Planktonic and Biofilm-Associated Pseudomonas aeruginosa and Staphylococcus epidermidis Elicit Differential Human Peripheral Blood Cell Responses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Biofilm Formation
2.3. Biofilm Mass Quantification
2.4. PBMC Separation
2.5. Co-Culture of PBMC with Intact Biofilms, Disrupted Biofilms or Planktonic Bacteria
2.6. Evaluation of Cell Viability by Trypan Blue Dye Exclusion Test
2.7. Immunofluorescence Staining for Cell Surface and Activation Markers
2.8. Quantification of Cytokines in Culture Supernatants
2.9. Characterization of P. aeruginosa and S. epidermidis Biofilms in the Adopted Co-Culture Conditions
2.10. Statistical Analysis
3. Results
3.1. Bacterial Cell Number at Time 0 and PBMC Vitality after 24 h Co-Culture with Bacteria
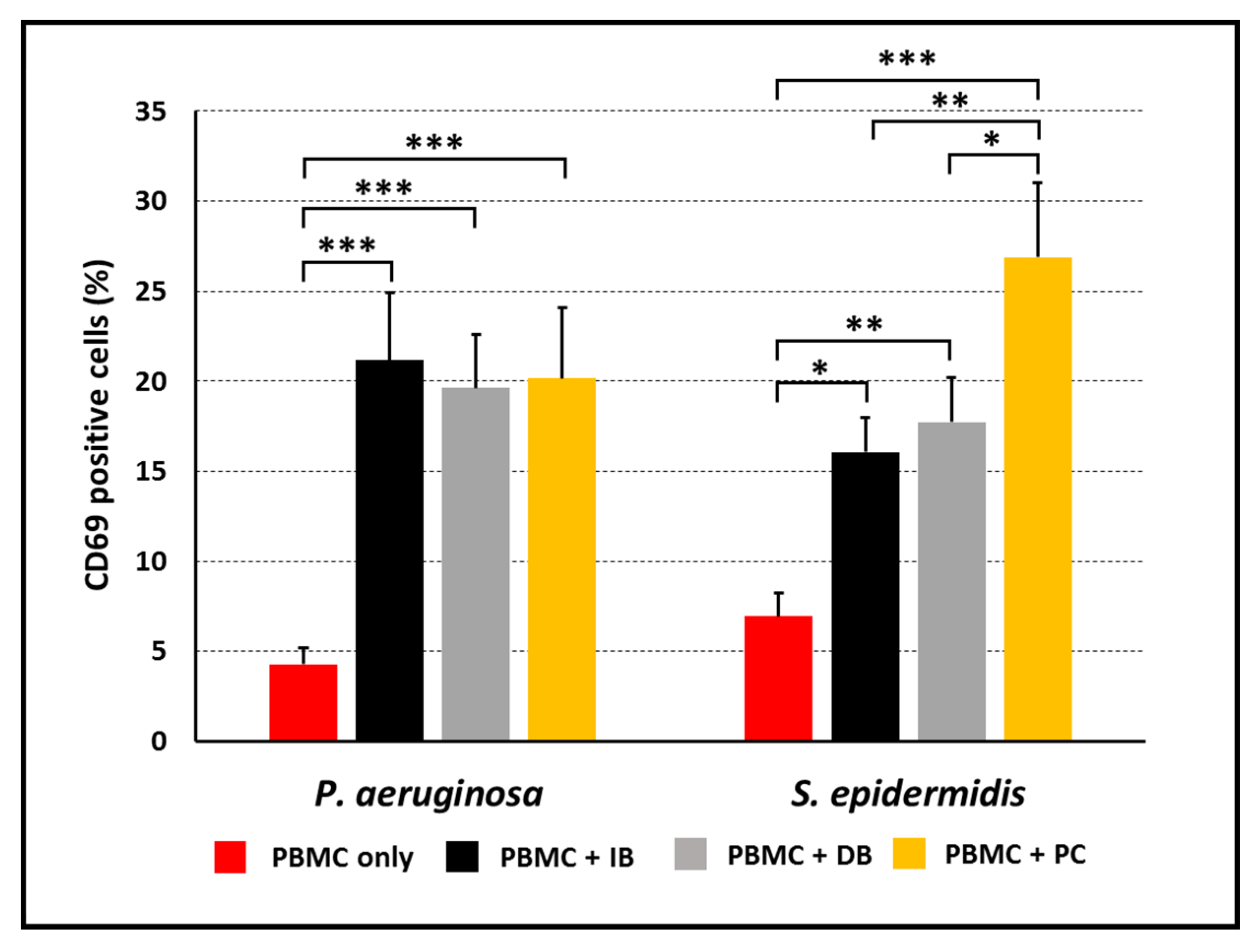
3.2. Activation of PBMC Stimulated with P. aeruginosa or S. epidermidis in Planktonic or Biofilm Mode of Growth
3.3. Activated Cell-Subsets Following Stimulation with P. aeruginosa or S. epidermidis in Planktonic or Biofilm Mode of Growth
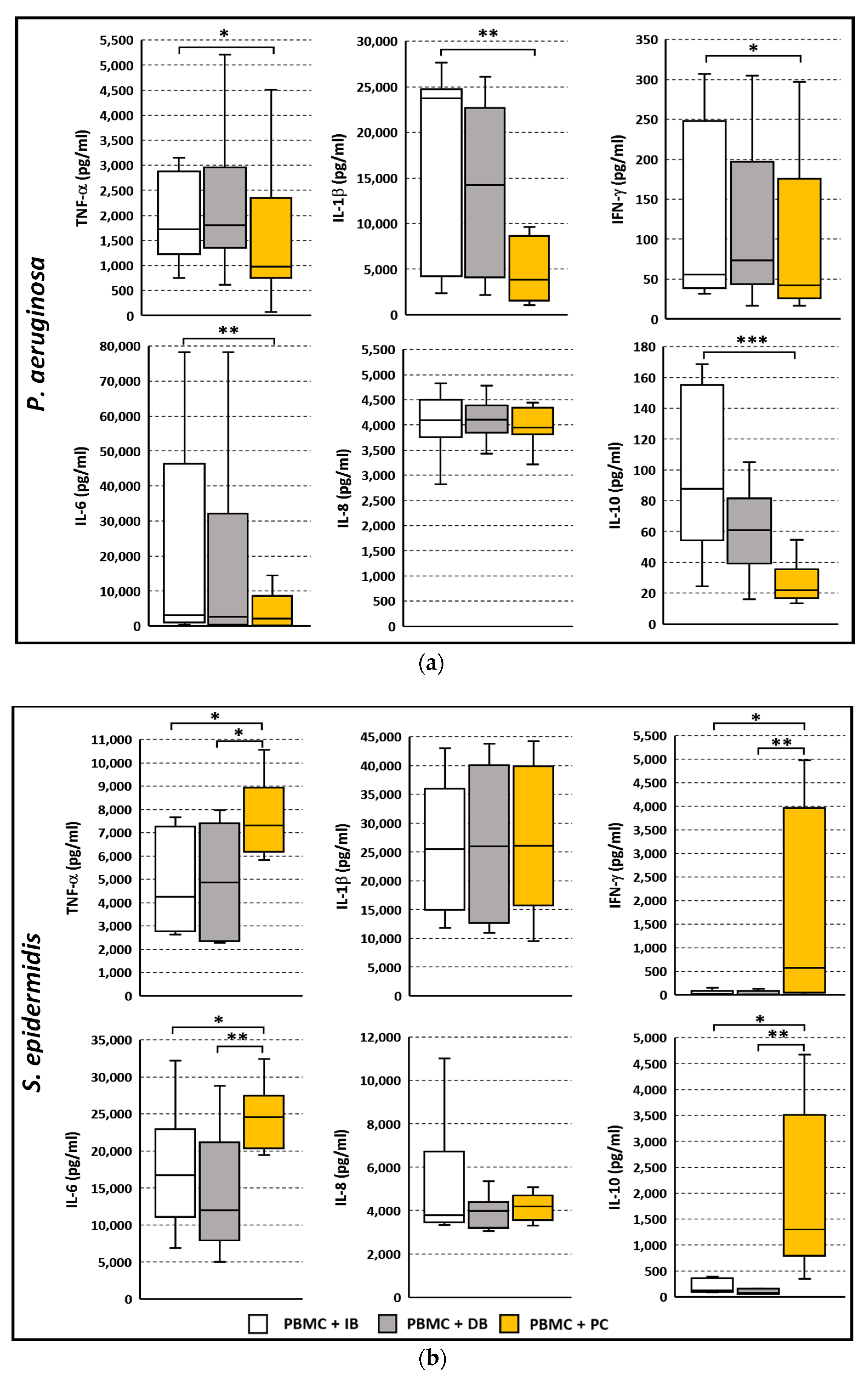
3.4. Cytokine Secretion Profile Following Stimulation with P. aeruginosa or S. epidermidis in Planktonic or Biofilm Mode of Growth
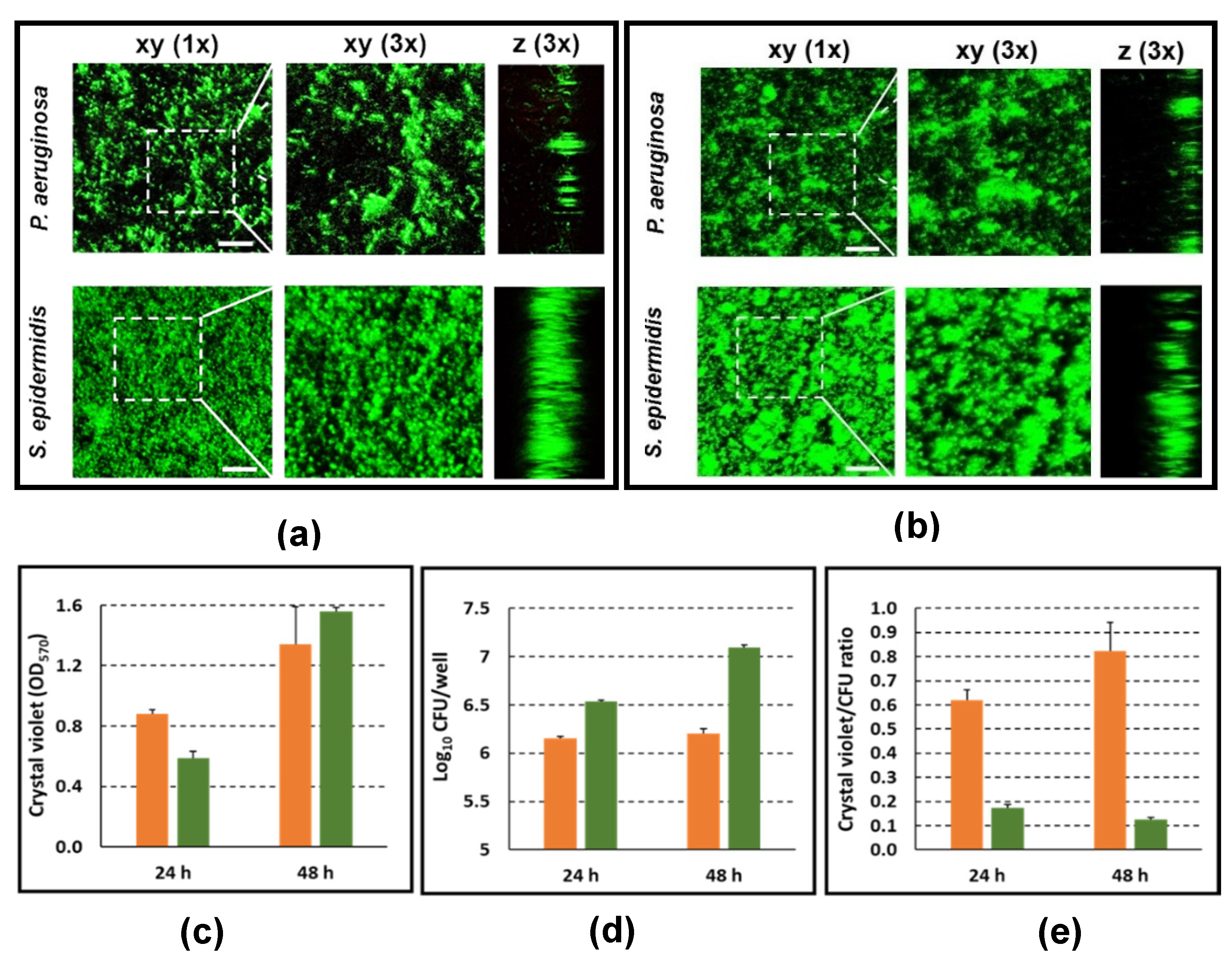
3.5. Characterization of P. aeruginosa and S. epidermidis Biofilms in Terms of Biofilm-Architecture, Biofilm Biomass, and Biofilm-Associated Viable Bacteria Count
3.6. Live Imaging of PBMC Interacting with P. aeruginosa and S. epidermidis Biofilms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, M.; Hall-Stoodley, L.; Allan, R.N.; Faust, S.N. New approaches to the treatment of biofilm-related infections. J. Infect. 2014, 69, 47–52. [Google Scholar] [CrossRef]
- Khan, F.; Pham, D.T.N.; Oloketuyi, S.F.; Kim, Y.M. Antibiotics application strategies to control biofilm formation in pathogenic bacteria. Curr. Pharm. Biotechnol. 2020, 21, 270–286. [Google Scholar] [CrossRef]
- Campoccia, D.; Mirzaei, R.; Montanaro, L.; Arciola, C.R. Hijacking of immune defences by biofilms: A multifront strategy. Biofouling 2019, 35, 1055–1074. [Google Scholar] [CrossRef] [PubMed]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta 2016, 1858, 1044–1060. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef]
- Rybtke, M.; Hultqvist, L.D.; Givskov, M.; Tolker-Nielsen, T. Pseudomonas aeruginosa biofilm infections: Community structure, antimicrobial tolerance and immune response. J. Mol. Biol. 2015, 427, 3628–3645. [Google Scholar] [CrossRef]
- Speziale, P.; Pietrocola, G.; Foster, T.J.; Geoghegan, J.A. Protein-based biofilm matrices in Staphylococci. Front. Cell. Infect. Microbiol. 2014, 4, 171. [Google Scholar] [CrossRef] [Green Version]
- Brancatisano, F.L.; Maisetta, G.; Di Luca, M.; Esin, S.; Bottai, D.; Bizzarri, R.; Campa, M.; Batoni, G. Inhibitory effect of the human liver-derived antimicrobial peptide hepcidin 20 on biofilms of polysaccharide intercellular adhesin (PIA)-positive and PIA-negative strains of Staphylococcus epidermidis. Biofouling 2014, 30, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Ciornei, C.D.; Novikov, A.; Beloin, C.; Fitting, C.; Caroff, M.; Ghigo, J.M.; Cavaillon, J.M.; Adib-Conquy, M. Biofilm-forming Pseudomonas aeruginosa bacteria undergo lipopolysaccharide structural modifications and induce enhanced inflammatory cytokine response in human monocytes. Innate Immun. 2010, 16, 288–301. [Google Scholar] [CrossRef]
- Dötsch, A.; Eckweiler, D.; Schniederjans, M.; Zimmermann, A.; Jensen, V.; Scharfe, M.; Geffers, R.; Häussler, S. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS ONE 2012, 7, e31092. [Google Scholar] [CrossRef]
- Resch, A.; Rosenstein, R.; Nerz, C.; Götz, F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 2005, 71, 2663–2676. [Google Scholar] [CrossRef] [Green Version]
- Batoni, G.; Martinez-Pomares, L.; Esin, S. Editorial: Immune response to biofilms. Front. Immunol. 2021, 12, 696356. [Google Scholar] [CrossRef]
- Kaya, E.; Grassi, L.; Benedetti, A.; Maisetta, G.; Pileggi, C.; Di Luca, M.; Batoni, G.; Esin, S. In vitro interaction of Pseudomonas aeruginosa biofilms with human peripheral blood mononuclear cells. Front. Cell. Infect. Microbiol. 2020, 10, 187. [Google Scholar] [CrossRef]
- Piras, A.M.; Esin, S.; Benedetti, A.; Maisetta, G.; Fabiano, A.; Zambito, Y.; Batoni, G. Antibacterial, antibiofilm, and antiadhesive properties of different quaternized chitosan derivatives. Int. J. Mol. Sci. 2019, 20, 6297. [Google Scholar] [CrossRef] [Green Version]
- Esin, S.; Counoupas, C.; Aulicino, A.; Brancatisano, F.L.; Maisetta, G.; Bottai, D.; Di Luca, M.; Florio, W.; Campa, M.; Batoni, G. Interaction of Mycobacterium tuberculosis cell wall components with the human natural killer cell receptors NKp44 and toll-like receptor 2. Scand. J. Immunol. 2013, 77, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Di Luca, M.; Navari, E.; Esin, S.; Menichini, M.; Barnini, S.; Trampuz, A.; Casani, A.; Batoni, G. Detection of biofilms in biopsies from chronic rhinosinusitis patients: In vitro biofilm forming ability and antimicrobial susceptibility testing in biofilm mode of growth of isolated bacteria. Adv. Exp. Med. Biol. 2018, 1057, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zeng, W.; Huang, X.; Wang, S.; Zheng, J.; Pan, M.; Wang, Y. Peripheral CD19hi B cells exhibit activated phenotype and functionality in promoting IgG and IgM production in human autoimmune diseases. Sci. Rep. 2017, 7, 13921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faure, E.; Kwong, K.; Nguyen, D. Pseudomonas aeruginosa in chronic lung infections: How to adapt within the host? Front. Immunol. 2018, 9, 2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuong, C.; Otto, M. Staphylococcus epidermidis infections. Microbes Infect. 2002, 4, 481–489. [Google Scholar] [CrossRef]
- Leid, J.G.; Shirtliff, M.E.; Costerton, J.W.; Stoodley, P. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect. Immun. 2002, 70, 6339–6345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Secor, P.R.; James, G.A.; Fleckman, P.; Olerud, J.E.; McInnerney, K.; Stewart, P.S. Staphylococcus aureus biofilm and planktonic cultures differentially impact gene expression, mapk phosphorylation, and cytokine production in human keratinocytes. BMC Microbiol. 2011, 11, 143. [Google Scholar] [CrossRef] [Green Version]
- Brady, R.A.; Mocca, C.P.; Plaut, R.D.; Takeda, K.; Burns, D.L. Comparison of the immune response during acute and chronic Staphylococcus aureus infection. PLoS ONE 2018, 13, e0195342. [Google Scholar] [CrossRef] [Green Version]
- Spiliopoulou, A.I.; Kolonitsiou, F.; Krevvata, M.I.; Leontsinidis, M.; Wilkinson, T.S.; Mack, D.; Anastassiou, E.D. Bacterial adhesion, intracellular survival and cytokine induction upon stimulation of mononuclear cells with planktonic or biofilm phase Staphylococcus epidermidis. FEMS Microbiol. Lett. 2012, 330, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.H.; Park, M.D.; Otto, M. Host Response to Staphylococcus epidermidis Colonization and Infections. Front. Cell. Infect. Microbiol. 2017, 7, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito-Taki, T.; Tanabe, M.J.; Mochizuki, H.; Matsumoto, T.; Nakano, M.; Takada, H.; Tsujimoto, M.; Kotani, S.; Kusumoto, S.; Shiba, T.; et al. Polyclonal B cell activation by cell wall preparations of gram-positive bacteria. In vitro responses of spleen cells obtained from Balb/c, nu/nu, nu/+, C3H/He, C3H/HeJ and hybrid (DBA/N x Balb/c)F1 mice. Microbiol. Immunol. 1980, 24, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, T.K.; Kobayashi, S.D.; Freedman, B.; Porter, A.R.; Voyich, J.M.; Otto, M.; Schneewind, O.; DeLeo, F.R. Interaction of Staphylococci with human B cells. PLoS ONE 2016, 11, e0164410. [Google Scholar] [CrossRef]
- Mauch, R.M.; Jensen, P.Ø.; Moser, C.; Levy, C.E.; Høiby, N. Mechanisms of humoral immune response against Pseudomonas aeruginosa biofilm infection in cystic fibrosis. J. Cyst. Fibros. 2018, 17, 143–152. [Google Scholar] [CrossRef] [Green Version]
- Chalifour, A.; Jeannin, P.; Gauchat, J.F.; Blaecke, A.; Malissard, M.; N’Guyen, T.; Thieblemont, N.; Delneste, Y. Direct bacterial protein PAMP recognition by human NK cells involves TLRs and triggers alpha-defensin production. Blood 2004, 104, 1778–1783. [Google Scholar] [CrossRef] [Green Version]
- Esin, S.; Batoni, G.; Counoupas, C.; Stringaro, A.; Brancatisano, F.L.; Colone, M.; Maisetta, G.; Florio, W.; Arancia, G.; Campa, M. Direct binding of human NK cell natural cytotoxicity receptor NKp44 to the surfaces of mycobacteria and other bacteria. Infect. Immun. 2008, 76, 1719–1727. [Google Scholar] [CrossRef] [Green Version]
- Sultan, N.; Cağlar, K.; Aybay, C.; Imir, T. Effect of a Staphylococcus epidermidis-extracted slime factor on human natural killer cell activity. Microbios 1997, 91, 89–95. [Google Scholar] [PubMed]
- Peoples, J.D.; Cheung, S.; Nesin, M.; Lin, H.; Tatad, A.M.; Hoang, D.; Perlman, J.M.; Cunningham-Rundles, S. Neonatal cord blood subsets and cytokine response to bacterial antigens. Am. J. Perinatol. 2009, 26, 647–657. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Katz, J.; Zhang, P.; Ashtekar, A.R.; Gaddis, D.E.; Fan, M.; Michalek, S.M. Contribution of a Streptococcus mutans antigen expressed by a Salmonella vector vaccine in dendritic cell activation. Infect. Immun. 2011, 79, 3792–3800. [Google Scholar] [CrossRef] [Green Version]
- Cerca, F.; França, Â.; Pérez-Cabezas, B.; Carvalhais, V.; Ribeiro, A.; Azeredo, J.; Pier, G.; Cerca, N.; Vilanova, M. Dormant bacteria within Staphylococcus epidermidis biofilms have low inflammatory properties and maintain tolerance to vancomycin and penicillin after entering planktonic growth. J. Med. Microbiol. 2014, 63, 1274–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubota, K. Innate IFN-gamma production by subsets of natural killer cells, natural killer T cells and gamma delta T cells in response to dying bacterial-infected macrophages. Scand. J. Immunol. 2010, 71, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Jesaitis, A.J.; Franklin, M.J.; Berglund, D.; Sasaki, M.; Lord, C.I.; Bleazard, J.B.; Duffy, J.E.; Beyenal, H.; Lewandowski, Z. Compromised host defense on Pseudomonas aeruginosa biofilms: Characterization of neutrophil and biofilm interactions. J. Immunol. 2003, 171, 4329–4339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, K.Y.; Park, M.D.; Otto, M. Immune evasion mechanisms of Staphylococcus epidermidis biofilm infection. Front. Microbiol. 2018, 9, 359. [Google Scholar] [CrossRef] [PubMed]
- Kristian, S.A.; Birkenstock, T.A.; Sauder, U.; Mack, D.; Götz, F.; Landmann, R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J. Infect. Dis. 2008, 197, 1028–1035. [Google Scholar] [CrossRef] [Green Version]
- Cerca, N.; Jefferson, K.K.; Oliveira, R.; Pier, G.B.; Azeredo, J. Comparative antibody-mediated phagocytosis of Staphylococcus epidermidis cells grown in a biofilm or in the planktonic state. Infect. Immun. 2006, 74, 4849–4855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heilmann, C.; Schweitzer, O.; Gerke, C.; Vanittanakom, N.; Mack, D.; Gotz, F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 1996, 20, 1083–1091. [Google Scholar] [CrossRef]
- Lee, J.; Monk, I.R.; Gonçalves da Silva, A.; Seemann, T.; Chua, K.; Kearns, A.; Hill, R.; Woodford, N.; Bartels, M.D.; Strommenger, B.; et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 2018, 3, 1175–1185. [Google Scholar] [CrossRef]
- Fredheim, E.G.; Granslo, H.N.; Flægstad, T.; Figenschau, Y.; Rohde, H.; Sadovskaya, I.; Mollnes, T.E.; Klingenberg, C. Staphylococcus epidermidis polysaccharide intercellular adhesin activates complement. FEMS Immunol. Med. Microbiol. 2011, 63, 269–280. [Google Scholar] [CrossRef]
- Sainz-Mejías, M.; Jurado-Martín, I.; McClean, S. Understanding Pseudomonas aeruginosa-host interactions: The ongoing quest for an efficacious vaccine. Cells 2020, 9, 2617. [Google Scholar] [CrossRef]
- Wu, L.; Estrada, O.; Zaborina, O.; Bains, M.; Shen, L.; Kohler, J.E.; Patel, N.; Musch, M.W.; Chang, E.B.; Fu, Y.X.; et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science 2005, 309, 774–777. [Google Scholar] [CrossRef] [Green Version]
- Meduri, G.U.; Kanangat, S.; Stefan, J.; Tolley, E.; Schaberg, D. Cytokines IL-1beta, IL-6, and TNF-alpha enhance in vitro growth of bacteria. Am. J. Respir. Crit. CareMed. 1999, 160, 961–967. [Google Scholar] [CrossRef]
- Alhede, M.; Bjarnsholt, T.; Jensen, P.O.; Phipps, R.K.; Moser, C.; Christophersen, L.; Christensen, L.D.; van Gennip, M.; Parsek, M.; Høiby, N.; et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 2009, 155, 3500–3508. [Google Scholar] [CrossRef] [Green Version]
- Walker, T.S.; Tomlin, K.L.; Worthen, G.S.; Poch, K.R.; Lieber, J.G.; Saavedra, M.T.; Fessler, M.B.; Malcolm, K.C.; Vasil, M.L.; Nick, J.A. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 2005, 73, 3693–3701. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaya, E.; Batoni, G.; Di Luca, M.; Apolloni, E.; Mazzoni, A.; Maisetta, G.; Esin, S. Planktonic and Biofilm-Associated Pseudomonas aeruginosa and Staphylococcus epidermidis Elicit Differential Human Peripheral Blood Cell Responses. Microorganisms 2021, 9, 1846. https://doi.org/10.3390/microorganisms9091846
Kaya E, Batoni G, Di Luca M, Apolloni E, Mazzoni A, Maisetta G, Esin S. Planktonic and Biofilm-Associated Pseudomonas aeruginosa and Staphylococcus epidermidis Elicit Differential Human Peripheral Blood Cell Responses. Microorganisms. 2021; 9(9):1846. https://doi.org/10.3390/microorganisms9091846
Chicago/Turabian StyleKaya, Esingül, Giovanna Batoni, Mariagrazia Di Luca, Eleonora Apolloni, Alessandro Mazzoni, Giuseppantonio Maisetta, and Semih Esin. 2021. "Planktonic and Biofilm-Associated Pseudomonas aeruginosa and Staphylococcus epidermidis Elicit Differential Human Peripheral Blood Cell Responses" Microorganisms 9, no. 9: 1846. https://doi.org/10.3390/microorganisms9091846
APA StyleKaya, E., Batoni, G., Di Luca, M., Apolloni, E., Mazzoni, A., Maisetta, G., & Esin, S. (2021). Planktonic and Biofilm-Associated Pseudomonas aeruginosa and Staphylococcus epidermidis Elicit Differential Human Peripheral Blood Cell Responses. Microorganisms, 9(9), 1846. https://doi.org/10.3390/microorganisms9091846









