Pochonia chlamydosporia Isolate PC-170-Induced Expression of Marker Genes for Defense Pathways in Tomatoes Challenged by Different Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Material and Growth Conditions
2.2. Colonization of PC-170 on the Roots of Tomatoes
2.2.1. Inoculation Methods
2.2.2. The Detection of Colonization
2.2.3. PC-170 and Meloidogyne incognita Interact in Tomatoes
2.2.4. Direct Interaction
2.2.5. Split-Root Experiments
2.2.6. Mutant Tomato Lines
2.2.7. PC-170 and Pathogens Interact in Tomatoes
2.2.8. Quantitative RT-PCR Analysis
2.2.9. Statistical Analysis
3. Result
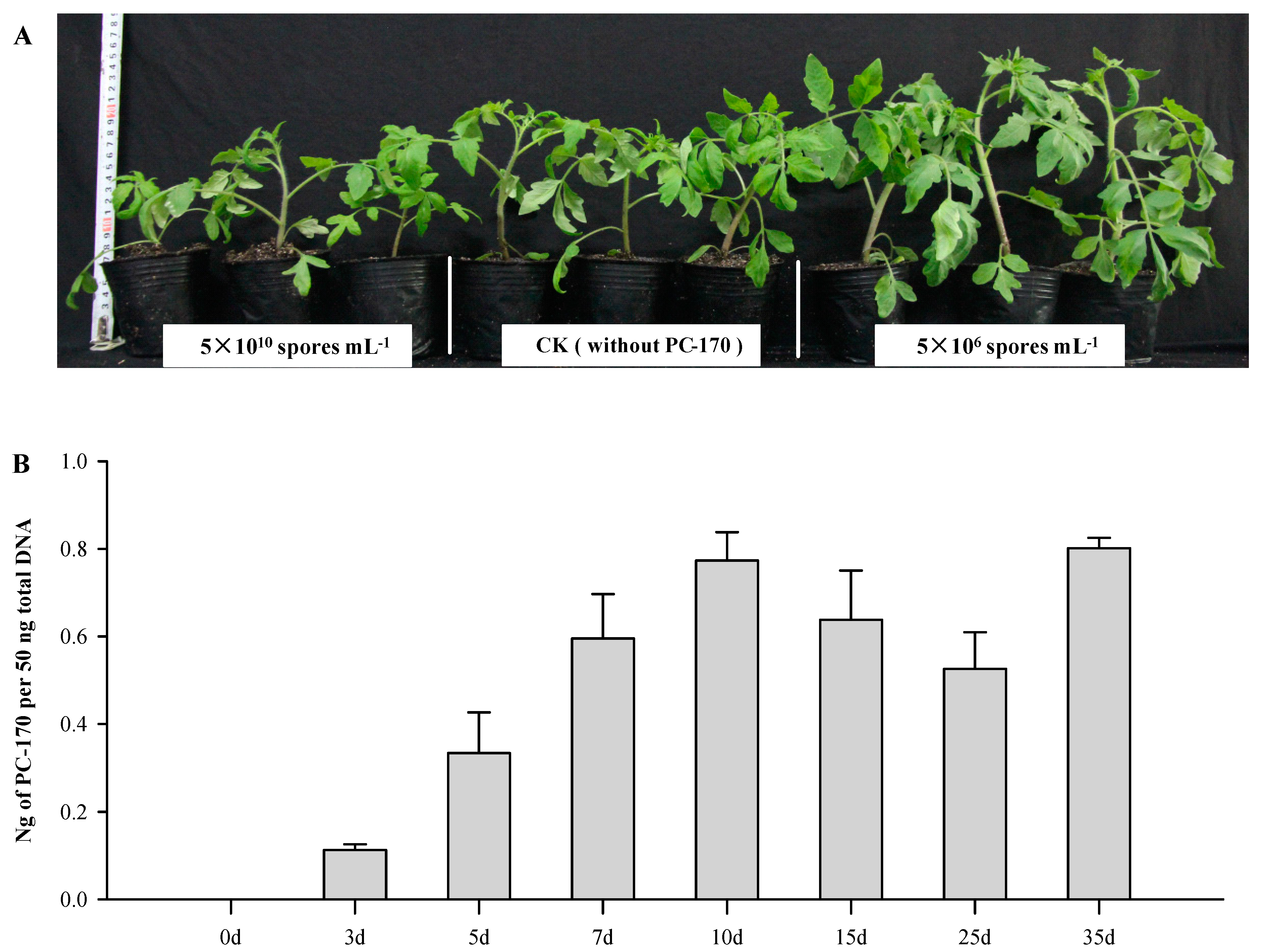
3.1. Pochonia chlamydosporia PC-170 Colonizes Tomato Roots and Promotes Growth
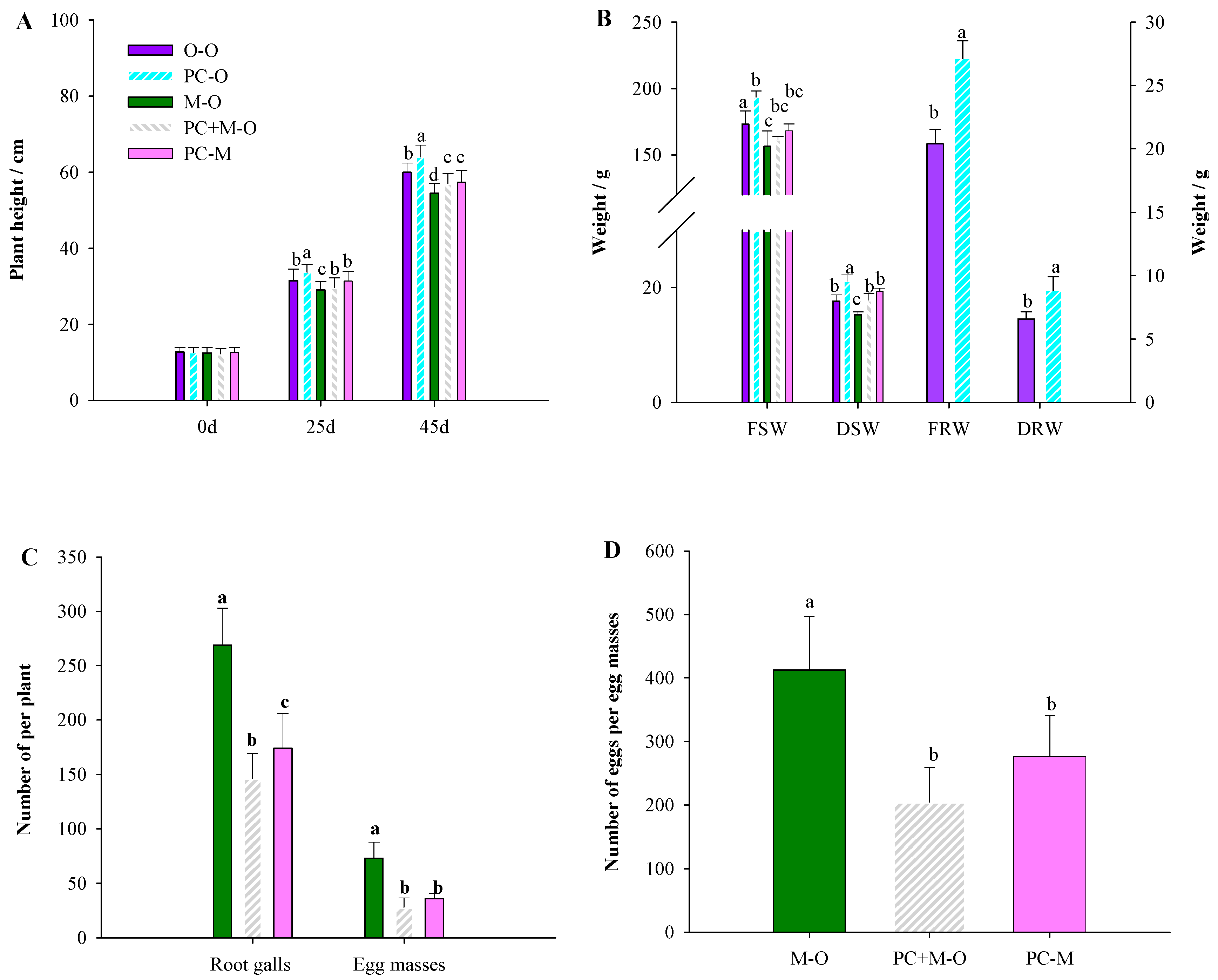
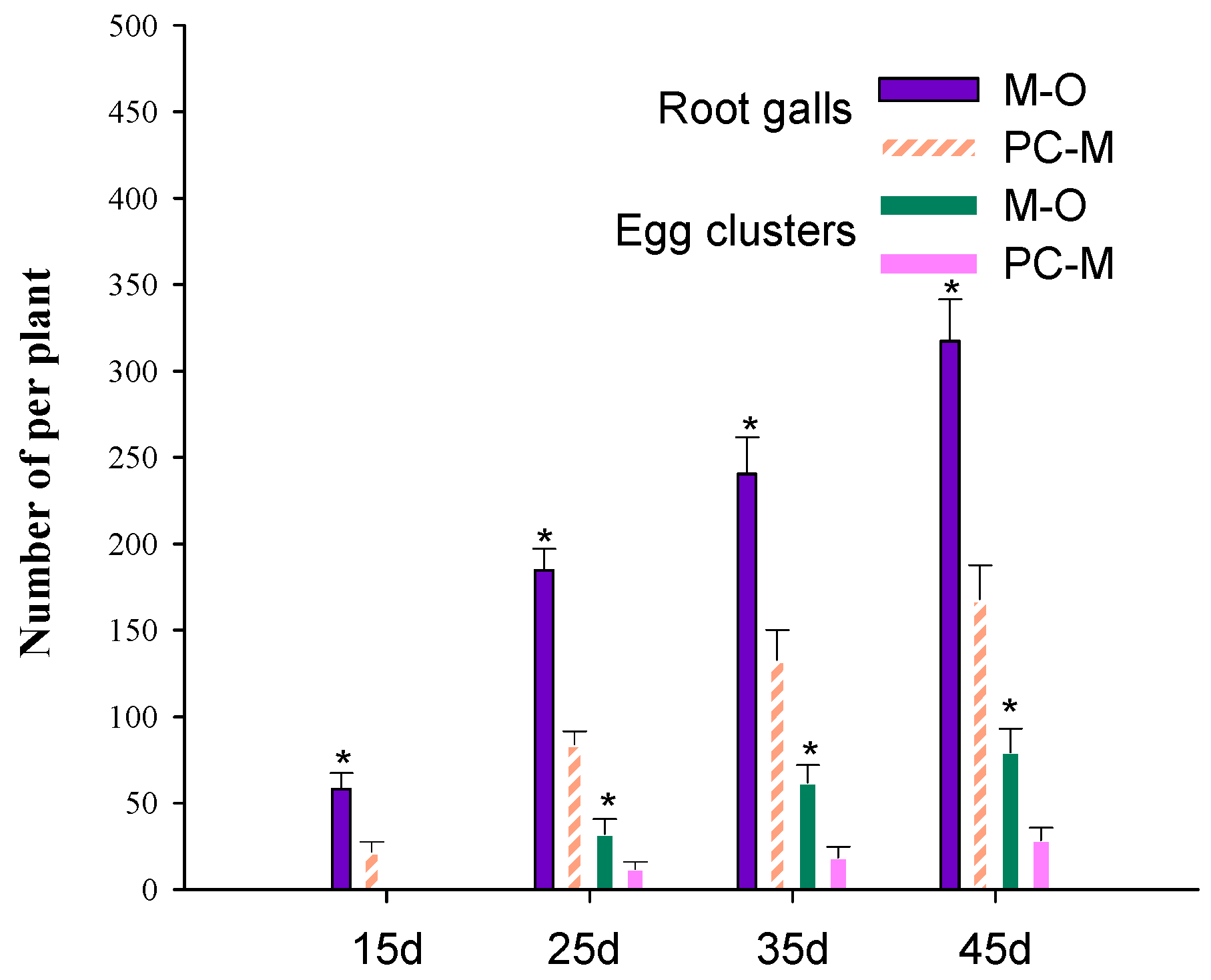
3.2. PC-170 Improves Resistance to Meloidogyne incognita
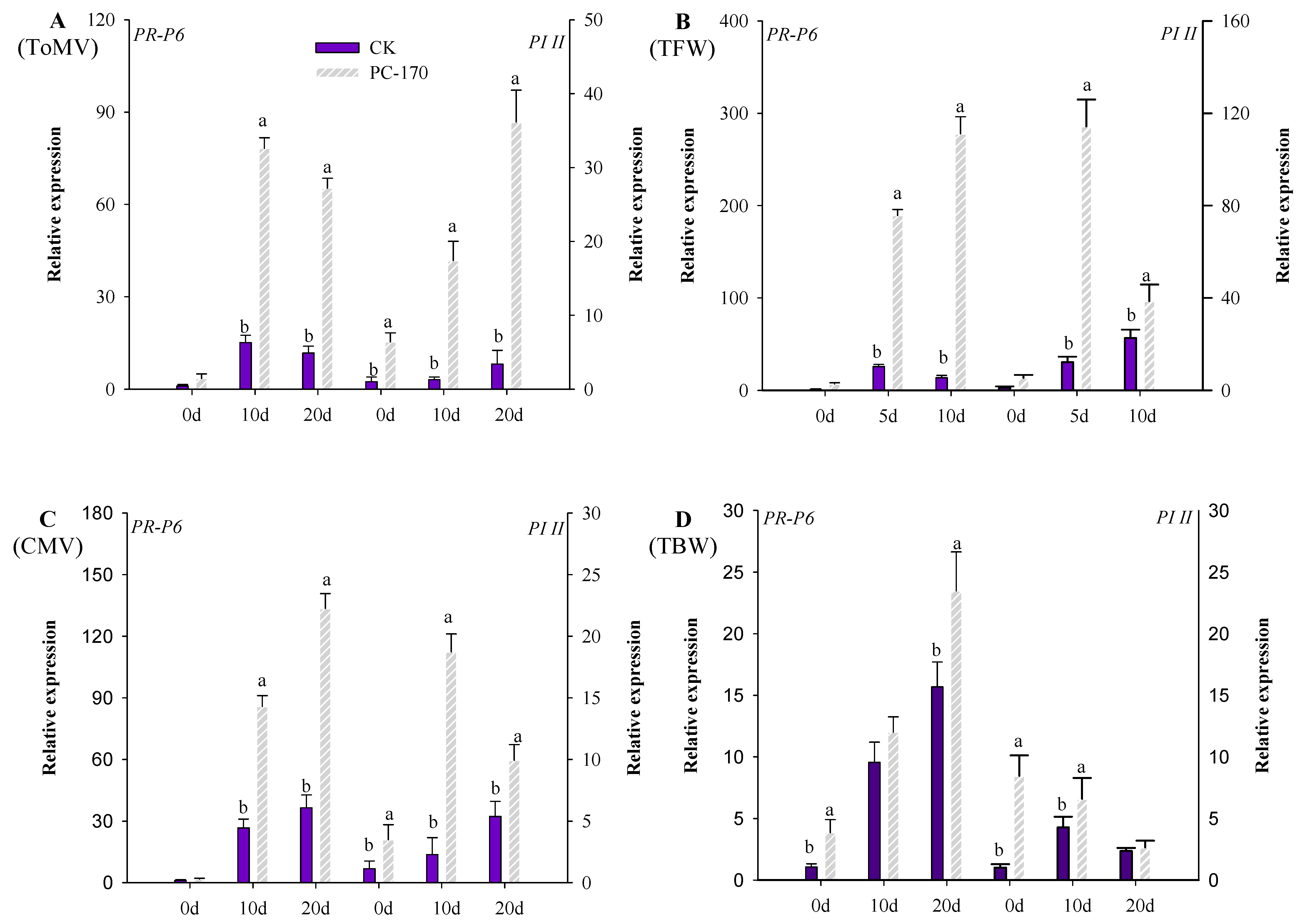
3.3. PC-170 Improves Resistance to Different Pathogens in Tomatoes
3.4. PC-170 Induces Systemic Resistance in Tomatoes
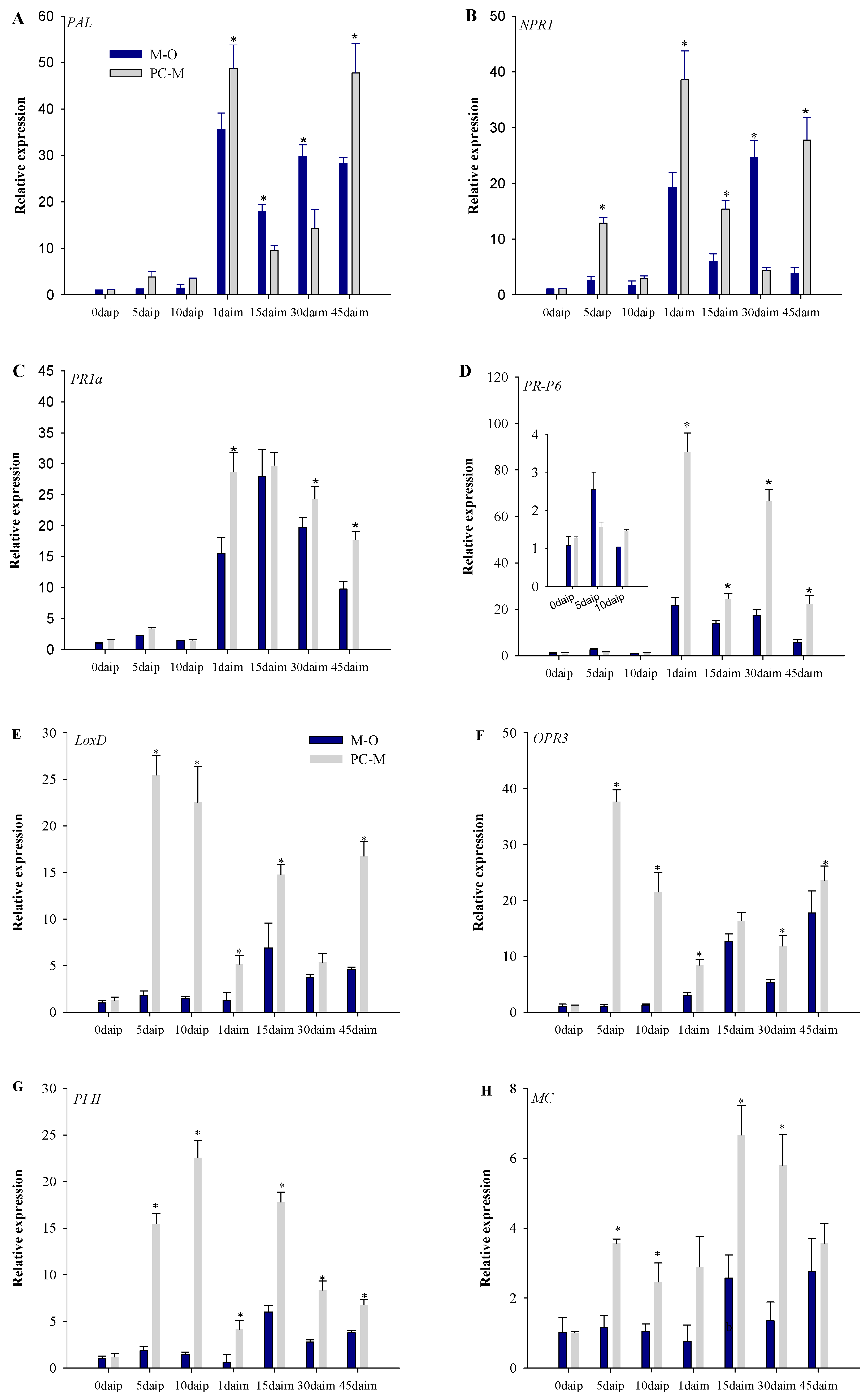
3.5. Expression of Defense Pathway Marker Genes in Different Interactions
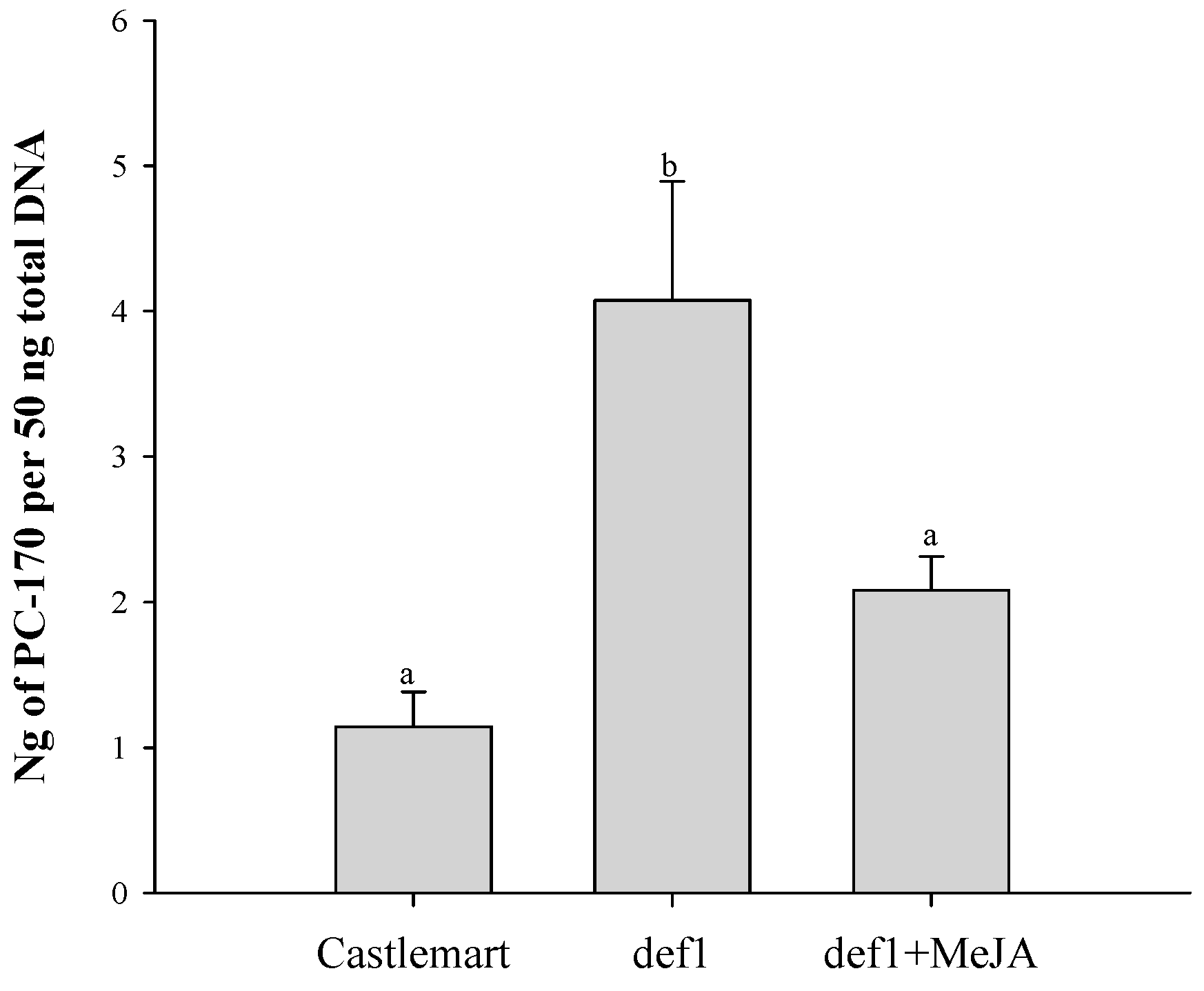
3.6. PC-170 and Meloidogyne incognita Interact in Mutant Tomato Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.E.; Mesarich, C.H.; Thomma, B.P. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 2015, 53, 541–563. [Google Scholar] [CrossRef]
- Van der Burgh, A.M.; Joosten, M. Plant Immunity: Thinking Outside and Inside the Box. Trends Plant Sci. 2019, 24, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Biere, A.; Goverse, A. Plant-Mediated Systemic Interactions Between Pathogens, Parasitic Nematodes, and Herbivores Above- and Belowground. Annu. Rev. Phytopathol. 2016, 54, 499–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezawa, T.; Saito, K. How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol. 2018, 220, 1116–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almario, J.; Jeena, G.; Wunder, J.; Langen, G.; Zuccaro, A.; Coupland, G.; Bucher, M. Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc. Natl. Acad. Sci. USA 2017, 114, E9403–E9412. [Google Scholar] [CrossRef] [Green Version]
- Larriba, E.; Jaime, M.D.; Nislow, C.; Martin-Nieto, J.; Lopez-Llorca, L.V. Endophytic colonization of barley (Hordeum vulgare) roots by the nematophagous fungus Pochonia chlamydosporia reveals plant growth promotion and a general defense and stress transcriptomic response. J. Plant Res. 2015, 128, 665–678. [Google Scholar] [CrossRef]
- Gams, W.; Zare, R. A revision of Verticillium sect. Prostrata. III. Generic classification. Nova Hedwig. 2001, 72, 329–337. [Google Scholar] [CrossRef]
- Escudero, N.; Lopez-Llorca, L.V. Effects on plant growth and root-knot nematode infection of an endophytic GFP transformant of the nematophagous fungus Pochonia chlamydosporia. Symbiosis 2012, 57, 33–42. [Google Scholar] [CrossRef]
- Tobin, J.D.; Haydock, P.P.J.; Hare, M.C.; Woods, S.R.; Crump, D.H. Effect of the fungus Pochonia chlamydosporia and fosthiazate on the multiplication rate of potato cyst nematodes (Globodera pallida and G. rostochiensis) in potato crops grown under UK field conditions. Biol. Control 2008, 46, 194–201. [Google Scholar] [CrossRef]
- Silva, S.D.; Carneiro, R.; Faria, M.; Souza, D.A.; Monnerat, R.G.; Lopes, R.B. Evaluation of Pochonia chlamydosporia and Purpureocillium lilacinum for Suppression of Meloidogyne enterolobii on Tomato and Banana. J. Nematol. 2017, 49, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Riggs, R.D.; Crippen, D. Isolation, Selection, and Efficacy of Pochonia chlamydosporia for Control of Rotylenchulus reniformis on Cotton. Phytopathology 2005, 95, 890–893. [Google Scholar] [CrossRef] [Green Version]
- Dallemole-Giaretta, R.; Freitas, L.G.; Lopes, E.A.; Pereira, O.L.; Zooca, R.J.F.; Ferraz, S. Screening of Pochonia chlamydosporia Brazilian isolates as biocontrol agents of Meloidogyne javanica. Crop Prot. 2012, 42, 102–107. [Google Scholar] [CrossRef]
- Viggiano, J.R.; de Freitas, L.G.; Lopes, E.A. Use of Pochonia chlamydosporia to control Meloidogyne javanica in cucumber. Biol. Control 2014, 69, 72–77. [Google Scholar] [CrossRef]
- Maciá-Vicente, J.G.; Rosso, L.C.; Ciancio, A.; Jansson, H.B.; Lopez-Llorca, L.V. Colonisation of barley roots by endophytic Fusarium equiseti and Pochonia chlamydosporia: Effects on plant growth and disease. Ann. Appl. Biol. 2009, 155, 391–401. [Google Scholar] [CrossRef]
- Bordallo, J.J.; Lopez-Llorca, L.V.; Jansson, H.-B.; Salinas, J.; Persmark, L.; Asensio, L. Colonization of plant roots by egg-parasitic and nematode-trapping fungi. New Phytol. 2001, 154, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Manzanilla-López, R.H.; Esteves, I.; Powers, S.J.; Kerry, B.R. Effects of crop plants on abundance of Pochonia chlamydosporia and other fungal parasites of root-knot and potato cyst nematodes. Ann. Appl. Biol. 2011, 159, 118–129. [Google Scholar] [CrossRef]
- Zavala-Gonzalez, E.A.; Rodriguez-Cazorla, E.; Escudero, N.; Aranda-Martinez, A.; Martinez-Laborda, A.; Ramirez-Lepe, M.; Vera, A.; Lopez-Llorca, L.V. Arabidopsis thaliana root colonization by the nematophagous fungus Pochonia chlamydosporia is modulated by jasmonate signaling and leads to accelerated flowering and improved yield. New Phytol. 2017, 213, 351–364. [Google Scholar] [CrossRef] [Green Version]
- Zavala-Gonzalez, E.A.; Escudero, N.; Lopez-Moya, F.; Aranda-Martinez, A.; Exposito, A.; Ricaño-Rodríguez, J.; Naranjo-Ortiz, M.A.; Ramírez-Lepe, M.; Lopez-Llorca, L.V. Some isolates of the nematophagous fungus Pochonia chlamydosporia promote root growth and reduce flowering time of tomato. Ann. Appl. Biol. 2015, 166, 472–483. [Google Scholar] [CrossRef]
- Monfort, E.; Lopez-Llorca, L.V.; Jansson, H.B.; Salinas, J.; Park, J.O.; Sivasithamparam, K. Colonisation of seminal roots of wheat and barley by egg-parasitic nematophagous fungi and their effects on Gaeumannomyces graminis var. tritici and development of root-rot. Soil Biol. Biochem. 2005, 37, 1229–1235. [Google Scholar] [CrossRef]
- Rosso, L.C.; Colagiero, M.; Salatino, N.; Ciancio, A. Observations on the effect of trophic conditions on Pochonia chlamydosporiagene expression. Ann. Appl. Biol. 2014, 164, 232–243. [Google Scholar] [CrossRef]
- Ghahremani, Z.; Escudero, N.; Saus, E.; Gabaldon, T.; Javier Sorribas, F. Pochonia chlamydosporia Induces Plant-Dependent Systemic Resistance to Meloidogyne incognita. Front. Plant Sci. 2019, 10, 945. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.; Qin, F.; Shen, B.; Shi, Q.; Liu, C.; Zhang, X.; Jiao, Y.; Lu, J.; Gao, Y.; Suarez-Fernandez, M.; et al. Genome and secretome analysis of Pochonia chlamydosporia provide new insight into egg-parasitic mechanisms. Sci. Rep. 2018, 8, 1123–1139. [Google Scholar] [CrossRef]
- Lopez-Llorca, L.V.; Bordallo, J.J.; Salinas, J.; Monfort, E.; Lopez-Serna, M.L. Use of light and scanning electron microscopy to examine colonisation of barley rhizosphere by the nematophagous fungus Verticillium chlamydosporium. Int. Res. Rev. J. Microsc. 2002, 33, 61–67. [Google Scholar] [CrossRef]
- Blazquez, M.A.; Nelson, D.C.; Weijers, D. Evolution of Plant Hormone Response Pathways. Annu. Rev. Plant Biol. 2020, 71, 327–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wondafrash, M.; Van Dam, N.M.; Tytgat, T.O. Plant systemic induced responses mediate interactions between root parasitic nematodes and aboveground herbivorous insects. Front. Plant Sci. 2013, 4, 87. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favery, B.; Quentin, M.; Jaubert-Possamai, S.; Abad, P. Gall-forming root-knot nematodes hijack key plant cellular functions to induce multinucleate and hypertrophied feeding cells. J. Insect Physiol. 2016, 84, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Gheysen, G.; Mitchum, M.G. Phytoparasitic Nematode Control of Plant Hormone Pathways. Plant Physiol. 2019, 179, 1212–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kammerhofer, N.; Radakovic, Z.; Regis, J.M.; Dobrev, P.; Vankova, R.; Grundler, F.M.; Siddique, S.; Hofmann, J.; Wieczorek, K. Role of stress-related hormones in plant defence during early infection of the cyst nematode Heterodera schachtii in Arabidopsis. New Phytol. 2015, 207, 778–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshioka, K.; Kachroo, P.; Tsui, F.; Sharma, S.B.; Shah, J.; Klessig, D.F. Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 2001, 226, 447–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uknes, S.; Dincher, S.; Friedrich, L.; Negrotto, D.; Williams, S.; Thompson-Taylor, H.; Potter, S.; Ward, E.; Ryals, J. Regulation of Pathogenesis-Related Protein-1a Gene Expression in Tobacco. Plant Cell 1993, 5, 159–169. [Google Scholar]
- Pallas, J.A.; Paiva, N.L.; Lamb, C.; Dixon, R. Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J. 1996, 10, 281–293. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Metraux, J.P.; Brown, R.; Kazan, K.; et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef] [Green Version]
- Wasternack, C.; Stenzel, I.; Hause, B.; Hause, G.; Kutter, C.; Maucher, H.; Neumerkel, J.; Feussner, I.; Miersch, O. The wound response in tomato--role of jasmonic acid. J Plant Physiol. 2006, 163, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, H.A.; Araujo Filho, J.V.; Freitas, L.G.; Castillo, P.; Rubio, M.B.; Hermosa, R.; Monte, E. Tomato progeny inherit resistance to the nematode Meloidogyne javanica linked to plant growth induced by the biocontrol fungus Trichoderma atroviride. Sci. Rep. 2017, 7, 40216–40229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Medina, A.; Fernandez, I.; Lok, G.B.; Pozo, M.J.; Pieterse, C.M.; Van Wees, S.C. Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 2017, 213, 1363–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolba, S.R.T.; Rosso, L.C.; Pentimone, I.; Colagiero, M.; Moustafa, M.M.A.; Elshawaf, I.I.S.; Bubici, G.; Prigigallo, M.I.; Ciancio, A. Root Endophytism by Pochonia chlamydosporia Affects Defense-Gene Expression in Leaves of Monocot and Dicot Hosts under Multiple Biotic Interactions. Plants 2021, 10, 718. [Google Scholar] [CrossRef]
- Khatabi, B.; Molitor, A.; Lindermayr, C.; Pfiffi, S.; Durner, J.; von Wettstein, D.; Kogel, K.H.; Schafer, P. Ethylene supports colonization of plant roots by the mutualistic fungus Piriformospora indica. PLoS ONE 2012, 7, e35502. [Google Scholar] [CrossRef]
- Pentimone, I.; Lebron, R.; Hackenberg, M.; Rosso, L.C.; Colagiero, M.; Nigro, F.; Ciancio, A. Identification of tomato miRNAs responsive to root colonization by endophytic Pochonia chlamydosporia. Appl. Microbiol. Biotechnol. 2018, 102, 907–919. [Google Scholar] [CrossRef]
- Lin, R.; Liu, C.; Shen, B.; Bai, M.; Ling, J.; Chen, G.; Mao, Z.; Cheng, X.; Xie, B. Analysis of the complete mitochondrial genome of Pochonia chlamydosporia suggests a close relationship to the invertebrate-pathogenic fungi in Hypocreales. BMC Microbiol. 2015, 15, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B. The Functional Research of Extracellular Proteases Related to Nematode Egg Parasitism in Pochonia chlamydosporia. Ph.D. Thesis, Hunan Agricultural University, Changsha, China, 1 June 2015. [Google Scholar]
- Howe, G.A.; Lightner, J.; Browse, J.; Ryan, C.A. An Octadecanoid Pathway Mutant (JL5) of Tomato 1s Compromised in Signaling for Defense against lnsect Attack. Plant Cell 1996, 8, 2067–2077. [Google Scholar] [PubMed] [Green Version]
- Brading, P.; Hammond-Kosack, K.; Parr, A.; Jones, J. Salicylic acid is not required for Cf-2- and Cf-9-dependent resistance of tomato to Cladosporium fulvum. Plant J. 2000, 23, 305–318. [Google Scholar] [CrossRef] [Green Version]
- Murphy, J.F.; Reddy, M.S.; Ryu, C.M.; Kloepper, J.W.; Li, R. Rhizobacteria-Mediated Growth Promotion of Tomato Leads to Protection Against Cucumber mosaic virus. Phytopathology 2003, 93, 1301–1307. [Google Scholar] [CrossRef] [Green Version]
- Song, W.T.; Zhou, L.G.; Yang, C.Z.; Cao, X.D.; Zhang, L.Q.; Liu, X.L. Tomato Fusarium wilt and its chemical control strategies in a hydroponic system. Crop. Prot. 2004, 23, 243–247. [Google Scholar] [CrossRef]
- Tans-Kersten, J.; Brown, D.; Allen, C. Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant Microbe Interact. 2004, 17, 686–695. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Raez, J.A.; Verhage, A.; Fernandez, I.; Garcia, J.M.; Azcon-Aguilar, C.; Flors, V.; Pozo, M.J. Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. J. Exp. Bot. 2010, 61, 2589–2601. [Google Scholar] [CrossRef] [Green Version]
- El Oirdi, M.; El Rahman, T.A.; Rigano, L.; El Hadrami, A.; Rodriguez, M.C.; Daayf, F.; Vojnov, A.; Bouarab, K. Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 2011, 23, 2405–2421. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Meng, G.; Wang, M.; Qian, Z.; Zhang, Y.; Yang, W. Tomato SlPUB24 enhances resistance to Xanthomonas euvesicatoria pv. perforans race T3. Hortic. Res. 2021, 8, 30. [Google Scholar] [CrossRef]
- Omomowo, O.I.; Babalola, O.O. Bacterial and Fungal Endophytes: Tiny Giants with Immense Beneficial Potential for Plant Growth and Sustainable Agricultural Productivity. Microorganisms 2019, 7, 481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interact. 2011, 25, 139–150. [Google Scholar] [CrossRef] [Green Version]
- Esteves, I.; Peteira, B.; Atkins, S.D.; Magan, N.; Kerry, B. Production of extracellular enzymes by different isolates of Pochonia chlamydosporia. Mycol. Res. 2009, 113, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Morton, C.O.; Mauchline, T.H.; Kerry, B.R.; Hirsch, P.R. PCR-based DNA fingerprinting indicates host-related genetic variation in the nematophagous fungus Pochonia chlamydosporia. Mycol. Res. 2003, 107, 198–205. [Google Scholar] [CrossRef]
- Lahrmann, U.; Strehmel, N.; Langen, G.; Frerigmann, H.; Leson, L.; Ding, Y.; Scheel, D.; Herklotz, S.; Hilbert, M.; Zuccaro, A. Mutualistic root endophytism is not associated with the reduction of saprotrophic traits and requires a noncompromised plant innate immunity. New Phytol. 2015, 207, 841–857. [Google Scholar] [CrossRef]
- Lagunas, B.; Schafer, P.; Gifford, M.L. Housing helpful invaders: The evolutionary and molecular architecture underlying plant root-mutualist microbe interactions. Exp. Bot. 2015, 66, 2177–2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hause, B.; Schaarschmidt, S. The role of jasmonates in mutualistic symbioses between plants and soil-born microorganisms. Phytochemistry 2009, 70, 1589–1599. [Google Scholar] [CrossRef]
- Miche, L.; Battistoni, F.; Gemmer, S.; Belghazi, M.; Reinhold-Hurek, B. Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Mol. Plant-Microbe Interact. 2006, 19, 502–511. [Google Scholar] [CrossRef] [Green Version]
- Tejeda-Sartorius, M.; Martinez de la Vega, O.; Delano-Frier, J.P. Jasmonic acid influences mycorrhizal colonization in tomato plants by modifying the expression of genes involved in carbohydrate partitioning. Plant Physiol. 2008, 133, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Dias-Arieira, C.R.; Santana, S.D.; de Freitas, L.G.; da Cunha, T.P.L.; Biela, F.; Puerari, H.H.; Chiamolera, F.M. Efficiency of Pochonia chlamydosporia in Meloidogyne incognita control in lettuce crop (Lactuca sativa L.). J. Food Agric. Environ. 2011, 9, 561–563. [Google Scholar]
- Macia-Vicente, J.G.; Jansson, H.B.; Mendgen, K.; Lopez-Llorca, L.V. Colonization of barley roots by endophytic fungi and their reduction of take-all caused by Gaeumannomyces graminis var. tritici. Can. J. Microbiol. 2008, 54, 600–609. [Google Scholar] [CrossRef] [Green Version]
- Sahebani, N.; Hadavi, N. Biological control of the root-knot nematode Meloidogyne javanica by Trichoderma harzianum. Soil Biol. Biochem. 2008, 40, 2016–2020. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrocka, J.; Malolepsza, U. Diversity in plant systemic resistance induced by Trichoderma. Biol. Control 2013, 67, 149–156. [Google Scholar] [CrossRef]
- Salas-Marina, M.A.; Silva-Flores, M.A.; Uresti-Rivera, E.E.; Castro-Longoria, E.; Herrera-Estrella, A.; Casas-Flores, S. Colonization of Arabidopsis roots by Trichoderma atroviride promotes growth and enhances systemic disease resistance through jasmonic acid/ethylene and salicylic acid pathways. European Journal of Plant Pathology 2011, 131, 15–26. [Google Scholar] [CrossRef]
- Rotenberg, D.; Thompson, T.S.; German, T.L.; Willis, D.K. Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods 2006, 138, 49–59. [Google Scholar] [CrossRef]
- Sarmento, R.A.; Lemos, F.; Bleeker, P.M.; Schuurink, R.C.; Pallini, A.; Oliveira, M.G.; Lima, E.R.; Kant, M.; Sabelis, M.W.; Janssen, A. A herbivore that manipulates plant defence. Ecol. Lett. 2011, 14, 229–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gayoso, C.; Pomar, F.; Novo-Uzal, E.; Merino, F.; de Ilarduya, O.M. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant. Biol. 2010, 10, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uppalapati, S.R.; Ayoubi, P.; Weng, H.; Palmer, D.A.; Mitchell, R.E.; Jones, W.; Bender, C.L. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant. J. 2005, 42, 201–217. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, X.; Zhao, J.-L.; Bai, M.; Ping, X.-X.; Li, Y.-L.; Yang, Y.-H.; Mao, Z.-C.; Yang, G.-S.; Xie, B.-Y. Pochonia chlamydosporia Isolate PC-170-Induced Expression of Marker Genes for Defense Pathways in Tomatoes Challenged by Different Pathogens. Microorganisms 2021, 9, 1882. https://doi.org/10.3390/microorganisms9091882
Zhuang X, Zhao J-L, Bai M, Ping X-X, Li Y-L, Yang Y-H, Mao Z-C, Yang G-S, Xie B-Y. Pochonia chlamydosporia Isolate PC-170-Induced Expression of Marker Genes for Defense Pathways in Tomatoes Challenged by Different Pathogens. Microorganisms. 2021; 9(9):1882. https://doi.org/10.3390/microorganisms9091882
Chicago/Turabian StyleZhuang, Xia, Jian-Long Zhao, Miao Bai, Xing-Xing Ping, Yan-Lin Li, Yu-Hong Yang, Zhen-Chuan Mao, Guo-Shun Yang, and Bing-Yan Xie. 2021. "Pochonia chlamydosporia Isolate PC-170-Induced Expression of Marker Genes for Defense Pathways in Tomatoes Challenged by Different Pathogens" Microorganisms 9, no. 9: 1882. https://doi.org/10.3390/microorganisms9091882
APA StyleZhuang, X., Zhao, J.-L., Bai, M., Ping, X.-X., Li, Y.-L., Yang, Y.-H., Mao, Z.-C., Yang, G.-S., & Xie, B.-Y. (2021). Pochonia chlamydosporia Isolate PC-170-Induced Expression of Marker Genes for Defense Pathways in Tomatoes Challenged by Different Pathogens. Microorganisms, 9(9), 1882. https://doi.org/10.3390/microorganisms9091882








