Novel Thermotolerant Amylase from Bacillus licheniformis Strain LB04: Purification, Characterization and Agar-Agarose
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Collection
2.3. Microbial Strains and Medium Composition
2.4. Screening of Amylase Producing Bacteria and Growth Conditions
2.5. Morphological, Biochemical, and Physiological Characterization
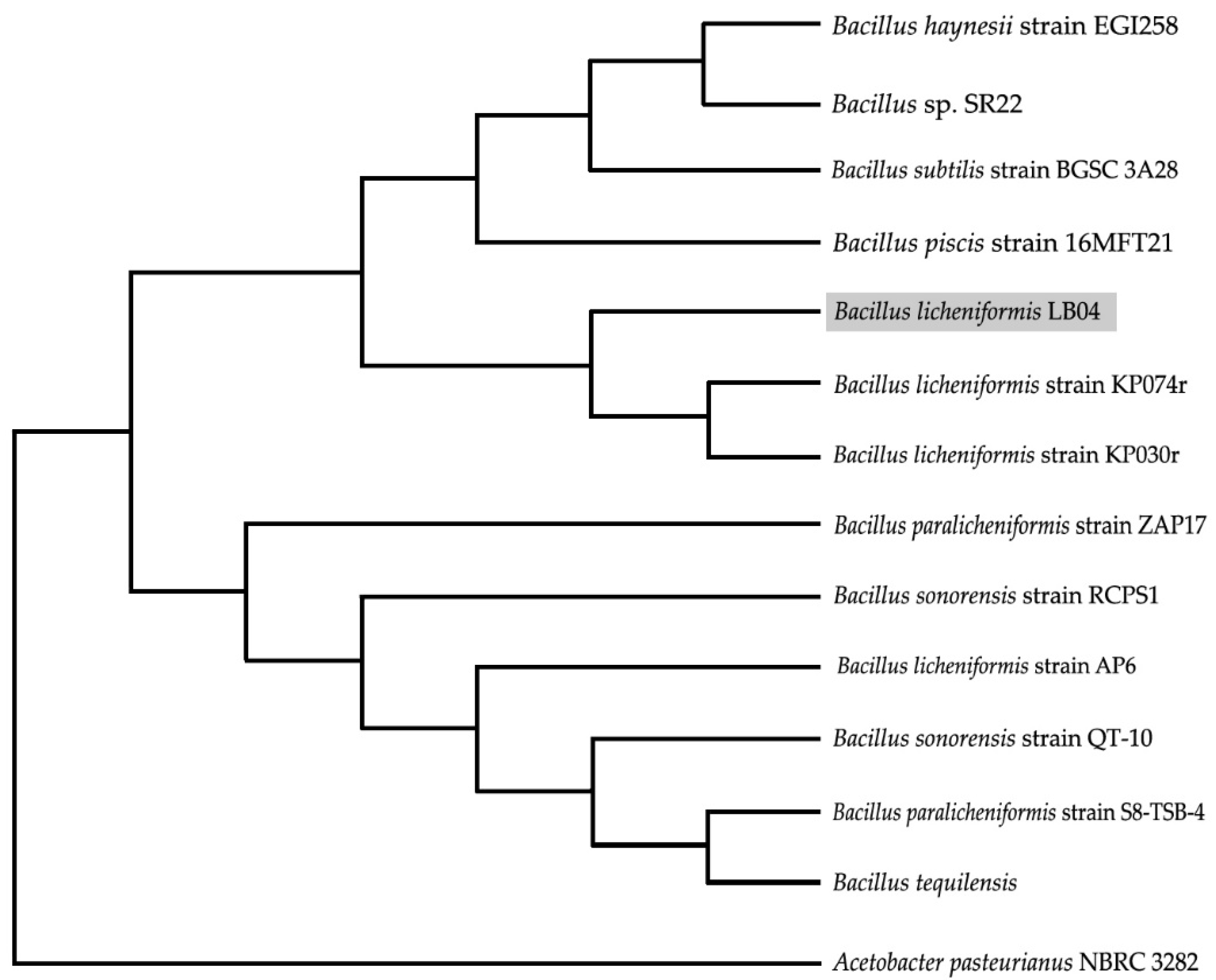
2.6. Molecular Phylogenetic Analyses
2.7. Amylase Activity Assay
2.8. Preparation of Crude Extract for Purification of α-Amylase
2.9. Purification of α-Amylase LB04 by Ion Exchange Chromatography
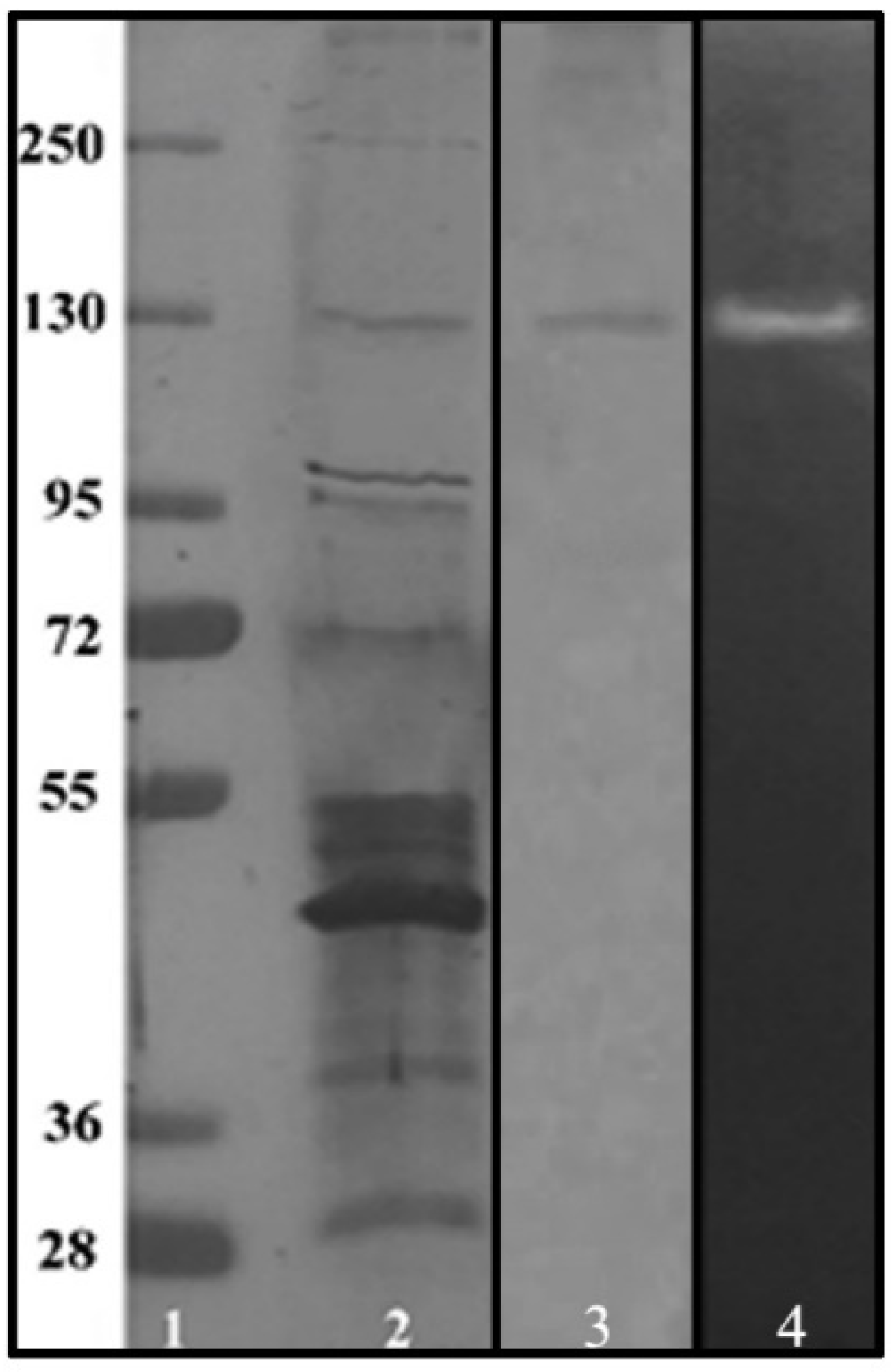
2.10. Electrophoretic Analysis of Purified Enzyme and Zymography
2.11. Characterization of Enzymatic Properties of Purified α-Amylase LB04
2.11.1. Thermostability and Acidic Resistance on α-Amylase Activity
2.11.2. Effects of Calcium Ions on the Enzymatic Activity of α-Amylase
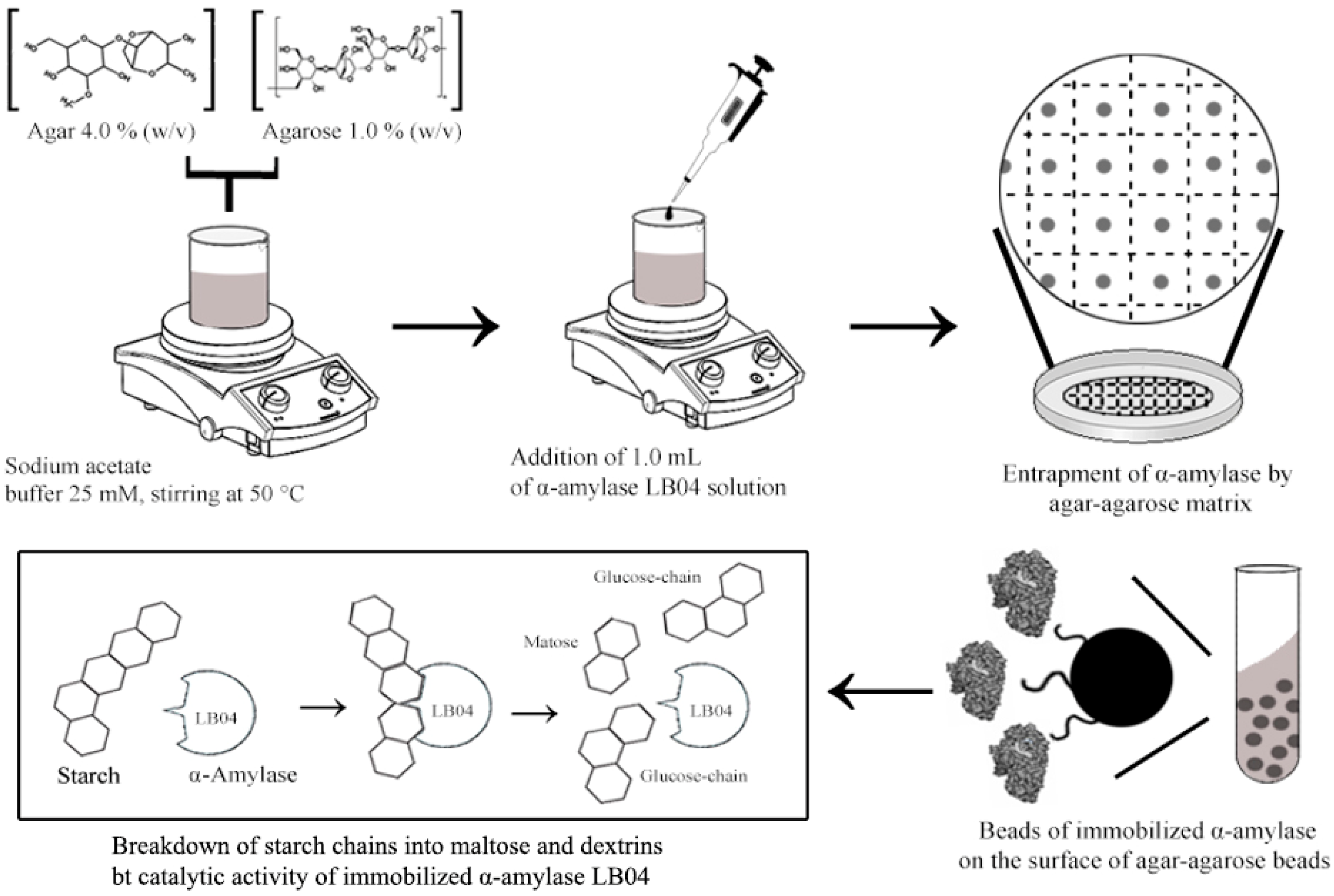
2.12. Immobilization of α-Amylase LB04 on Agar-Agarose Beads
2.13. Immobilized Enzyme Assay
3. Results
3.1. Identification and Starch-Degrading Activity of the Strain LB04
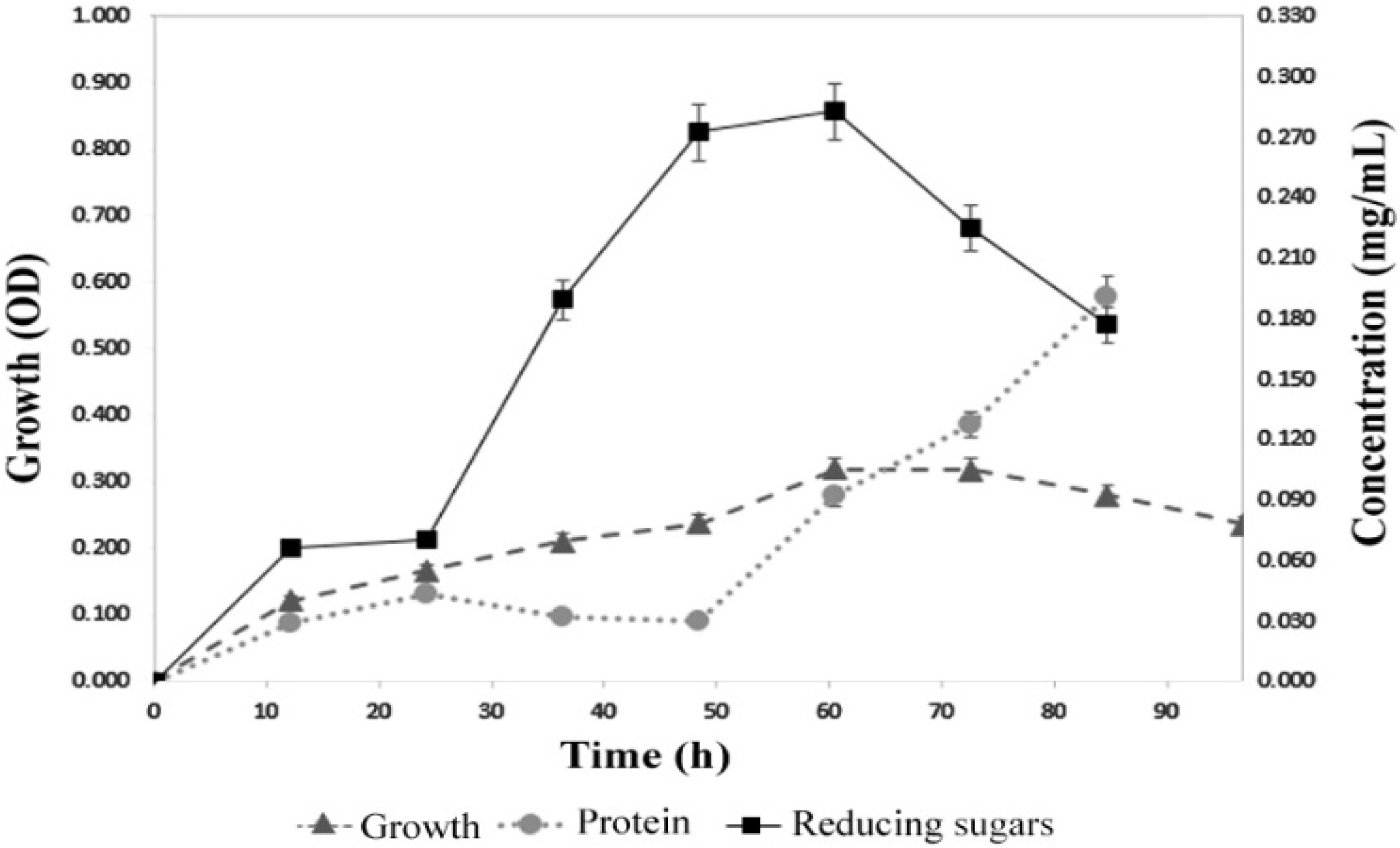
3.2. Growth Rate of Bacillus licheniformis Strain LB04
3.3. Production of α-Amylase by B. licheniformis Strain LB04
3.4. Purification of α-Amylase
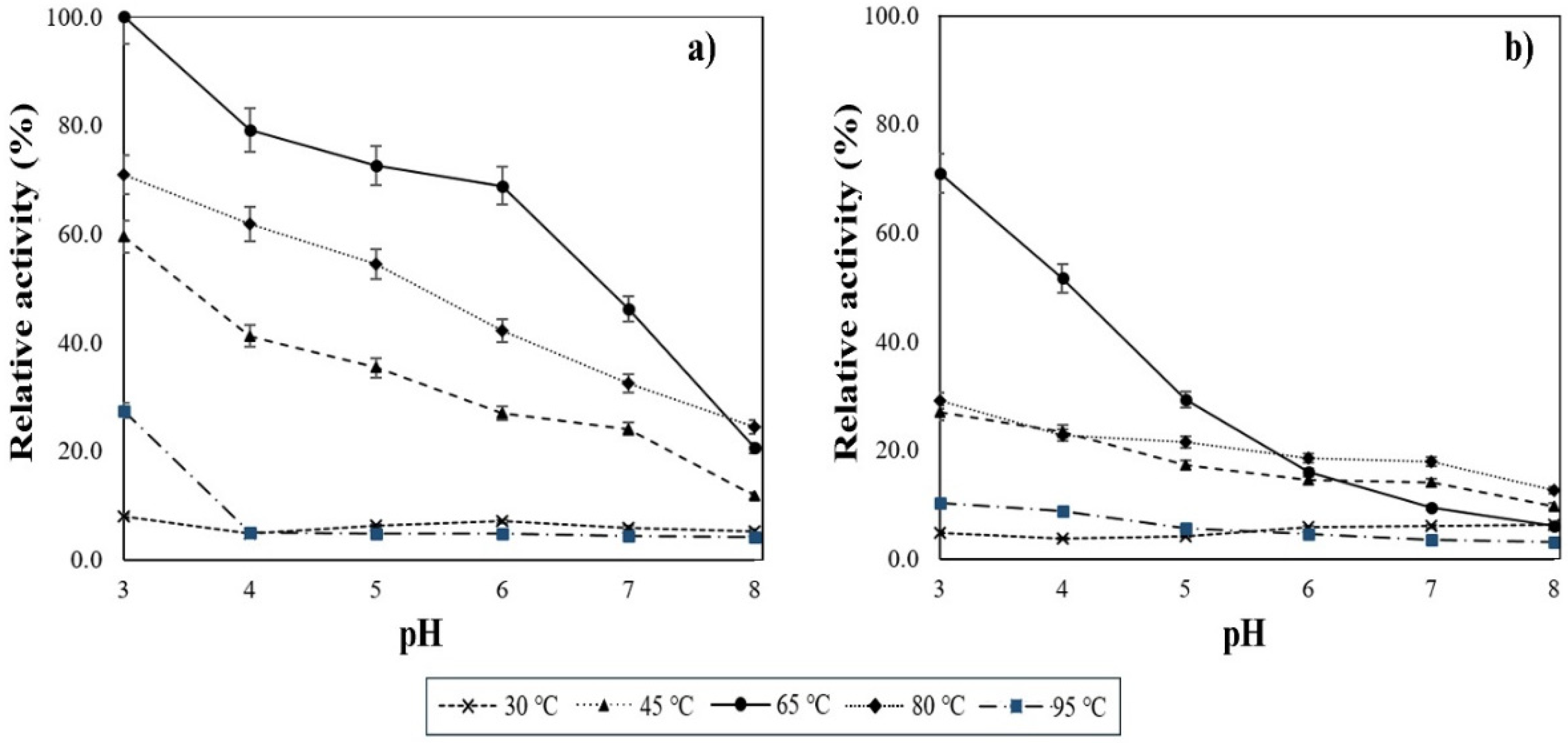
3.5. High Temperature and pH Ranges of Activity for the α-Amylase Produced by Strain LB04
3.6. Influence of Calcium Ions on the Enzymatic Activity
3.7. Immobilization of α-Amylase on Agar-Agarose Beads
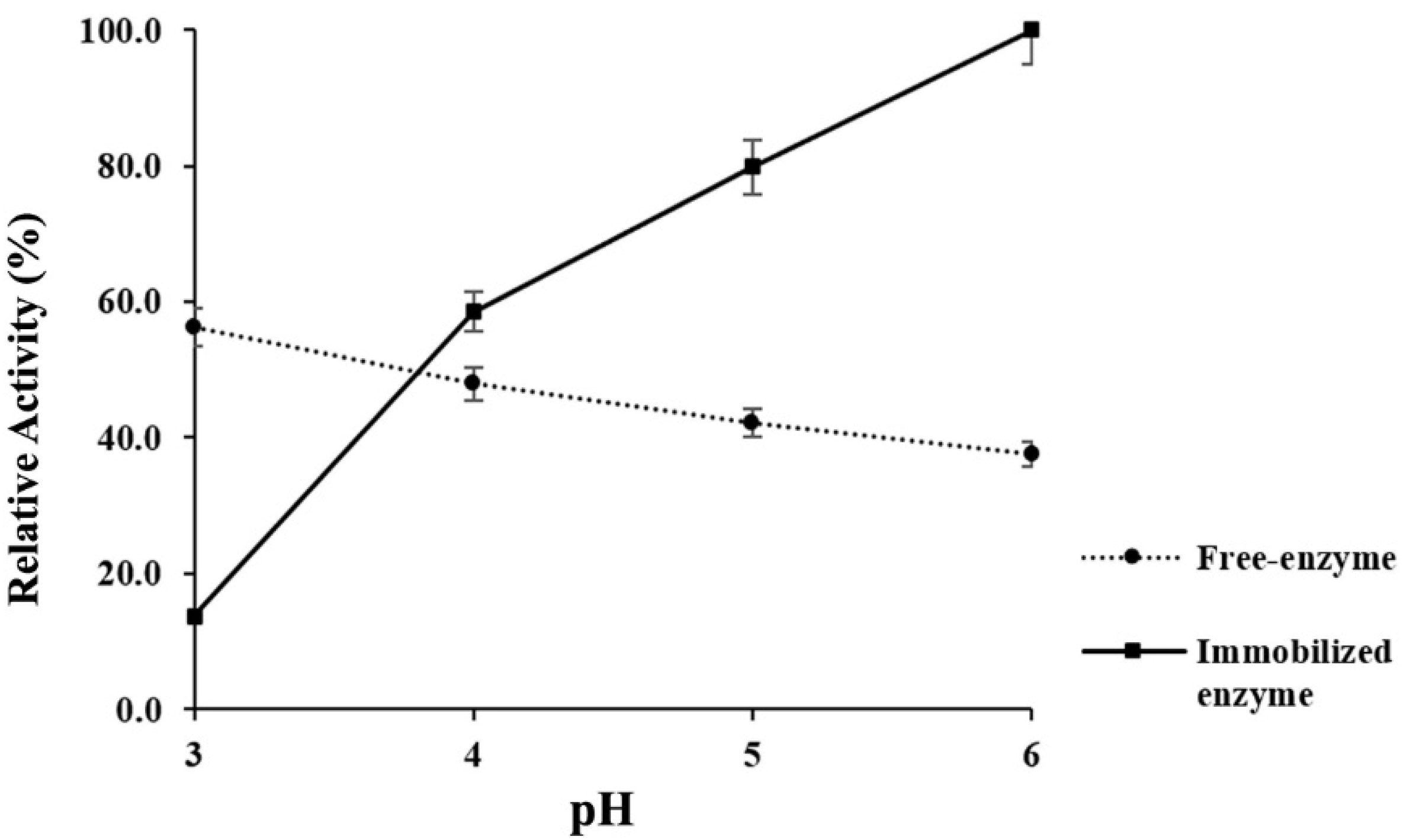
3.8. Acidic Stability of Immobilized α-Amylase
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robertson, G.; Wong, D.; Lee, C.; Wagschal, K.; Smith, M.R.; Orts, W.J. Native or raw starch digestion: A key step in energy efficient biorefining of grain. J. Agric. Food Chem. 2006, 54, 353–365. [Google Scholar] [CrossRef]
- Peng, H.; Chen, M.; Yi, L.; Zhang, X.; Wang, M.; Xiao, Y.; Zhang, N. Identification and characterization of a novel raw-starch degrading α-amylase (AmyASS) from the marine fish pathogen Aeromonas salmonicida ssp. salmonicid. J. Mol. Catal. B Enzym 2015, 119, 71–77. [Google Scholar] [CrossRef]
- Straksys, A.; Kochane, T.; Budriene, S. Catalytic properties of maltogenic α-amylase from Bacillus stearothermophilus immobilized onto poly(urethane urea) microparticles. Food Chem. 2016, 211, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.; Saini, H.; Dahiya, A. Amylases: Characteristics and industrial applications. J. Pharmacogn. Phytochem. 2017, 6, 1865–1871. [Google Scholar]
- Simair, A.A.; Qureshi, A.S.; Khushk, I.; Ali, C.H.; Lashari, S.; Bhutto, M.A.; Mangrio, G.S.; Lu, C. Production and Partial Characterization of α-Amylase Enzyme from Bacillus sp. BCC 01-50 and Potential Applications. BioMed Res. Int. 2017, 2017, 5259–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alpha-Amylase Baking Enzyme Market to Reach USD 347.7 Million by 2026|Reports and Data. Available online: https://www.globenewswire.com/newsrelease/2019/04/25/1809974/0/en/Alpha-Amylase-Baking-Enzyme-Market-To-Reach-USD-347-7-Million-By-2026-Reports-And-Data.html (accessed on 25 April 2019).
- Barrera, G.; Tadini, C.; León, A.; Ribotta, P. Use of alpha-amylase and amyloglucosidase combinations to minimize the bread quality problems caused by high levels of damaged starch. J. Food Sci. Technol. 2016, 53, 3675–3684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purhagen, J.; Sjöö, M.; Eliasson, A. Starch affecting anti-staling agents and their function in freestanding and pan-baked bread. Food Hydrocoll. 2011, 25, 1656–1666. [Google Scholar] [CrossRef]
- Sen, S.; Jana, A.; Bandyopadhyay, P.; Das-Mohapatra, P.; Raut, S. Thermostable amylase production from hot spring isolate Exiguobacterium sp.: A promising agent for natural detergents. Sustain. Chem. Pharm. 2016, 3, 59–68. [Google Scholar] [CrossRef]
- Sundarram, A.; Murthy, T. α-Amylase Production and Applications: A Review. J. Appl. Environ. Microbiol. 2014, 2, 166–175. [Google Scholar]
- Bouacem, K.; Laribi-Habchi, H.; Mechri, S.; Hacene, H.; Jaouadi, B.; Bouanane-Darenfed, A. Biochemical characterization of a novel thermostable chitinase from Hydrogenophilus hirschii strain KB-DZ44. Int. J. Biol. Macromol. 2018, 106, 338–350. [Google Scholar] [CrossRef]
- Allala, F.; Bouacem, K.; Boucherba, N.; Azzouz, Z.; Mechri, S.; Sahnoun, M.; Benallaoua, S.; Hacene, H.; Jaouadi, B.; Bouanane-Darenfed, A. Purification, biochemical, and molecular characterization of a novel extracellular thermostable and alkaline α-amylase from Tepidimonas fonticaldi strain HB23. Int. J. Biol. Macromol. 2019, 132, 558–574. [Google Scholar] [CrossRef]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Mohamed, S.; Khan, A.; Al-Bar, A.; El-Shishtawy, R. Immobilization of Trichoderma harzianum α-amylase on treated wool: Optimization and characterization. Molecules 2014, 19, 8027–8038. [Google Scholar] [CrossRef]
- Mohamed, S.; Al-Harbi, M.; Almulaiky, Y.; Ibrahim, I.; Salah, H.; El-Badry, M.; Abdel-Aty, A.; Fahmy, A.; El-Shishtawy, R. Immobilization of Trichoderma harzianum α-amylase on PPyAgNp/Fe3O4-nanocomposite: Chemical and physical properties. Art. Cells Nanomed. Biotechnol. 2018, 46, S201–S206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, O.; Jaiswal, N. Immobilization of a thermostable α-amylase on agarose and agar matrices and its application in starch stain removal. World Appl. Sci. J. 2011, 13, 572–577. [Google Scholar]
- Blanco de la Cruz, L. Glycerol Bioconversion by Thermotolerant Microorganism Isolated from Northeast Mexico. Master’s Thesis, Nuevo Leon Autonomous University, Nuevo Leon, Mexico, 2016. [Google Scholar]
- Minor, V. Ein Neues Verfahren zu der Klinischen Untersuchung der Schweißabsonderung. Dtsch. Z. Nervenheilkd 1928, 101, 302–308. [Google Scholar] [CrossRef]
- Deljou, A.; Arezi, I. Production of thermostable extracellular α-amylase by a moderate thermophilic Bacillus licheniformis-AZ2 isolated from Qinarje Hot spring (Ardebil prov. of Iran). Period. Biol. 2016, 118, 405–416. [Google Scholar]
- Miller, G. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Upadhyay, M.; Sharma, R.; Pandey, A.; Rajak, R. An improved zymographic method for detection of amylolytic enzymes of fungi on polyacrylamide gels. Mycologist 2005, 19, 138–140. [Google Scholar] [CrossRef]
- Gamal, R.; Abou-Taleb, K.; Abd-Elhalem, B. Isolation, Identification and Production of Amylases from Thermophilic Spore Forming Bacilli Using Starch Raw Materials Under Submerged Culture. AASCIT J. Biosci. 2017, 3, 52–68. [Google Scholar]
- El-Sheshtawy, H.; Aiad, I.; Osman, M.; Kobisy, A.; El-Sheshtawy, H.; Abo-Elnasr, A. Production of biosurfactant from Bacillus licheniformis for microbial enhanced oil recovery and inhibition the growth of sulfate reducing bacteria. Egypt. J. Pet. 2015, 24, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Hiteshi, K.; Didwal, G.; Gupta, R. Production optimization of α-amylase from Bacillus licheniformis. J. Adv. Res. Biol. Pharm. Res. 2016, 1, 1–14. [Google Scholar]
- Alariya, S.; Sethi, S.; Gupta, S.; Lal-Gupta, B. Amylase activity of a starch degrading bacteria isolated from soil. Arch. Appl. Sci. Res. 2013, 5, 15–24. [Google Scholar]
- Chaudhary, M.; Rana, N.; Vaidya, D.; Ghabru, A.; Rana, K.; Dipta, B. Immobilization of Amylase by Entrapment Method in Different Natural Matrix. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 1097–1103. [Google Scholar] [CrossRef]
- Esfahanibolandbalaie, Z.; Rostami, K.; Mirdamadi, S. Some studies of α-amylase production using Aspergillus oryzae. Pak. J. Biol. Sci. 2008, 11, 2553–2559. [Google Scholar] [CrossRef] [PubMed]
- Nathan, S.; Nair, M. Engineering a repression-free catabolite-enhanced expression system for a thermophilic alpha-amylase from Bacillus licheniformis. J. Biotechnol. 2013, 168, 394–402. [Google Scholar] [CrossRef]
- Russell, J.; Baldwin, R. Substrate preferences in rumen bacteria: Evidence of catabolite regulatory mechanisms. Appl. Environ. Microbiol. 1978, 36, 319–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maktouf, S.; Kamoun, A.; Moulis, C.; Remaud-Simeon, M.; Châabouni, S. A new raw-starch-digesting α-amylase: Production under solid-state fermentation on crude millet and biochemical characterization. J. Microbiol. Biotechnol. 2013, 23, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Burgess-Cassler, A.; Imam, S.; Gould, J. High-molecular-weight amylase activities from bacteria degrading starch-plastic films. Appl. Environ. Microbiol. 1991, 57, 612–614. [Google Scholar] [CrossRef] [Green Version]
- Abou-Dobara, M.; El-Sayed, A.; El-Fallal, A.; Omar, M. Production and partial characterization of high molecular weight extracellular α-amylase from Thermoactinomyces vulgaris isolated from Egyptian soil. Pol. J. Microbiol. 2011, 60, 65–71. [Google Scholar] [CrossRef]
- Timilsina, P.; Pandey, G.; Shrestha, A.; Ojha, M.; Karki, T. Purification and characterization of a noble thermostable algal starch liquefying α-amylase from Aeribacillus pallidus BTPS-2 isolated from geothermal spring of Nepal. Biotechnol. Rep. 2020, 28, e00551. [Google Scholar] [CrossRef]
- Behal, A.; Singh, J.; Sharma, M.; Puri, P.; Batra, N. Characterization of alkaline α-amylase from Bacillus sp. AB 04. Int. J. Agric. Biol. 2006, 8, 80–83. [Google Scholar]
- Pinto, E.; Dorn, M.; Feltes, B. The tale of a versatile enzyme: α-amylase evolution, structure, and potential biotechnological applications for the bioremediation of n-alkanes. Chemosphere 2020, 250, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Matpan-Bekler, F.; Acer, Ö.; Güven, K. Co-Production of Thermostable, Calcium-Independent α-Amylase and Alkali-Metallo Protease from Newly Isolated Bacillus licheniformis Dv3. Innov. Rom. Food Biotechnol. 2015, 16, 21–30. [Google Scholar]
- Du, R.; Qiaozhi, S.; Qiaoge, Z.; Zhao, F.; Kim, R.; Zhou, Z.; Han, Y. Purification and characterization of novel thermostable and Ca-independent α-amylase produced by Bacillus amyloliquefaciens BH072. Int. J. Biol. Macromol. 2018, 115, 1151–1156. [Google Scholar] [CrossRef]
- Hmidet, N.; Bayoudh, A.; Berrin, J.; Kanoun, S.; Juge, N.; Nasri, M. Purification and biochemical characterization of a novel α-amylase from Bacillus licheniformis NH1. Cloning, nucleotide sequence and expression of amyN gene in Escherichia coli. Process Biochem. 2008, 43, 499–510. [Google Scholar] [CrossRef]
- Wu, H.; Tian, X.; Dong, Z.; Zhang, Y.; Huang, L.; Liu, X.; Jin, P.; Wang, Z. Engineering of Bacillus amyloliquefaciens α-Amylase with Improved Calcium Independence and Catalytic Efficiency by Error-Prone PCR. Starch-Stärke 2018, 70, 3–4. [Google Scholar] [CrossRef]
- Li, Z.; Duan, X.; Chen, S.; Wu, J. Improving the reversibility of thermal denaturation and catalytic efficiency of Bacillus licheniformis α-amylase through stabilizing a long loop in domain B. PLoS ONE 2017, 12, e0173187. [Google Scholar] [CrossRef]
- Huang, Y.; Krauss, G.; Cottaz, S.; Driguez, H.; Lipps, G. A highly acid-stable and thermostable endo-β-glucanase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochem. J. 2005, 385, 581–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talekar, S.; Chavare, S. Optimization of immobilization of α-amylase in alginate gel and its comparative biochemical studies with free α-amylase. Recent Res. Sci. Technol. 2012, 4, 1–5. [Google Scholar]
- Sharma, M.; Sharma, V.; Majumdar, D. Entrapment of α-Amylase in Agar Beads for Biocatalysis of Macromolecular Substrate. Int. Sch. Res. Not. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Tavano, O.; Fernandez-Lafuente, R.; Goulart, A.; Monti, R. Optimization of the immobilization of sweet potato amylase using glutaraldehyde-agarose support. Characterization of the immobilized enzyme. Process Biochem. 2013, 48, 1054–1058. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Huang, L.; Jia, L.; Gui, S.; Fu, Y.; Zheng, D.; Guo, W.; Lu, F. Improvement of the acid stability of B. licheniformis alpha amylase by site-directed mutagenesis. Process Biochem. 2017, 58, 174–180. [Google Scholar] [CrossRef]
- Kalita, T.; Sangma, S.; Bez, G.; Ambasht, P. Immobilization of Acid Phosphatase in Agar-agar and Gelatin: Comparative Characterization. J. Sci. Res. 2020, 64, 193–200. [Google Scholar]
- Mesbah, N.; Wiegel, J. Improvement of Activity and Thermostability of Agar-Entrapped, Thermophilic, Haloalkaliphilic Amylase AmyD8. Catal. Lett. 2018, 148, 2665–2674. [Google Scholar] [CrossRef]
- Fernandez-Caresani, J.; Dallegrave, A.; dos Santos, J. Amylases immobilization by sol–gel entrapment: Application for starch hydrolysis. J. Sol-Gel Sci. Technol. 2020, 94, 229–240. [Google Scholar] [CrossRef]
- Mardani, T.; Khibani, M.; Mokarram, R.; Hamishehkar, H. Immobilization of α-amylase on chitosan-montmorillonite nanocomposite beads. Int. J. Biol. Macromol. 2018, 120, 354–360. [Google Scholar] [CrossRef]

| Characterization of the LB04 Strain | ||
|---|---|---|
| Test | LB04 | |
| Colony character | Color | White |
| Colony Shape | Irregular | |
| Edge | Undulate | |
| Elevation | Flat | |
| Morphology | Gram character | + |
| Shape | Rod | |
| Arrangement | Single | |
| Motility | − | |
| Biochemical profile | Amylase | + |
| Protease | + | |
| Lipase | + | |
| Catalase | + | |
| Oxidase | − | |
| Indole | − | |
| Gas production | − | |
| SH2 production | − | |
| Glucose | + | |
| Fructose | − | |
| Lactose | − | |
| Environmental tolerance | Temp. tolerance range | 20–90 °C |
| pH tolerance range | 2.0–10.0 | |
| Step | Total Activity (U) | Total Proteins (mg) | Specific Activity (U/mg) | Purification (Fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 14,450 ± 142.7 | 44.0 | 328.4 ± 6.5 | 1.0 | 100 |
| Ammonium sulfate precipitation (65%) | 6647 ± 98.2 | 10.0 | 664.7 ± 10.6 | 2.0 | 46 |
| Ammonium sulfate precipitation (85%) | 4018 ± 34.9 | 2.8 | 1435 ± 27.8 | 4.4 | 28 |
| DEAE-Sepharose | 3092 ± 12.1 | 0.8 | 3865 ± 47.2 | 11.8 | 21.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva-Salinas, A.; Rodríguez-Delgado, M.; Gómez-Treviño, J.; López-Chuken, U.; Olvera-Carranza, C.; Blanco-Gámez, E.A. Novel Thermotolerant Amylase from Bacillus licheniformis Strain LB04: Purification, Characterization and Agar-Agarose. Microorganisms 2021, 9, 1857. https://doi.org/10.3390/microorganisms9091857
Silva-Salinas A, Rodríguez-Delgado M, Gómez-Treviño J, López-Chuken U, Olvera-Carranza C, Blanco-Gámez EA. Novel Thermotolerant Amylase from Bacillus licheniformis Strain LB04: Purification, Characterization and Agar-Agarose. Microorganisms. 2021; 9(9):1857. https://doi.org/10.3390/microorganisms9091857
Chicago/Turabian StyleSilva-Salinas, Anaid, Melissa Rodríguez-Delgado, Jesús Gómez-Treviño, Ulrico López-Chuken, Clarita Olvera-Carranza, and Edgar Allan Blanco-Gámez. 2021. "Novel Thermotolerant Amylase from Bacillus licheniformis Strain LB04: Purification, Characterization and Agar-Agarose" Microorganisms 9, no. 9: 1857. https://doi.org/10.3390/microorganisms9091857
APA StyleSilva-Salinas, A., Rodríguez-Delgado, M., Gómez-Treviño, J., López-Chuken, U., Olvera-Carranza, C., & Blanco-Gámez, E. A. (2021). Novel Thermotolerant Amylase from Bacillus licheniformis Strain LB04: Purification, Characterization and Agar-Agarose. Microorganisms, 9(9), 1857. https://doi.org/10.3390/microorganisms9091857






