
References
- Bolin, B.; Arrhenius, E. Nitrogen-an essential life factor and a growing environmental hazard. Report from Nobel Symposium No. 38. Ambio 1977, 6, 96–105. [Google Scholar]
- Bohlool, B.B.; Ladha, J.K.; Garrity, D.P.; George, T. Biological nitrogen-fixation for sustainable agriculture-a perspective. Plant Soil 1992, 141, 1–11. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T.; Gillis, M.; Hoste, B.; Vancanneyt, M.; Kersters, K.; De Ley, J. Azoarcus gen. nov., nitrogen-fixing proteobacteria associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth) and description of two species Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int. J. Syst. Bacteriol. 1993, 43, 574–584. [Google Scholar] [CrossRef]
- Reinhold, B.; Hurek, T.; Niemann, E.-G.; Fendrik, I. Close association of Azospirillum and diazotrophic rods with different root zones of Kallar grass. Appl. Environ. Microbiol. 1986, 52, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Hurek, T.; Handley, L.; Reinhold-Hurek, B.; Piché, Y. Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Mol. Plant-Microbe Interact. 2002, 15, 233–242. [Google Scholar] [CrossRef]
- Egener, T.; Hurek, T.; Reinhold-Hurek, B. Use of green fluorescent protein to detect expression of nif genes of Azoarcus sp. BH72, a grass-associated diazotroph, on rice roots. Mol. Plant-Microbe Interact. 1998, 11, 71–75. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Egener, T.; Hurek, T.; Reinhold-Hurek, B. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant-Microbe Interact. 1999, 12, 813–819. [Google Scholar] [CrossRef]
- Chen, M.H.; Sheu, S.Y.; James, E.K.; Young, C.C.; Chen, W.M. Azoarcus olearius sp. nov., a nitrogen-fixing bacterium isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 3755–3761. [Google Scholar] [CrossRef]
- Faoro, H.; Rene Menegazzo, R.; Battistoni, F.; Gyaneshwar, P.; do Amaral, F.P.; Taule, C.; Rausch, S.; Goncalves Galvao, P.; de Los Santos, C.; Mitra, S.; et al. The oil-contaminated soil diazotroph Azoarcus olearius DQS-4T is genetically and phenotypically similar to the model grass endophyte Azoarcus sp. BH72. Environ. Microbiol. Rep. 2017, 9, 223–238. [Google Scholar] [CrossRef]
- Sarkar, A.; Reinhold-Hurek, B. Transcriptional profiling of nitrogen fixation and the role of NifA in the diazotrophic endophyte Azoarcus sp. strain BH72. PLoS ONE 2014, 9, e86527. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant Biol. 2011, 14, 435–443. [Google Scholar] [CrossRef]
- Bladergroen, M.R.; Badelt, K.; Spaink, H.P. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 2003, 16, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Beust, A.; Sappa, P.K.; Völker, U.; Dinse, T.; Herglotz, J.; Reinhold-Hurek, B. Two functionally deviating Type 6 secretion systems occur in the nitrogen-fixing endophyte Azoarcus olearius BH72. Front. Microbiol. 2019, 10, 459. [Google Scholar] [CrossRef] [PubMed]
- Egener, T.; Martin, D.E.; Sarkar, A.; Reinhold-Hurek, B. Role of a ferrodoxine gene cotranscribed with the nifHDK operon in N2 fixation and nitrogenase “switch off” of Azoarcus sp. strain BH72. J. Bacteriol. 2001, 183, 3752–3760. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martin, D.; Hurek, T.; Reinhold-Hurek, B. Occurrence of three PII-like signal transmitter proteins in the diazotroph Azoarcus sp. BH72. Mol. Microbiol. 2000, 38, 276–288. [Google Scholar] [CrossRef][Green Version]
- Oetjen, J.; Reinhold-Hurek, B. Characterization of the DraT/DraG-system for posttranslational regulation of nitrogenase in the endophytic beta-proteobacterium Azoarcus sp. BH72. J. Bacteriol. 2009, 191, 3726–3735. [Google Scholar] [CrossRef][Green Version]
- Sarkar, A.; Köhler, J.; Hurek, T.; Reinhold-Hurek, B. A novel regulatory role of the Rnf complex of Azoarcus sp. strain BH72. Mol. Microbiol. 2012, 83, 408–422. [Google Scholar] [CrossRef]
- Yang, Z.M.; Han, Y.L.; Ma, Y.; Chen, Q.H.; Zhan, Y.H.; Lu, W.; Cai, L.; Hou, M.S.; Chen, S.F.; Yan, Y.L.; et al. Global investigation of an engineered nitrogen-fixing Escherichia coli strain reveals regulatory coupling between host and heterologous nitrogen-fixation genes. Sci. Rep.-UK 2018, 8, 10928. [Google Scholar] [CrossRef] [PubMed]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.G.; Liguori, M.J. Comparison of RNA-Seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front. Genet. 2018, 9, 636. [Google Scholar] [CrossRef]
- Dienstbier, A.; Amman, F.; Stipl, D.; Petrackova, D.; Vecerek, B. Comparative integrated omics analysis of the hfq regulon in Bordetella pertussis. Int. J. Mol. Sci. 2019, 20, 3073. [Google Scholar] [CrossRef] [PubMed]
- Silue, N.; Marcantonio, E.; Campbell-Valois, F.X. RNA-Seq analysis of the T3SA regulon in Shigella flexneri reveals two new chromosomal genes upregulated in the on-state. Methods 2020, 176, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, G.; Linde, S.C.; Coolon, J.D. Genome-wide effect of tetracycline, doxycycline and 4-epidoxycycline on gene expression in Saccharomyces cerevisiae. Yeast 2020, 37, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Fung-Leung, W.P.; Bittner, A.; Ngo, K.; Liu, X.J. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef]
- Hamilton, T.L.; Ludwig, M.; Dixon, R.; Boyd, E.S.; Dos Santos, P.C.; Setubal, J.C.; Bryant, D.A.; Dean, D.R.; Peters, J.W. Transcriptional profiling of nitrogen fixation in Azotobacter vinelandii. J. Bacteriol. 2011, 193, 4477–4486. [Google Scholar] [CrossRef]
- Shi, H.W.; Wang, L.Y.; Li, X.X.; Liu, X.M.; Hao, T.Y.; He, X.J.; Chen, S.F. Genome-wide transcriptome profiling of nitrogen fixation in Paenibacillus sp. WLY78. BMC Microbiol. 2016, 16, 25. [Google Scholar] [CrossRef]
- Reinhold, B.; Hurek, T.; Fendrik, I. Strain-specific chemotaxis of Azospirillum spp. J. Bacteriol. 1985, 162, 190–195. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Forstner, K.U.; Vogel, J.; Sharma, C.M. READemption-a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 2014, 30, 3421–3423. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Sarkar, A.; Marszalkowska, M.; Schäfer, M.; Pees, T.; Klingenberg, H.; Macht, F.; Reinhold-Hurek, B. Global expression analysis of the response to microaerobiosis reveals an important cue for endophytic establishment of Azoarcus sp. BH72. Env. Microbiol. 2017, 19, 198–217. [Google Scholar] [CrossRef]
- Luque-Almagro, V.M.; Manso, I.; Sullivan, M.J.; Rowley, G.; Ferguson, S.J.; Moreno-Vivian, C.; Richardson, D.J.; Gates, A.J.; Roldan, M.D. Transcriptional and translational adaptation to aerobic nitrate anabolism in the denitrifier Paracoccus denitrificans. Biochem. J. 2017, 474, 1769–1787. [Google Scholar] [CrossRef] [PubMed]
- Luque-Almagro, V.M.; Gates, A.J.; Moreno-Vivian, C.; Ferguson, S.J.; Richardson, D.J.; Roldan, M.D. Bacterial nitrate assimilation: Gene distribution and regulation. Biochem Soc. T 2011, 39, 1838–1843. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Vivián, C.; Flores, E. Nitrate assimilation in bacteria. In Biology of the Nitrogen Cycle; Bothe, H., Ferguson, S.J., Newton, W.E., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 263–282. [Google Scholar]
- Sarkar, A. Studies on the regulation of genes related to nitrogen fixation and N-assimilation in Azoarcus sp. strain BH72: The role of NtrBC. Ph. D. Thesis, University of Bremen, Bremen, Germany, 2003. [Google Scholar]
- Norman, J.S.; Friesen, M.L. Complex N acquisition by soil diazotrophs: How the ability to release exoenzymes affects N fixation by terrestrial free-living diazotrophs. ISME J. 2017, 11, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.E.; Reinhold-Hurek, B. Distinct roles of PII-like signal transmitter proteins and amtB in regulation of nif gene expression, nitrogenase activity, and posttranslational modification of NifH in Azoarcus sp. strain BH72. J. Bacteriol. 2002, 184, 2251–2259. [Google Scholar] [CrossRef] [PubMed]
- Mus, F.; Tseng, A.; Dixon, R.; Peters, J.W. Diazotrophic growth allows Azotobacter vinelandii to overcome the deleterious effects of a glnE deletion. Appl. Environ. Microbiol. 2017, 83, e00808-17. [Google Scholar] [CrossRef]
- Krause, A.; Ramakumar, A.; Bartels, D.; Battistoni, F.; Bekel, T.; Boch, J.; Böhm, M.; Friedrich, F.; Hurek, T.; Krause, L.; et al. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 2006, 24, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Yu, R.; Schellhorn, H. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 2011, 79, 375–386. [Google Scholar] [CrossRef]
- Zhao, K.; Liu, M.; Burgess, R.R. Promoter and regulon analysis of nitrogen assimilation factor, σ54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 2010, 38, 1273–1283. [Google Scholar] [CrossRef]
- Buschart, A.; Sachs, S.; Chen, X.; Herglotz, J.; Krause, A.; Reinhold-Hurek, B. Flagella mediate endophytic competence rather than act as MAMPS in rice-Azoarcus sp. strain BH72 interactions. Mol. Plant-Microbe Interact. 2012, 25, 191–199. [Google Scholar] [CrossRef]
- Brown, D.R.; Barton, G.; Pan, Z.; Buck, M.; Wigneshweraraj, S. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat. Commun. 2014, 5, 4115. [Google Scholar] [CrossRef]
- Hauberg-Lotte, L.; Klingenberg, H.; Scharf, C.; Böhm, M.; Plessl, J.; Friedrich, F.; Völker, U.; Becker, A.; Reinhold-Hurek, B. Environmental factors affecting the expression of pilAB as well as the proteome and transcriptome of the grass endophyte Azoarcus sp. strain BH72. PLoS ONE 2012, 7, e30421. [Google Scholar] [CrossRef][Green Version]
- Duncan, M.J.; Fraenkel, D.G. Alpha-ketoglutarate dehydrogenase mutant of Rhizobium meliloti. J. Bacteriol. 1979, 137, 415–419. [Google Scholar] [CrossRef]
- Dymov, S.I.; Meek, D.J.J.; Steven, B.; Driscoll, B.T. Insertion of transpoon Tn5tac1 in the Sinorhizobium meliloti malate dehydrogenase (mdh) gene results in conditional polar effects on downstream TCA cycle genes. Mol. Plant-Microbe Interact. 2004, 17, 1318–1327. [Google Scholar] [CrossRef] [PubMed]
- McDermott, T.R.; Kahn, M.L. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 1992, 174, 4790–4797. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, A.; Contador, C.A.; Fan, K.; Lam, H.M. Interaction and regulation of carbon, nitrogen, and phosphorus metabolisms in root nodules of legumes. Front. Plant Sci. 2018, 9, 1860. [Google Scholar] [CrossRef] [PubMed]
- Vemuri, G.N.; Altman, E.; Sangurdekar, D.P.; Khodursky, A.B.; Eiteman, M.A. Overflow metabolism in Escherichia coli during steady-state growth: Transcriptional regulation and effect of the redox ratio. Appl. Environ. Microbiol. 2006, 72, 3653–3661. [Google Scholar] [CrossRef]
- Han, K.; Lim, H.C.; Hong, J. Acetic-acid formation in Escherichia-coli fermentation. Biotechnol. Bioeng. 1992, 39, 663–671. [Google Scholar] [CrossRef]
- Majewski, R.A.; Domach, M.M. Simple constrained-optimization view of acetate overflow in E. coli. Biotechnol. Bioeng. 1990, 35, 732–738. [Google Scholar] [CrossRef]
- Mandon, K.; Michel-Reydellet, N.; Encarnacion, S.; Kaminski, P.A.; Leija, A.; Cevallos, M.A.; Elmerich, C.; Mora, J. Poly-ß-hydroxybutyrate turnover in Azorhizobium caulinodans is required for growth and affects nifA expression. J. Bacteriol. 1998, 180, 5070–5076. [Google Scholar] [CrossRef]
- Senior, P.J.; Dawes, E.A. The regulation of poly-ß-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem. J. 1973, 134, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.J.; Masakapalli, S.K.; Karunakaran, R.; Webb, I.U.C.; Green, R.; Watmough, N.J.; Kruger, N.J.; Ratcliffe, R.G.; Poole, P.S. Lipogenesis and redox balance in nitrogen-fixing pea bacteroids. J. Bacteriol. 2016, 198, 2864–2875. [Google Scholar] [CrossRef] [PubMed]
- Hurek, T.; Reinhold, B.; Fendrik, I.; Niemann, E.G. Root-zone-specific oxygen tolerance of Azospirillum spp. and diazotrophic rods closely associated with Kallar grass. Appl. Environ. Microbiol. 1987, 53, 163–169. [Google Scholar] [CrossRef]
- Yoshida, K.; Takemoto, Y.; Sotsuka, T.; Tanaka, K.; Takenaka, S. PhaP phasins play a principal role in poly-ß-hydroxybutyrate accumulation in free-living Bradyrhizobium japonicum. BMC Microbiol. 2013, 13, 290. [Google Scholar] [CrossRef]
- Taga, M.E.; Walker, G.C. Sinorhizobium meliloti requires a cobalamin-dependent ribonucleotide reductase for symbiosis with its plant host. Mol. Plant-Microbe Interact. 2010, 23, 1643–1654. [Google Scholar] [CrossRef]
- Evans, H.J.; Kliewer, M. Vitamin B12 compounds in relation to the requirements of cobalt for higher plants and nitrogen-fixing organisms. Ann. N. Y. Acad. Sci. 1964, 112, 735–755. [Google Scholar] [CrossRef]
- Okuda, A.; Yamaguchi, M. Nitrogen fixing microorganisms in paddy soils. VI. Vitamin B12 activity in nitrogen fixing blue green algae. Soil Sci. Plant Nutr. 1960, 6, 76–85. [Google Scholar] [CrossRef]
- Phillips, D.A.; Joseph, C.M.; Yang, G.P.; Martinez-Romero, E.; Sanborn, J.R.; Volpin, H. Identification of lumichrome as a Sinorhizobium enhancer of alfalfa root respiration and shoot growth. Proc. Natl. Acad. Sci. USA 1999, 96, 12275–12280. [Google Scholar] [CrossRef] [PubMed]
- Streit, W.R.; Joseph, C.M.; Phillips, D.A. Biotin and other water-soluble vitamins are key growth factors for alfalfa root colonization by Rhizobium meliloti 1021. Mol. Plant-Microbe Interact. 1996, 9, 330–338. [Google Scholar] [CrossRef]
- Garcia-Angulo, V.A. Overlapping riboflavin supply pathways in bacteria. Crit. Rev. Microbiol. 2017, 43, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Wang, Y.; Wang, Z.; Wang, G.; Liu, D.; Fu, J.; Chen, T.; Zhao, X. Deregulation of purine pathway in Bacillus subtilis and its use in riboflavin biosynthesis. Microb. Cell Fact. 2014, 13, 101. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Tang, W.; Zhang, D. Manipulation of purine metabolic networks for riboflavin production in Bacillus subtilis. Acs Omega 2020, 5, 29140–29146. [Google Scholar] [CrossRef]
- Van Soom, C.; de Wilde, P.; Vanderleyden, J. HoxA is a transcriptional regulator for expression of the hup structural genes in free-living Bradyrhizobium japonicum. Mol. Microbiol. 1997, 23, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Durmowicz, M.C.; Maier, R.J. Roles of HoxX and HoxA in biosynthesis of hydrogenase in Bradyrhizobium japonicum. J. Bacteriol. 1997, 179, 3676–3682. [Google Scholar] [CrossRef]
- Rey, F.E.; Oda, Y.; Harwood, C.S. Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J. Bacteriol. 2006, 188, 6143–6152. [Google Scholar] [CrossRef] [PubMed]
- Brewin, N.J. Hydrogenase and energy efficiency in nitrogen fixing symbionts. In Genes involved in Microbe-Plant Interactions; Verma, D.P.S., Hohn, T., Eds.; Springer: Vienna, Austria, 1984. [Google Scholar]
- Mouncey, N.J.; Mitchenall, L.A.; Pau, R.N. The modE gene product mediates molybdenum-dependent expression of genes for the high-affinity molybdate transporter and modG in Azotobacter vinelandii. Microbiology 1996, 142, 1997–2004. [Google Scholar] [CrossRef]
- McNicholas, P.M.; Chiang, R.C.; Gunsalus, R.P. The Escherichia coli modE gene: Effect of modE mutations on molybdate dependent modA expression. FEMS Microbiol. Lett. 1996, 145, 117–123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grunden, A.M.; Ray, R.M.; Rosentel, J.K.; Healy, F.G.; Shanmugam, K.T. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J. Bacteriol. 1996, 178, 735–744. [Google Scholar] [CrossRef]
- Kutsche, M.; Leimkuhler, S.; Angermuller, S.; Klipp, W. Promoters controlling expression of the alternative nitrogenase and the molybdenum uptake system in Rhodobacter capsulatus are activated by NtrC, independent of σ54, and repressed by molybdenum. J. Bacteriol. 1996, 178, 2010–2017. [Google Scholar] [CrossRef] [PubMed]
- Delgado, M.J.; Tresierra-Ayala, A.; Talbi, C.; Bedmar, E.J. Functional characterization of the Bradyrhizobium japonicum modA and modB genes involved in molybdenum transport. Microbiology 2006, 152, 199–207. [Google Scholar] [CrossRef]
- Figueira, R.; Brown, D.R.; Ferreira, D.; Eldridge, M.J.G.; Burchell, L.; Pan, Z.; Helaine, S.; Wigneshweraraj, S. Adaptation to sustained nitrogen starvation by Escherichia coli requires the eukaryote-like serine/threonine kinase YeaG. Sci. Rep.-UK 2015, 5, 17524. [Google Scholar] [CrossRef] [PubMed]
- Veening, J.W.; Smits, W.K.; Kuipers, O.P. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 2008, 62, 193–210. [Google Scholar] [CrossRef]
- Akiva, E.; Copp, J.N.; Tokuriki, N.; Babbitt, P.C. Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily. Proc. Natl. Acad. Sci. USA 2017, 114, E9549–E9558. [Google Scholar] [CrossRef]
- Georg, J.; Hess, W.R. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Thomason, M.K.; Storz, G. Bacterial antisense RNAs: How many are there, and what are they doing? Annu. Rev. Genet. 2010, 44, 167–188. [Google Scholar] [CrossRef] [PubMed]
- Saberi, F.; Kamali, M.; Najafi, A.; Yazdanparast, A.; Moghaddam, M.M. Natural antisense RNAs as mRNA regulatory elements in bacteria: A review on function and applications. Cell Mol. Biol. Lett. 2016, 21, 6. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solaiyappan Mani, S.; Reinhold-Hurek, B. RNA-Seq Provides New Insights into the Gene Expression Changes in Azoarcus olearius BH72 under Nitrogen-Deficient and Replete Conditions beyond the Nitrogen Fixation Process. Microorganisms 2021, 9, 1888. https://doi.org/10.3390/microorganisms9091888
Solaiyappan Mani S, Reinhold-Hurek B. RNA-Seq Provides New Insights into the Gene Expression Changes in Azoarcus olearius BH72 under Nitrogen-Deficient and Replete Conditions beyond the Nitrogen Fixation Process. Microorganisms. 2021; 9(9):1888. https://doi.org/10.3390/microorganisms9091888
Chicago/Turabian StyleSolaiyappan Mani, Shanmugam, and Barbara Reinhold-Hurek. 2021. "RNA-Seq Provides New Insights into the Gene Expression Changes in Azoarcus olearius BH72 under Nitrogen-Deficient and Replete Conditions beyond the Nitrogen Fixation Process" Microorganisms 9, no. 9: 1888. https://doi.org/10.3390/microorganisms9091888
APA StyleSolaiyappan Mani, S., & Reinhold-Hurek, B. (2021). RNA-Seq Provides New Insights into the Gene Expression Changes in Azoarcus olearius BH72 under Nitrogen-Deficient and Replete Conditions beyond the Nitrogen Fixation Process. Microorganisms, 9(9), 1888. https://doi.org/10.3390/microorganisms9091888





