Whole Genome Sequencing of Methicillin-Resistant Staphylococcus epidermidis Clinical Isolates Reveals Variable Composite SCCmec ACME among Different STs in a Tertiary Care Hospital in Oman
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Genomic DNA Extraction and Whole Genome Sequencing (WGS)
2.3. Mutlilocus Sequence Typing (MLST) and Whole Genome Single Nucleotide Polymorphism (SNP) Phylogeny Tree
2.4. Identification of SCCmec, ACME, and Acquired Antimicrobial Resistance Genes
2.5. Antibiotic and Copper Susceptibility
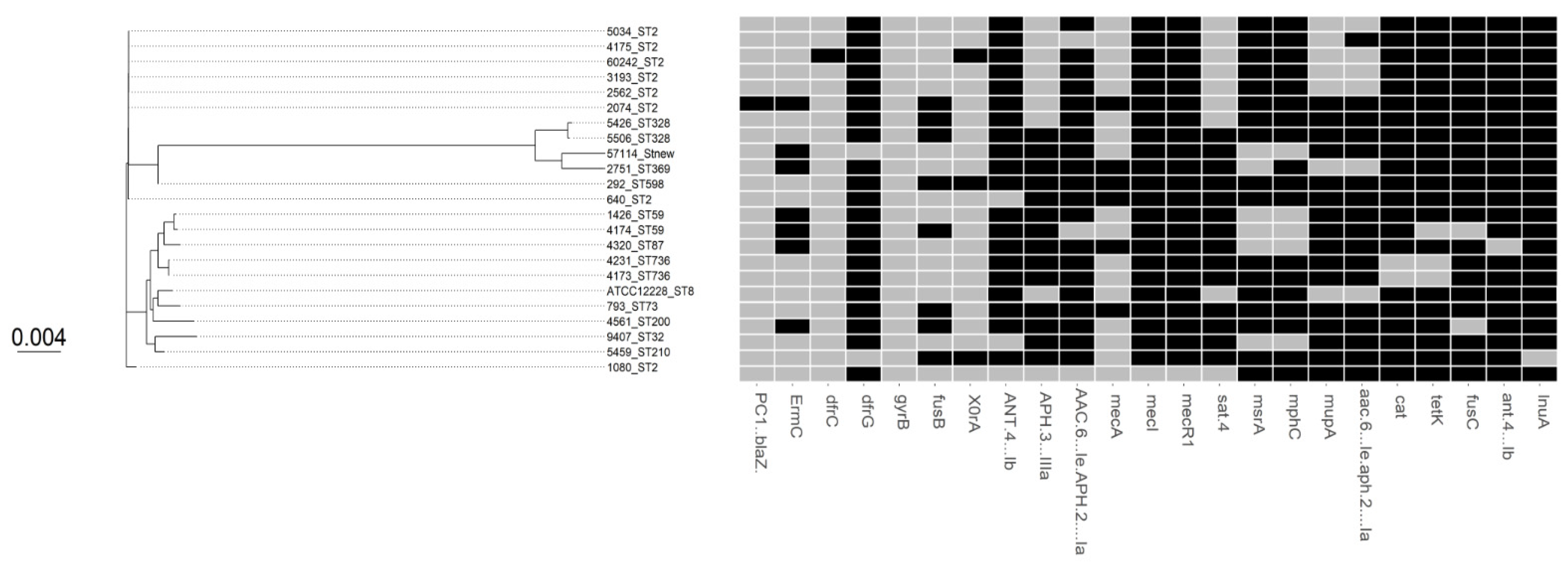
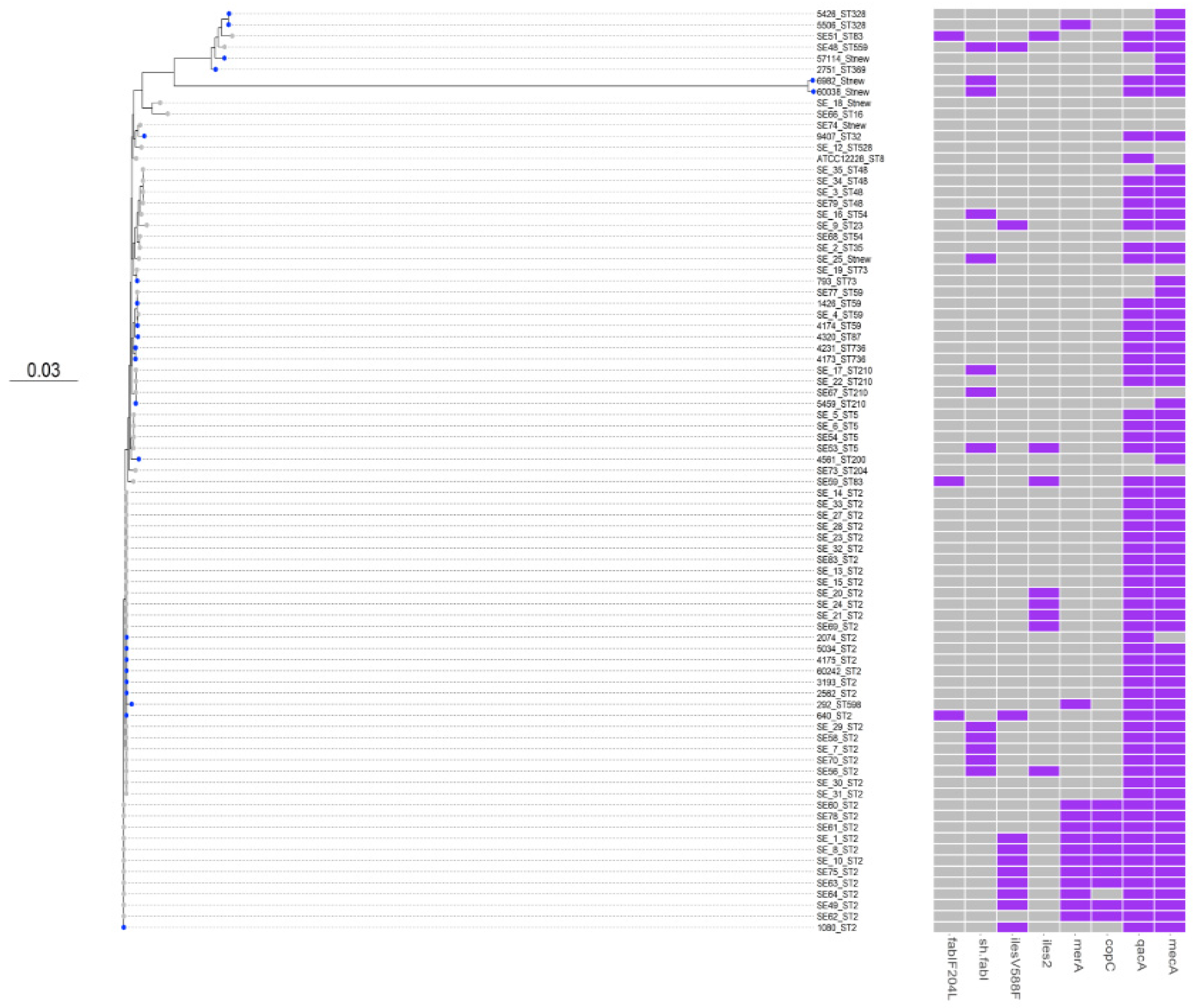
3. Results
3.1. Comparative Phylogenetic Tree Analysis
3.2. SCCmec and ACME Types
3.3. Antimicrobial Susceptibility and Virulence Conferring Genes
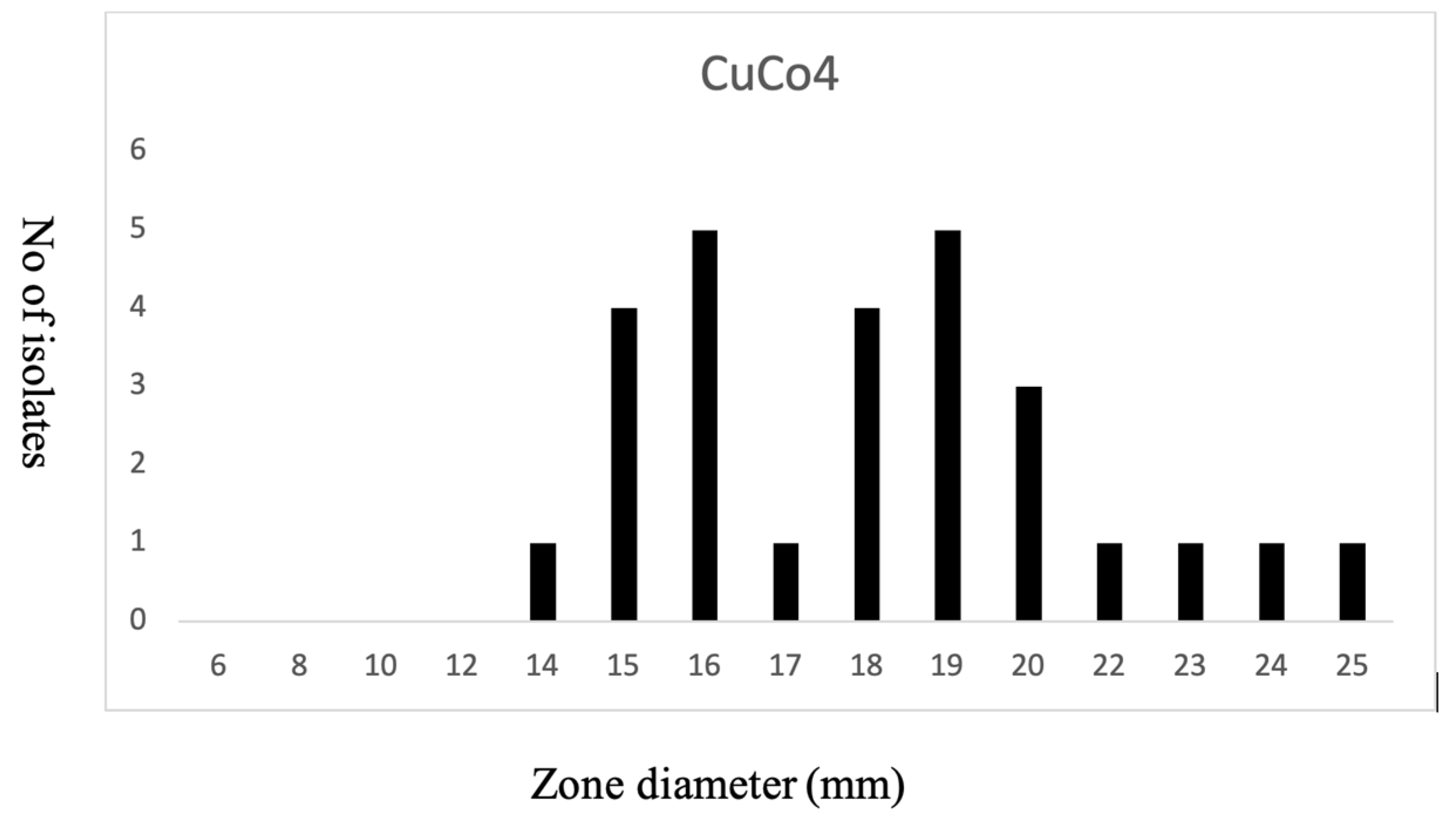
3.4. Biocides Susceptibility
3.5. Copper Susceptibility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brescó, M.S.; Harris, L.G.; Thompson, K.; Stanic, B.; Morgenstern, M.; O’Mahony, L.; Richards, R.G.; Moriarty, T.F. Pathogenic Mechanisms and Host Interactions in Staphylococcus Epidermidis Device-Related Infection. Front. Microbiol. 2017, 8, 1401. [Google Scholar] [CrossRef] [Green Version]
- Otto, M. Staphylococcus Epidermidis—The “accidental” Pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Almebairik, N.; Zamudio, R.; Ironside, C.; Joshi, C.; Ralph, J.D.; Roberts, A.P.; Gould, I.M.; Morrissey, J.A.; Hijazi, K.; Oggioni, M.R. Genomic Stability of Composite SCCmec ACME and COMER-Like Genetic Elements in Staphylococcus Epidermidis Correlates With Rate of Excision. Front. Microbiol. 2020, 11, 166. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.H.; Monk, I.R.; da Silva, A.G.; Seemann, T.; Chua, K.Y.L.; Kearns, A.; Hill, R.; Woodford, N.; Bartels, M.D.; Strommenger, B.; et al. Global Spread of Three Multidrug-Resistant Lineages of Staphylococcus Epidermidis. Nat. Microbiol. 2018, 3, 1175–1185. [Google Scholar] [CrossRef]
- Dhawan, B.; Rao, C.; Udo, E.E.; Gadepalli, R.; Vishnubhatla, S.; Kapil, A. Dissemination of Methicillin-Resistant Staphylococcus Aureus SCCmec Type IV and SCCmec Type v Epidemic Clones in a Tertiary Hospital: Challenge to Infection Control. Epidemiol. Infect. 2015, 143, 343–353. [Google Scholar] [CrossRef] [Green Version]
- Diep, B.A.; Stone, G.G.; Basuino, L.; Graber, C.J.; Miller, A.; des Etages, S.A.; Jones, A.; Palazzolo-Ballance, A.M.; Perdreau-Remington, F.; Sensabaugh, G.F.; et al. The Arginine Catabolic Mobile Element and Staphylococcal Chromosomal Cassette Mec Linkage: Convergence of Virulence and Resistance in the USA300 Clone of Methicillin-Resistant Staphylococcus Aureus. J. Infect. Dis. 2008, 197, 1523–1530. [Google Scholar] [CrossRef] [Green Version]
- Planet, P.J.; Diaz, L.; Kolokotronis, S.O.; Narechania, A.; Reyes, J.; Xing, G.; Rincon, S.; Smith, H.; Panesso, D.; Ryan, C.; et al. Parallel Epidemics of Community-Associated Methicillin-Resistant Staphylococcus Aureus USA300 Infection in North and South America. J. Infect. Dis. 2015, 212, 1874–1882. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, A.M.; McManus, B.A.; Coleman, D.C. First Description of Novel Arginine Catabolic Mobile Elements (ACMEs) Types IV and V Harboring a Kdp Operon in Staphylococcus Epidermidis Characterized by Whole Genome Sequencing. Infect. Genet. Evol. 2018, 61, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Planet, P.J.; Larussa, S.J.; Dana, A.; Smith, H.; Xu, A.; Ryan, C.; Uhlemann, A.C.; Boundy, S.; Goldberg, J.; Narechania, A.; et al. Emergence of the Epidemic Methicillin-Resistant Staphylococcus Aureus Strain USA300 Coincides with Horizontal Transfer of the Arginine Catabolic Mobile Element and SpeG-Mediated Adaptations for Survival on Skin. mBio 2013, 4, e00889-13. [Google Scholar] [CrossRef] [Green Version]
- McManus, B.A.; O’Connor, A.M.; Egan, S.A.; Flanagan, P.R.; Coleman, D.C. First Description of Arginine Catabolic Mobile Element (ACME) Type VI Harboring the Kdp Operon Only in Staphylococcus Epidermidis Using Short and Long Read Whole Genome Sequencing: Further Evidence of ACME Diversity. Infect. Genet. Evol. 2019, 71, 51–53. [Google Scholar] [CrossRef]
- O’Connor, A.M.; McManus, B.A.; Kinnevey, P.M.; Brennan, G.I.; Fleming, T.E.; Cashin, P.J.; O’Sullivan, M.; Polyzois, I.; Coleman, D.C. Significant Enrichment and Diversity of the Staphylococcal Arginine Catabolic Mobile Element ACME in Staphylococcus Epidermidis Isolates from Subgingival Peri-Implantitis Sites and Periodontal Pockets. Front. Microbiol. 2018, 9, 1588. [Google Scholar] [CrossRef]
- Zapotoczna, M.; Riboldi, G.P.; Moustafa, A.M.; Dickson, E.; Narechania, A.; Morrissey, J.A.; Planet, P.J.; Holden, M.T.G.; Waldron, K.J.; Geoghegan, J.A. Mobile-Genetic-Element-Encoded Hypertolerance to Copper Protects Staphylococcus Aureus from Killing by Host Phagocytes. mBio 2018, 9, e00550-18. [Google Scholar] [CrossRef] [Green Version]
- Purves, J.; Thomas, J.; Riboldi, G.P.; Zapotoczna, M.; Tarrant, E.; Andrew, P.W.; Londoño, A.; Planet, P.J.; Geoghegan, J.A.; Waldron, K.J.; et al. A Horizontally Gene Transferred Copper Resistance Locus Confers Hyper-Resistance to Antibacterial Copper Toxicity and Enables Survival of Community Acquired Methicillin Resistant Staphylococcus Aureus USA300 in Macrophages. Environ. Microbiol. 2018, 20, 1576–1589. [Google Scholar] [CrossRef] [Green Version]
- Fones, H.; Preston, G.M. The Impact of Transition Metals on Bacterial Plant Disease. FEMS Microbiol. Rev. 2013, 37, 495–519. [Google Scholar] [CrossRef] [Green Version]
- Chaturvedi, K.S.; Henderson, J.P. Pathogenic Adaptations to Host-Derived Antibacterial Copper. Front. Cell. Infect. Microbiol. 2014, 4, 3. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Chang, F.M.J.; Giedroc, D.P. Copper Transport and Trafficking at the Host-Bacterial Pathogen Interface. Acc. Chem. Res. 2014, 47, 3605–3613. [Google Scholar] [CrossRef]
- Johnson, M.D.L.; Kehl-Fie, T.E.; Klein, R.; Kelly, J.; Burnham, C.; Mann, B.; Rosch, J.W. Role of Copper Efflux in Pneumococcal Pathogenesis and Resistance to Macrophage-Mediated Immune Clearance. Infect. Immun. 2015, 83, 1684–1694. [Google Scholar] [CrossRef] [Green Version]
- Hyre, A.N.; Kavanagh, K.; Kock, N.D.; Donati, G.L.; Subashchandrabose, S. Copper Is a Host Effector Mobilized to Urine during Urinary Tract Infection to Impair Bacterial Colonization. Infect. Immun. 2017, 85, e01041-16. [Google Scholar] [CrossRef] [Green Version]
- Stojanov, M.; Moreillon, P.; Sakwinska, O. Excision of Staphylococcal Cassette Chromosome Mec in Methicillin-Resistant Staphylococcus Aureus Assessed by Quantitative PCR Microbiology. BMC Res. Notes 2015, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Archer, G.L. Roles of CcrA and CcrB in Excision and Integration of Staphylococcal Cassette Chromosome Mec, a Staphylococcus Aureus Genomic Island. J. Bacteriol. 2010, 192, 3204–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosario-Cruz, Z.; Eletsky, A.; Daigham, N.S.; Al-Tameemi, H.; Swapna, G.V.T.; Kahn, P.C.; Szyperski, T.; Montelione, G.T.; Boyd, J.M. The CopBL Operon Protects Staphylococcus Aureus from Copper Toxicity: CopL Is an Extracellular Membrane–Associated Copper-Binding Protein. J. Biol. Chem. 2019, 294, 4027–4044. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koboldt, D.C.; Larson, D.E.; Wilson, R.K. Using Varscan 2 for Germline Variant Calling and Somatic Mutation Detection. Curr. Protoc. Bioinform. 2013, 44, 15.4.1–15.4.17. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Larsen, J.; Enright, M.C.; Godoy, D.; Spratt, B.G.; Larsen, A.R.; Skov, R.L. Multilocus Sequence Typing Scheme for Staphylococcus Aureus: Revision of the Gmk Locus. J. Clin. Microbiol. 2012, 50, 2538–2539. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Snippy: Fast Bacterial Variant Calling from NGS Reads [Internet]. Available online: www.github.com (accessed on 15 January 2020).
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Thi Hoang, D.; Chernomor, O.; von Haeseler, A.; Quang Minh, B.; Sy Vinh, L.; Rosenberg, M.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Kaya, H.; Hasman, H.; Larsen, J.; Stegger, M.; Johannesen, T.B.; Allesøe, R.L.; Lemvigh, C.K.; Aarestrup, F.M.; Lund, O.; Larsen, A.R. SCC Mec Finder, a Web-Based Tool for Typing of Staphylococcal Cassette Chromosome Mec in Staphylococcus Aureus Using Whole-Genome Sequence Data. mSphere 2018, 3, e00612-17. [Google Scholar] [CrossRef] [Green Version]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia Coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- Maria, A.; Tetzschner, M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia Coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef] [Green Version]
- Altschup, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 2006, 34, W32–W37. [Google Scholar] [CrossRef] [Green Version]
- Russell Editor, D.J. Multiple Sequence Alignment; Humana Press: New work, NY, USA, 2014; Volume 15, pp. 75–101. [Google Scholar]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and Precise Alignment of Raw Reads against Redundant Databases with KMA. BMC Bioinform. 2018, 19, 1–8. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, I.; Oggioni, M.R.; Knight, D.; Curiao, T.; Coque, T.; Kalkanci, A.; Martinez, J.L.; Baldassarri, L.; Orefici, G.; Yetiş, Ü.; et al. Evaluation of Epidemiological Cut-off Values Indicates That Biocide Resistant Subpopulations Are Uncommon in Natural Isolates of Clinically-Relevant Microorganisms. PLoS ONE 2014, 9, e86669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstein, M.P.; Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 20th Information Supplement CLSI Document M100-S20. Available online: https://kaldur.landspitali.is/focal/gaedahandbaekur/gnhsykla.nsf/5e27f2e5a88c898e00256500003c98c2/9c4f4955ccb9f8100025751a0046b075/$FILE/M100-S20%20Performance%20Standards%20for%20Antimicrobial%20Susceptibility%20Testing.pdf (accessed on 30 July 2021).
- Barbier, F.; Lebeaux, D.; Hernandez, D.; Delannoy, A.S.; Caro, V.; François, P.; Schrenzel, J.; Ruppé, E.; Gaillard, K.; Wolff, M.; et al. High Prevalence of the Arginine Catabolic Mobile Element in Carriage Isolates of Methicillin-Resistant Staphylococcus Epidermidis. J. Antimicrob. Chemother. 2011, 66, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Onishi, M.; Urushibara, N.; Kawaguchiya, M.; Ghosh, S.; Shinagawa, M.; Watanabe, N.; Kobayashi, N. Prevalence and Genetic Diversity of Arginine Catabolic Mobile Element (ACME) in Clinical Isolates of Coagulase-Negative Staphylococci: Identification of ACME Type I Variants in Staphylococcus Epidermidis. Infect. Genet. Evol. 2013, 20, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Soroush, S.; Jabalameli, F.; Taherikalani, M.; Amirmozafari, N.; Imani Fooladi, A.A.; Asadollahi, K.; Beigverdi, R.; Emaneini, M. Investigation of Biofilm Formation Ability, Antimicrobial Resistance and the Staphylococcal Cassette Chromosome Mec Patterns of Methicillin Resistant Staphylococcus Epidermidis with Different Sequence Types Isolated from Children. Microb. Pathog. 2016, 93, 126–130. [Google Scholar] [CrossRef]
- Ito, T.; Katayama, Y.; Hiramatsu, K. Cloning and Nucleotide Sequence Determination of the Entire Mec DNA of Pre-Methicillin-Resistant Staphylococcus Aureus N315. Antimicrob. Agents Chemother. 1999, 43, 1449–1458. [Google Scholar] [CrossRef] [Green Version]
- Ahman, J.; Matuschek, E.; Webster, C.; Kahlmeter, G. Methicillin Resistance in Coagulase-Negative Staphylococci-Can. We Use the Same Screening Breakpoints for All Species? ESCMID eLibrary, 2017 ECCMID Conference Vienna. Available online: https://www.escmid.org/escmid_publications/escmid_elibrary/?q=Methicillin+Resistance+in+Coagulase-Negative+Staphylococci-Can&id=2173&L=0&x=0&y=0&tx_solr%5Bfilter%5D%5B0%5D=main_category%253ABacterial%2BSusceptibility%2B%2526%2BResistance&tx_solr%5Bfilter%5D%5B1%5D=author%253AJenny%2BAhman (accessed on 30 July 2021).
- Ciusa, M.L.; Furi, L.; Knight, D.; Decorosi, F.; Fondi, M.; Raggi, C.; Coelho, J.R.; Aragones, L.; Moce, L.; Visa, P.; et al. A Novel Resistance Mechanism to Triclosan That Suggests Horizontal Gene Transfer and Demonstrates a Potential Selective Pressure for Reduced Biocide Susceptibility in Clinical Strains of Staphylococcus Aureus. Int. J. Antimicrob. Agents 2012, 40, 210–220. [Google Scholar] [CrossRef] [Green Version]
- Grossoehme, N.; Kehl-Fie, T.E.; Ma, Z.; Adams, K.W.; Cowart, D.M.; Scott, R.A.; Skaar, E.P.; Giedroc, D.P. Control of Copper Resistance and Inorganic Sulfur Metabolism by Paralogous Regulators in Staphylococcus Aureus. J. Biol. Chem. 2011, 286, 13522–13531. [Google Scholar] [CrossRef] [Green Version]
- Hijazi, K.; Mukhopadhya, I.; Abbott, F.; Milne, K.; Al-Jabri, Z.J.; Oggioni, M.R.; Gould, I.M. Susceptibility to Chlorhexidine amongst Multidrug-Resistant Clinical Isolates of Staphylococcus Epidermidis from Bloodstream Infections. Int. J. Antimicrob. Agents 2016, 48, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Miragaia, M.; de Lencastre, H.; Perdreau-Remington, F.; Chambers, H.F.; Higashi, J.; Sullam, P.M.; Lin, J.; Wong, K.I.; King, K.A.; Otto, M.; et al. Genetic Diversity of Arginine Catabolic Mobile Element in Staphylococcus Epidermidis. PLoS ONE 2009, 4, e7722. [Google Scholar] [CrossRef] [Green Version]
- Schwan, W.R.; Warrener, P.; Keunz, E.; Kendall Stover, C.; Folger, K.R. Mutations in the CueA Gene Encoding a Copper Homeostasis P-Type ATPase Reduce the Pathogenicity of Pseudomonas Aeruginosa in Mice. Int. J. Med. Microbiol. 2005, 295, 237–242. [Google Scholar] [CrossRef]
- Shafeeq, S.; Yesilkaya, H.; Kloosterman, T.G.; Narayanan, G.; Wandel, M.; Andrew, P.W.; Kuipers, O.P.; Morrissey, J.A. The Cop Operon Is Required for Copper Homeostasis and Contributes to Virulence in Streptococcus Pneumoniae. Mol. Microbiol. 2011, 81, 1255–1270. [Google Scholar] [CrossRef] [Green Version]
- Ladomersky, E.; Khan, A.; Shanbhag, V.; Cavet, J.S.; Chan, J.; Weisman, G.A.; Petris, M.J. Host and Pathogen Copper-Transporting P-Type ATPases Function Antagonistically during Salmonella Infection. Infect. Immun. 2017, 85, e00351-17. [Google Scholar] [CrossRef] [Green Version]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I. The Isoleucyl-TRNA Synthetase Mutation V588F Conferring Mupirocin Resistance in Glycopeptide-Intermediate Staphylococcus Aureus Is Not Associated with a Significant Fitness Burden. J. Antimicrob. Chemother. 2004, 53, 102–104. [Google Scholar] [CrossRef]
- Furi, L.; Haigh, R.; al Jabri, Z.J.H.; Morrissey, I.; Ou, H.Y.; León-Sampedro, R.; Martinez, J.L.; Coque, T.M.; Oggioni, M.R. Dissemination of Novel Antimicrobial Resistance Mechanisms through the Insertion Sequence Mediated Spread of Metabolic Genes. Front. Microbiol. 2016, 7, 1008. [Google Scholar] [CrossRef] [Green Version]
| Isolate | Date | MLST | SCCmec Type | ACME Operons & (Type) |
|---|---|---|---|---|
| 5034 | November 2018 | 2 | IV (2B) | arc & opp3 (I) |
| 4175 | November 2018 | 2 | IV (2B) | arc & opp3 (I) |
| 60242 | August 2018 | 2 | IV (2B) | arc & opp3 (I) |
| 3193 | August 2018 | 2 | IV (2B) | arc & opp3 (I) |
| 2562 | August 2018 | 2 | IV (2B) | arc & opp3 (I) |
| 2074 | September 2018 | 2 | - | arc & opp3 (I) |
| 5426 | December 2018 | 328 | IV (2B&5) | arc & opp3 (I) |
| 5506 | November 2018 | 328 | V (5C2) | arc & opp3 (I) |
| 57114 | July 2018 | new | Iva (2B) | arc & opp3 (I) |
| 2751 | October 2018 | 369 | IVa (2B) | arc (II) |
| 292 | January 2019 | 598 | IV (2B&5) | arc & opp3 (I) |
| 640 | October 2018 | 2 | - | arc & opp3 (I) |
| 1426 | October 2018 | 59 | - | arc & opp3 (I) |
| 4174 | November 2018 | 59 | Iva (2B) | arc (II) |
| 4320 | November 2018 | 87 | kdp, arc & opp3 (V) | |
| 4231 | November 2018 | 736 | Iva (2B) | kdp, arc & opp3 (V) |
| 4173 | October 2018 | 736 | Iva (2B) | kdp, arc & opp3 (V) |
| 793 | December 2018 | 73 | - | arc & opp3 (I) |
| 4561 | November 2018 | 200 | - | arc & opp3 (I) |
| 9407 | September 2018 | 32 | - | kdp, arc & opp3 (V) |
| 5459 | January 2019 | 210 | V (5C2) | kdp, arc & opp3 (V) |
| 1080 | August 2018 | 2 | II (2A) | arc & opp3 (I) |
| 60038 | August 2018 | new | Iva (2B) | opp3 (III) |
| 6982 | November 2018 | new | - | arc & opp3 (I) |
| Antibiotic | No of Isolates (%) | ||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Penicillin G (P) | 0 (0%) | - | 24 (100%) |
| Amoxycillin/clavulanic acid (AMC) | 14(58%) | - | 10 (42%) |
| Oxacillin (OX) | 2(8%) | - | 22 (92%) |
| Erythromycin (E) | 2 (8%) | 0 (0%) | 22 (92%) |
| Cefoxitin (FOX) | 4 (16%) | - | 20 (83%) |
| Ciprofloxacin (CIP) | 9 (38%) | 1 (4%) | 14 (58%) |
| Gentamicin (CN) | 13 (54%) | 1 (4%) | 10 (42%) |
| Clindamycin (DA) | 13 (54%) | 0 (0%) | 11 (46%) |
| Rifampicin (RD) | 23 (96%) | 0 (0%) | 1 (4%) |
| Chloramphenicol (C) | 22 (92%) | 0 (0%) | 2 (8%) |
| Tigecycline (TGC) | 24 (100%) | - | 0 (0%) |
| * Vancomycin (VA, MIC) | 24 (100%) | - | 0 (0%) |
| ** Teicoplanin (TEC MIC) | 24 (100%) | - | 0 (0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Jabri, Z.; AL-Shabibi, Z.; AL-Bimani, A.; AL-Hinai, A.; AL-Shabibi, A.; Rizvi, M. Whole Genome Sequencing of Methicillin-Resistant Staphylococcus epidermidis Clinical Isolates Reveals Variable Composite SCCmec ACME among Different STs in a Tertiary Care Hospital in Oman. Microorganisms 2021, 9, 1824. https://doi.org/10.3390/microorganisms9091824
Al-Jabri Z, AL-Shabibi Z, AL-Bimani A, AL-Hinai A, AL-Shabibi A, Rizvi M. Whole Genome Sequencing of Methicillin-Resistant Staphylococcus epidermidis Clinical Isolates Reveals Variable Composite SCCmec ACME among Different STs in a Tertiary Care Hospital in Oman. Microorganisms. 2021; 9(9):1824. https://doi.org/10.3390/microorganisms9091824
Chicago/Turabian StyleAl-Jabri, Zaaima, Zahra AL-Shabibi, Atika AL-Bimani, Amal AL-Hinai, Ammar AL-Shabibi, and Meher Rizvi. 2021. "Whole Genome Sequencing of Methicillin-Resistant Staphylococcus epidermidis Clinical Isolates Reveals Variable Composite SCCmec ACME among Different STs in a Tertiary Care Hospital in Oman" Microorganisms 9, no. 9: 1824. https://doi.org/10.3390/microorganisms9091824
APA StyleAl-Jabri, Z., AL-Shabibi, Z., AL-Bimani, A., AL-Hinai, A., AL-Shabibi, A., & Rizvi, M. (2021). Whole Genome Sequencing of Methicillin-Resistant Staphylococcus epidermidis Clinical Isolates Reveals Variable Composite SCCmec ACME among Different STs in a Tertiary Care Hospital in Oman. Microorganisms, 9(9), 1824. https://doi.org/10.3390/microorganisms9091824






