Abstract
Strains of the food-borne pathogen Listeria (L.) monocytogenes have diverse virulence potential. This study focused on the virulence of three outbreak strains: the CC1 strain PF49 (serovar 4b) from a cheese-associated outbreak in Switzerland, the clinical CC2 strain F80594 (serovar 4b), and strain G6006 (CC3, serovar 1/2a), responsible for a large gastroenteritis outbreak in the USA due to chocolate milk. We analysed the genomes and characterized the virulence in vitro and in vivo. Whole-genome sequencing revealed a high conservation of the major virulence genes. Minor deviations of the gene contents were found in the autolysins Ami, Auto, and IspC. Moreover, different ActA variants were present. Strain PF49 and F80594 showed prolonged survival in the liver of infected mice. Invasion and intracellular proliferation were similar for all strains, but the CC1 and CC2 strains showed increased spreading in intestinal epithelial Caco2 cells compared to strain G6006. Overall, this study revealed long-term survival of serovar 4b strains F80594 and PF49 in the liver of mice. Future work will be needed to determine the genes and molecular mechanism behind the long-term survival of L. monocytogenes strains in organs.
1. Introduction
The Gram-positive bacterium Listeria monocytogenes is the causative agent of listeriosis, a rare, but severe food-borne infection in humans and animals. In healthy individuals, listeriosis is usually restricted to a non-invasive self-limited gastroenteritis, which typically occurs 24 h after ingestion of a large inoculum of bacteria and usually lasts two days [1]. In immunocompromised individuals and elderly, an invasive and systemic infection can occur within an incubation time of 20 to 30 days with a high mortality rate of 25–30%. Furthermore, pregnant women and neonates are a high-risk group. Invasive L. monocytogenes infection of non-pregnant individuals typically manifests as bacteremia/septicemia or as central nervous system infection including meningitis, meningoencephalitis and with lower prevalence also rhombencephalitis and brain abscess [2,3,4]. In pregnant women, L. monocytogenes can infect the fetus or unborn, leading to spontaneous abortion or stillbirth. Neonatal listeriosis may represent as sepsis or meningitis with severe consequences and a high case fatality of 20% [5].
There is evidence for the difference in the virulence and pathogenic potential of L. monocytogenes strains [6,7,8]. L. monocytogenes strains can be grouped into four major phylogenetic lineages (I, II, III and IV), and most clinical strains are of phylogenetic lineage I and II [9]. Of the 13 lineage-related serovars only three, namely 1/2a within lineage II, and 1/2b and 4b within lineage I, are responsible for the majority of the clinical cases of human (and animal) listeriosis [10]. Especially, serovar 4b strains cause over 50% of listeriosis cases worldwide in spite of being much less frequently found in food samples as compared to strains from serovar 1/2a and b [6,10]. Furthermore, L. monocytogenes strains can be assigned to multi-locus sequence types, which are grouped into clonal complexes (CC). Multi-locus sequence typing is based on the sequences of seven housekeeping genes. Recent advances in Listeria infection biology have shown that certain CCs such as CC1, CC2, CC4, and CC6 are more frequently associated with clinical cases, whereas others like CC9 and CC121 are mainly of food-borne origin [6,7,8,11].
The understanding of the pathophysiology of Listeria infection has made major steps forward, but there are still new insights to come. After ingestion and survival in the stomach, L. monocytogenes traverses the intestinal epithelial barriers via goblet cells and enterocytes into the lamina propria and spreads into the bloodstream through the lymph nodes to disseminate to the target organs of replication, the liver and spleen [2,12]. In case L. monocytogenes trespasses the liver barrier, the bacteria shuttle to secondary sites of clinical manifestation, the placenta, and the central nervous system. An arsenal of virulence factors mediates the complex pathogenicity of L. monocytogenes among them the internalins InlA and InlB, which are required for the entry into non-phagocytic cells [12,13,14]. Subsequent to the invasion step, L. monocytogenes is temporarily enclosed in a phagocytic vacuole, before it escapes into the host cell cytoplasm, which is mediated by the listeriolysin O (LLO) and the phospolipases (PlC) A and B. Once inside the host cell cytosol, L. monocytogenes can survive, replicate and move to the neighboring cell. The actin-assembly-inducing protein ActA is essential for intracellular motility and cell-to-cell spread. Additionally, there are numerous other virulence factors that e.g., hijack cellular processes, modulate the immune system reach even the eukaryotic nucleus and influence gene expression [12].
One puzzling issue regarding the pathogenicity of L. monocytogenes is the long incubation period of the invasive manifestation in humans and animals, varying from 24 h in cases showing gastroenteritis to 9 in cases with central nervous system infection and 27.5 days in cases with abortions, respectively [15]. This unusually long incubation period required for the manifestation of clinical systemic listeriosis implies that L. monocytogenes might persist in host tissues for extended periods without causing overt clinical signs. In a recently published paper, Vazquez-Boland and co-workers stipulated the hypothesis that hypervirulent strains, which survive longer in the liver and the spleen of the host, have a higher probability of secondary dissemination of the brain and the placenta [16]. Two hypervirulent 4b strains P14 and PF49, both assigned to CC1, were still detectable in the mouse liver three weeks post infection [16]. P14 was isolated from an adult patient with central nervous symptoms in Spain, and PF49 is an epidemic strain from a cheese-associated outbreak in Switzerland [17]. This observation is striking since most mouse studies have shown that the infection is cleared from the liver seven days post infection, when a sublethal dose of L. monocytogenes is administrated.
In this work, we follow-up this hypothesis using the serovar 4b strain PF49 (CC1) for an in-depth characterisation of its virulence properties. PF49 was associated with a large community-acquired epidemic of invasive listeriosis with a high proportion of brain infections (79%) [17]. We compared the genome of strain PF49 with strains F80594 (CC2) and G6006 (CC3). Strain F80594, also serovar 4b, has caused gastrointestinal listeriosis in a mainly healthy population [18] and strain G6006 (serovar 1/2b) was isolated during a large outbreak of gastroenteric listeriosis caused by the consumption of a heavily contaminated milk product [19] (Table 1). We characterized the virulence potential of these listeriosis strains in intestinal epithelial, hepatocytic, and phagocytic cells. We aimed to mimic the natural route of infection (translocation across the epithelium, infection of the liver, and contact with phagocytes in the bloodstream) and determined the pathogenicity in a mouse infection model. The laboratory isolate EGD was included as a control.

Table 1.
Strain information and epidemiological data.
2. Materials and Methods
2.1. Bacterial Strains
The strains were stored at −80 °C in 20% glycerol phosphate-buffered saline, pH 7.4 (PBS) and grown in brain-heart infusion (BHI, Merck, Darmstadt, Germany) broth or on agar at 37 °C. Inocula for in-vitro and in-vivo studies were prepared from early stationary phase cultures. Bacteria were harvested by centrifugation, washed in PBS, resuspended in glycerol-PBS and stored at −80 °C in 500 µL aliquots. The exact number of bacteria in the frozen aliquots was calculated by plate counting.
2.2. DNA Isolation, Genome Sequencing, and Analysis
The L. monocytogenes strains were cultivated under aerobic conditions at 37 °C in BHI with 125 rpm shaking and harvested by centrifugation. The resulting pellet was used for DNA isolation using the QIAGEN genomic-tip columns and buffers (QIAGEN), following the recommendations of the manufacturer. For G6006 and PF49, genome sequencing was performed using an Illumina GAII genome analyzer at the University of Veterinary Medicine Vienna, Austria. Sequencing was performed using paired-end sequencing technology and 100 bp read-length using Illumina standard protocols. For F80594, genome sequencing was performed using an Illumina HiSeq2000 sequencer at the Campus Science Support Facilities (CSF) Next-Generation Sequencing unit, Vienna, Austria, using paired-end sequencing technology and 100 bp read length. Three (F80594) and ten (G6006, PF49) million reads were used for a de novo assembly using ABySS [20] with a k-mer size of 64. For G6006, the assembly resulted in 28 contigs (>500 bp) with an average coverage of 319×. For PF49, the assembly resulted in 26 contigs (>500 bp), for which the average coverage was 330×. For F80594, the assembly resulted in 24 contigs with an average coverage of 114×. The draft genome sequences of the L. monocytogenes strains were annotated and analyzed using PATRIC, www.patricbrc.org (accessed on 15 June 2021) [21]. The presence and absence of key virulence genes was determined using BlastP searches using a list of 95 virulence proteins as recently described [22].
2.3. Mouse Infection Assays
Groups of female BALB/C mice weighing 18 to 22 g (six per strain and time point, Charles River) were infected intravenously (iv) with a sublethal dose (5 × 103 CFU) of L. monocytogenes and the bacterial load in the liver was monitored at time points 3, 5, 7, 10, 12, 14 and 20 days post infection (p.i.). At the specified time points, the mice were humanely sacrificed by exposing them to carbon dioxide. The liver was aseptically removed and homogenised in 10 mL PBS, and the number of CFU per organ was determined by standard plate counting. The mean 50% lethal doses (LD50) were tested using groups of five BALB/C mice, which were injected with 200 μL suspensions containing 103, 104, 105, 106, and 107 bacteria. The survival was monitored during a 7-day period. The control group was injected with 200 μL sterile PBS; LD50 values were determined according to the method of Reed and Muench [23].
2.4. In-Vitro Virulence Assays
Invasiveness, intracellular proliferation, and cell-to-cell spread were determined using human intestinal epithelial Caco-2, the mouse macrophage-like P388D1, and the mouse liver TIB-73 cell lines. Cells were cultivated in 24-well dishes (Greiner, Frickenhausen, Germany) in 1 mL of RPMI 1640 medium supplemented with 2 mM L-glutamine and 10% heat-inactivated foetal calf (FCS) serum under 5% CO2–95% O2 atmosphere at 37 °C. Cells were seeded in RPMI-complete medium 72 h prior to infection. The medium was changed 24 h before infection. A gentamicin protection assay was used to determine the invasiveness. Semiconfluent (80%) cell monolayers were infected with a L. monocytogenes suspension in RPMI at a multiplicity of infection of 20 for 45 min (P388D1 macrophages) and 60 min (Caco-2 and TIB-73 cells), washed three times with PBS and incubated for 30 min with fresh RPMI medium supplemented with 25 µg/mL gentamicin (Serva, Heidelberg, Germany) to kill extracellular bacteria. The cells were washed, lysed by the addition of 1 mL ice-cold distilled water followed by incubation on ice for 15 min, and the intracellular bacterial numbers were determined by plate counting using tryptic soy agar. The intracellular proliferation was determined by calculating the intracellular growth coefficient with the formula IGC = (CFU6h − CFUinvasion)/CFUinvasion. To measure the cell-to-cell spread, cells were infected with strains harbouring the plasmid pLSV16gfp-a. Transformation of L. monocytogenes PF49, F80594, G6006, and EGD was performed as described earlier [24]. The plasmid stability was confirmed for at least 36 h in cell culture medium supplemented with 7.5 µg tetracycline. Infection was performed as described above and the number of intracellular bacteria was counted after 6 h. For flow cytometry analysis, Caco-2 cells were detached from the dishes by incubation with 200 µL of trypsin solution (Trypsin-EDTA; Gibco Invitrogen Life Tech., Paisley, UK) for 5 min. The cells were then suspended in 1 mL PBS and transferred into plastic tubes. GFP fluorescence emitted by intracellular bacteria was measured by flow cytometry using an Epics Elite ESP cell sorter (Coulter, Krefeld, Germany) and the percentage of positively stained host cells was determined (488 nm line of an argon ion laser, green light channel: 525/10 nm bandpass filter, counts: 10,000 cells per sample per run). All experiments were repeated three times.
2.5. Statistics
Statistical analysis was performed using Excel16.0 and SPSS.24 software. The mean values and standard deviations (SD) were calculated. The Welch-test was used to determine variance homogeneity. The Tukey-HSD post hoc test (variance homogeneity) was used to determine significant differences between the IGC and the percentage of GFP positive cells of the different strains and the number of bacterial load in the liver at the different time points. p < 0.05 was considered as a statistically significant difference.
3. Results
3.1. Whole Genome Sequencing
For the 1983–1987 Swiss Vacherin cheese outbreak, the source of strain PF49, one additional complete genome was available (strain LL195) [25]. Additional to G6006, two genomes were available from strains of the milk outbreak in Illinois: FSL-R2-503 (G6054) and FSL-R2-502 (Table 2). The genome sequences of all listeriosis outbreak isolates revealed typical features of L. monocytogenes genomes. A general overview of the genomic properties and features is provided in Table 2. None of the strains contained a plasmid except for strain FSL-F2-502, which harboured a 57 kbp plasmid. The strains harboured between one and three prophages.

Table 2.
General genomic features of L. monocytogenes outbreak strains.
The Blast search using 95 L. monocytogenes virulence factors as query sequences, revealed a high conservation of the majority of virulence factors (same length and an amino acid identity higher than 90%). Among the major virulence factors, we observed only differences in the ActA sequence. F80594, G6006, FSL-R2-503 and FSL-R2-502 harboured an ActA4 variant which is comprised of 604 amino acids, whereas PF49 and LL195 had an ActA3 variant with 604 amino acids [26] (Table 3). Interestingly, PF49 harboured a truncated Vip protein (Lmo0320) resulting in 335 amino acids compared to 399 amino acids in EGD-e. Vip in the other strains had a length of 418 amino acids. G6006 and the other outbreak strains harboured the aut gene, encoding Auto, an autolysin which is important for L. monocytogenes virulence [27]. In contrast, Auto is absent from F80594 and PF49, which instead harbored IspC, another autolysin primarily found in serovar 4b strains [28]. Another notable difference between the strains studied here were different variants of Ami, an additional autolysin encoded by the lmo2588 gene [29,30]. G6006 encodes a full-length homolog of the EGD-e Ami consisting of 917 amino acids and sharing 99% amino acid identity, whereas F80954 and PF49 encoded a shorter version of Ami with 770 amino acids with 72% amino acid identity to the EGD-e Ami. The shorter Ami variants are due to a lower number of C-terminal GW repeats. Moreover, all strains harboured an Internalin J (InlJ1) variant, and the listeriolysin S locus was present in all strains except strain F80594.

Table 3.
Virulence-related genomic features of L. monocytogenes outbreak strains PF49, F80594 and G6006 included in this study. Other sequenced outbreak related strains (for the Swiss outbreak: LL195; the US outbreak: FSL-R2_503, FSL-R2-502) were analysed in addition. Amino acid (AA) identities are only shown when lower than 90%.
The additional outbreak strain genomes showed the same genetic features as the isolates included in the in-vitro and in-vivo assays with a few exceptions: there are possible truncations in InlJ and GtcA in LL195 compared to PF49 and a 57 kbp plasmid is only present in FSL-F2-502.
3.2. Animal Behaviour, Recovery of L. monocytogenes from the Liver and LD50
The behaviour of the mice was monitored visually daily post infection. Mice of groups infected with strain PF49, F80594, and EGD became apathetic three to five days after infection, displaying shaggy fur and reduced mobility. Upon day 5, all mice infected with these L. monocytogenes strains returned to normal behaviour. In contrast to the other strains, the mice infected with strain G6006 remained healthy throughout the observation period.
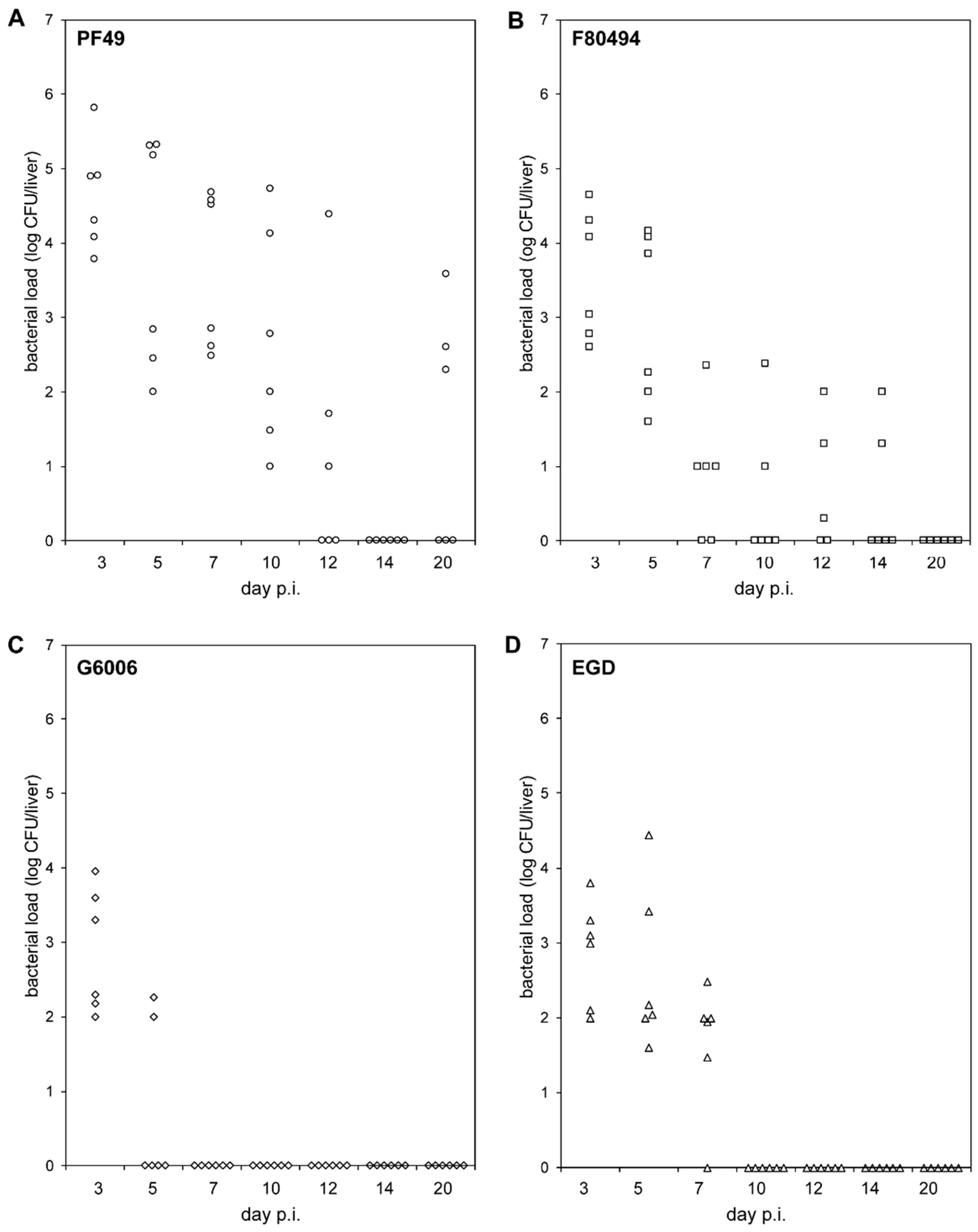
The two 4b strains, PF49 and F80594, showed different bacterial load patterns in the liver than strain G6006 and EGD (Figure 1, Table S1).
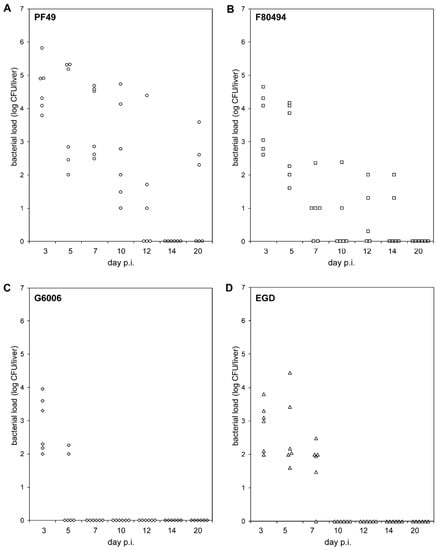
Figure 1.
In-vivo pathogenicity. Bacterial loads (log CFU) in the livers of BALB/C mice (n = 6) intravenously infected with strain PF49 ((A), circle), F80594 ((B), square), G6066 ((C), rhombus) and EGD ((D), triangle) 3, 5, 7, 10, 12, 14 and 20 days post infection (p.i.).
On day three (acute infection phase), infecting mice with strain PF49 resulted in a significantly higher bacterial load in the liver than with strain G6006 and EGD. On day 5, the bacterial load of mice infected with strain G6006 was significantly lower compared to the other three strains. PF49 and F80594 were still recovered at day 5 from the liver of all six animals in high numbers, whereas strain G6006 was only recovered from the livers of two animals in low numbers. At subsequent time points, strain G6006 was no longer detectable in the liver. Strain EGD was found at day 7 in the liver of six animals in low numbers. At later time points, EGD was not recovered from the liver. In contrast, strains PF49 and F80594 were still detectable in the liver up to day 20 and 14 p.i., respectively. These results demonstrate that PF49 and F80594 have an enhanced capacity to survive in vivo in the liver. Furthermore, the LD50 of strain G6006 and EGD (log CFU 5.83 ± 0 and 5.07 ± 0.3) was higher compared to strain PF49 and F80594 (log CFU 4.620 ± 0 and 4.66 ± 0.2).
3.3. Invasiveness, Intracellular Proliferation, and Spreading in Host Cells
To get insights into the invasiveness, intracellular proliferation, and spreading, we performed in-vitro assays.
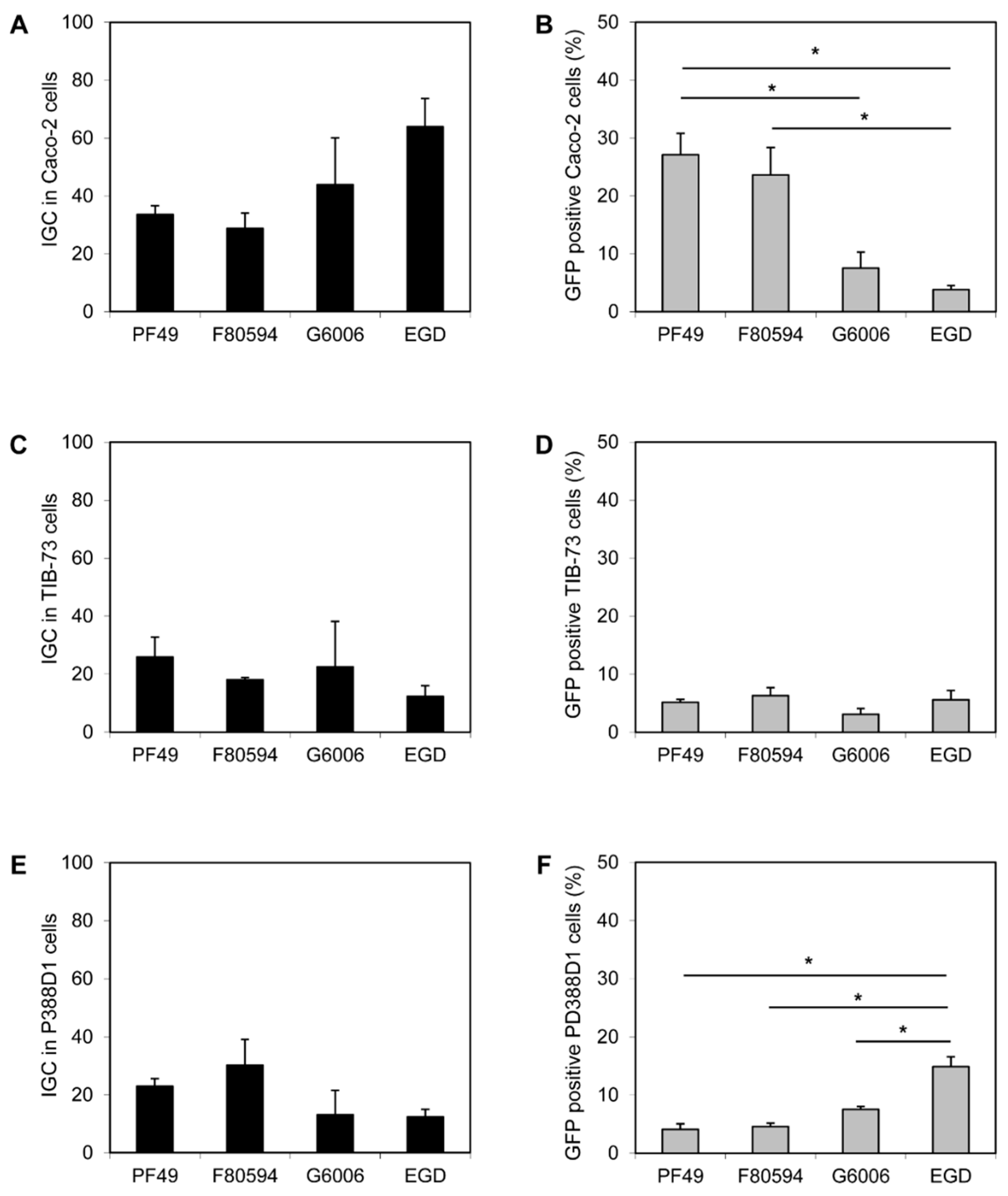
No difference in the invasion efficiency of all four strains (PF49, F80594, G6006, and EGD) was observed in Caco-2 and TIB-73 cells. Furthermore, the uptake of all strains by macrophage-like P388-D1 cells was equal (data not shown). In Caco-2 a higher intracellular proliferation (calculated as IGC) of strain G6006 and EGD was observed compared to strain PF49 and F80594; however, the effect was not statistically significant (Figure 2A). In contrast, a significantly higher number of GFP-positive Caco-2 cells were observed after infection with strain PF49 and F80594 (37.1 and 23.6%) compared to strain G6006 and EGD (7.5 and 3.8%, Figure 2B). In TIB-73 cells, no differences between the strains were observed for intracellular growth and cell-to-cell spread (Figure 2C,D). In macrophage-like P388D1 cells, the IGC was lower for strain G6006 and EGD than for strain PF49 and F80594. However, the differences were only small and not significant. In contrast, an increased cell-to-cell spread was observed for strain EGD compared to the epidemic strains (Figure 2E,F).
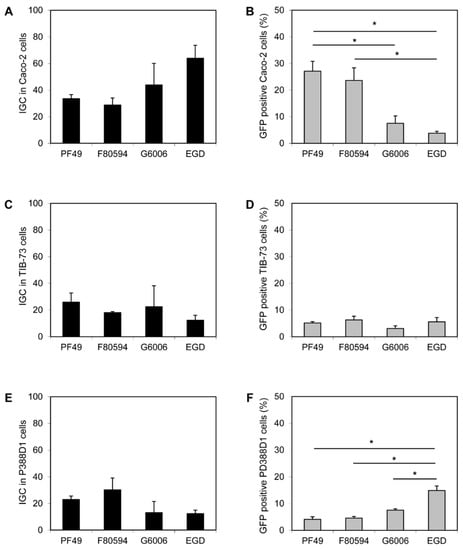
Figure 2.
In vitro virulence. Intracellular growth coefficient (IGC) in human intestinal epithelial Caco-2 (A), mouse liver TIB-73 (C) and mouse macrophage-like PD388D1 cells (E) and cell-to-cell spread, shown as percentage (%) of GFP positive Caco-2 (B), TIB-73 (D), PD388D1 cells (F). * statistically significant difference (p < 0.05).
4. Discussion
In this study, we combine genome analysis with in vitro virulence assays and mouse infection experiments for up to 20 days to unravel the pathogenicity of outbreak-associated L. monocytogenes strains. This study is thought to create some hypotheses why lineage 1 outbreak clones of serovar 4b might have led to a clinical outcome that was obviously very different from outbreaks of non-serovar 4b isolates. It further provides a hypothesis why some Listeria monocytogenes strains tend to persist in the murine body what was proven for PF49 and P14 [16].
The CC1 strain PF49 is an interesting isolate. Although the estimated infective dose was lower in the outbreak associated with PF49 than in the outbreak caused by G6006 (log CFU of 6 for strain PF49 and log CFU of 11 for strain G6006), the Swiss outbreak was remarkably severe [17,19]. A high percentage of meningoencephalitis cases was also recorded in the group of non-immunocompromised individuals what was not explainable [17]. The low LD50 value and the virulence properties proved PF49 to be a true hypervirulent isolate. This is in accordance with the findings of other studies that CC1 is overrepresented among clinical and epidemic strains, often isolated from patients with fever and no immunocompromising comorbidities [6,7,8]. Moreover, CC1 strains seem to be strongly associated with dairy products [31], which is consistent with the finding that soft cheese was the source of PF49. In contrast, the 4b strain F80594 was isolated from a small outbreak in a day-care facility in Denmark, in which three children (all of age two) were involved. The clinical manifestations were vomiting and fever. L. monocytogenes could be isolated from the blood of one patient [18]. The outcomes of the epidemiological follow-up investigations were limited. Neither an estimate of the ingested dose nor the exact food source are available. The isolate F80594 was assigned to CC2. Strains of CC2 were, like CC1 strains, more frequently isolated from clinical than from food-associated sources [6,11,32]. Moreover, CC2 strains are more frequently isolated from dairy than from meat and fish products [11,31]. The 1/2b serovar strain G6006 is assigned to CC3, an intermediate CC showing equal distribution among clinical and food isolates [6,31]. G6006 was responsible for a large outbreak of febrile gastroenteritis due to contaminated chocolate milk in the United States, including only one case with central nervous symptoms [19]. CC3 does not seem to be associated with a specific food type [11,31].
We detected differences in the colonization and clearance of the liver, and in the spreading capacity. In the mouse experiment, we detected that strains PF49 and F80494 showed significantly different colonization and clearance of the liver compared to strain G6006 and strain EGD. In line with the different clinical manifestations, the highest bacterial load was found in mice infected with strain PF49, followed by strain F80494 [17,18]. Genetic differences in virulence-associated genes between the two 4b strains, with an average nucleotide identity of 99.6%, were rare. PF49 harboured a different ActA variant than strains F80594 and G6006. F80594 and G6006 had an ActA4 variant, which is slightly longer than the ActA variant of strain PF49 and EGDe [26]. ActA is responsible for bacterial movement and cell-to-cell spread. Moreover, it has recently been shown to be also involved in aggregation as well as long-term colonization of the gut [33,34]. The effect of the different ActA variants on the virulence potential and pathogenicity of L. monocytogenes is not well understood. We can only speculate that the ActA3 variant of strain PF49 might cause the higher bacterial load at day three due to longer colonization of the gut.
Strains G6006 and EGD revealed lower bacterial loads in the liver at day three post infection, further decreasing at the following time points. This is the expected behaviour of L. monocytogenes in the liver of intravenously infected naïve wild-type mice. Clearance of strain G6006 was faster than that of strain EGD. On day seven, no bacteria were detectable in the liver. The decrease in the bacterial numbers is a consequence of an effective macrophage activation and protective immune response mediated by Th1 and CD8+ T-cells [35,36]. In contrast, bacteria were recovered from the liver of mice infected with the two 4b strains, up to day 14 for F80494 and day 20 for PF49, respectively. We speculate that some clones might escape the immune defence, thus leading to a more deferred style of infection. In addition to PF49, this phenomenon was found also in the CC1 strain P14, isolated from a patient with CNS manifestation during an outbreak in Spain [37]. Both 4b strains showed higher spreading in human intestinal epithelial Caco2 cells compared to strain G6066 and the lab strain EGD. However, these effects were only minor. Spreading in the mouse liver TIB-37 and macrophage-like PD388D1 cells was similar between the three outbreak strains. Only strain EGD showed significantly higher spreading capacities in the macrophage-like cells. No differences in the invasion efficiency, uptake by macrophage-like cells, and intracellular growth were observed.
For the sequence analysis, we have used a set of 95 L. monocytogenes virulence genes as query sequences for BlastP to identify virulence gene differences among the outbreak clones. As almost all of the major virulence genes were found highly similar to the reference sequences (>90% amino acid identity), we infer that the observed differences in virulence might be either due to posttranscriptional effects or unstudied metabolic pathways. However, some small differences in the virulence associated genes were observed. Notable is the presence/absence of the autolysins: IspC, known to be important for virulence [28], is only present in the two 4b strains, whereas G6006 harboured the autolysin Auto (Lmo1076). It has been speculated that IspC could be responsible for crossing the blood-brain barrier [28], which might contribute to the high percentage of central nervous system complications in the Swiss cheese outbreak. It is currently unclear if the presence of IspC might result in higher virulence in 4b strains, since Auto also has a role in virulence in L. monocytogenes [27]. Another genomic feature that might explain the difference in virulence in mice between the strains might be differences in the autolysin Ami, which is important for adhesion and internalization [29,30]. G6006 harbours an Ami homolog with high similarity (99% amino acid identity) and the same length (917 amino acids) as the functionally characterized EGD-e Ami. Interestingly, the Ami homologs in F80594 and PF49 are shorter (770 amino acids) and show lower amino acid identity (72%) to the EGD-e Ami. The lab strain EGD showed comparable virulence to G6006 in mice and similar spreading in Caco2 cells and Ami and Auto were most similar to that of G6006. Future studies are needed to identify the function and the virulence potential of the different autolysin homologs.
In this study, we showed that not only strains of CC1 [16], but also of CC2 have the potential to survive for extended time periods in the liver, enhancing the possibility for dissemination to the secondary organs. Experiments, e.g., transcriptome analysis of bacteria isolated from organs such as the liver, are needed to determine the molecular mechanism behind the long-term survival of some L. monocytogenes strains in this tissue. Additional studies including both a higher number of highly prevalent CCs overrepresented by clinical and epidemic strains and more strains per CC will be necessary to prove whether the higher pathogenicity and severe clinical manifestation is due to a prolonged survival capacity in the intestine, lymph nodes, liver, and spleen.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9081745/s1. Table S1. p-values comparing bacterial loads in the liver of animal infected with the different strains day 3 to 20 post infection (p.i.). p < 0.05 was considered as statistically significant difference (bold) as de-termined by the tukey-HSD posthoc test.
Author Contributions
Conceptualization, M.W., W.G. and J.A.V.-B.; methodology and investigation, M.W., J.S., S.S.-E. and K.R.; formal analysis, M.W., S.S.-E. and K.R.; resources, M.W., W.G.; writing—original draft preparation, M.W., S.S.-E., K.R. and J.A.V.-B.; writing—review and editing, M.W., S.S.-E., K.R.; visualization, K.R., M.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Austrian Jubilee Fund-ÖNB, grant number 9404. MW was funded by the Postgraduales Auslandsstipendium der Creditanstalt Austria and by the Austrian Science Fund (FWF), Schrödinger grant J-1694. Part of this work was created within a research project of the Austrian Competence Centre for Feed and Food Quality, Safety and Innovation (FFoQSI). The COMET-K1 competence centre FFoQSI is funded by the Austrian ministries BMK, BMDW and the Austrian provinces Lower Austria, Upper Austria and Vienna within the scope of COMET-Competence Centers for Excellent Technologies. The programme COMET is handled by the Austrian Research Promotion Agency FFG.
Institutional Review Board Statement
Animal experiments were conducted according to the regulations and guidelines in animal experimentation approved by the the Complutense University (animal facility registration no. 28079-I5ABC-M, Real Decreto 223/1988, Orden 13/10/1989, EU Directive 86/609/CEE).
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequences of the three strains sequenced in this study have been deposited in DDBJ/EMBL/GenBank under BioProject accession number PRJNA681816.
Acknowledgments
We thank Justin Daniels, Würzburg, Germany, and Bruno Gonzàlez-Zorn, Isabel Chico-Calero and Alberto Tierrez (all Madrid, Spain) for assistance in injecting the mice. Jürgen Kreft (Würburg, Germany) is gratefully thanked for fruitful discussion. L. monocytogenes strains PF49, F80594 and G6006 were kindly provided by J. Bille (Division of Infectious Diseases, University Hospital, Lausanne, Switzerland), P. Gerner-Smidt (Department of Gastrointestinal Infections, Division of Diagnostics, Statens Serum Institut, Copenhagen, Denmark) and B. Swaminathan (Centers for Disease Control and Prevention, Atlanta, Georgia, USA), respectively. L. monocytogenes EGD, serotype 1/2a, was provided by S. H. E. Kaufmann, Max-Planck Institute, Berlin, and was used throughout these studies as a control (referred to as EGD). Open Access Funding by the University of Veterinary Medicine Vienna.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Ooi, S.T.; Lorber, B. Gastroenteritis due to Listeria monocytogenes. Clin. Infect. Dis. 2005, 40, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Dominguez-Bernal, G.; Goebel, W.; Gonzalez-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef]
- Allerberger, F.; Wagner, M. Listeriosis: A resurgent foodborne infection. Clin. Microbiol. Infect. 2010, 16, 16–23. [Google Scholar] [CrossRef]
- Leclercq, A.; Charlier, C.; Lecuit, M. Global burden of listeriosis: The tip of the iceberg. Lancet. Infect. Dis. 2014, 14, 1027–1028. [Google Scholar] [CrossRef]
- Madjunkov, M.; Chaudhry, S.; Ito, S. Listeriosis during pregnancy. Arch. Gynecol. Obstet. 2017, 296, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Maury, M.M.; Tsai, Y.-H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Shah, M.K.; Burall, L.S.; Rakic-Martinez, M.; Datta, A.R. Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 2018, 102, 3475–3485. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chen, Y.; Gorski, L.; Ward, T.J.; Osborne, J.; Kathariou, S. Listeria monocytogenes source distribution analysis indicates regional heterogeneity and ecological niche preference among serotype 4b clones. MBio 2018, 9, e00396-18. [Google Scholar] [CrossRef]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Cartwright, E.J.; Jackson, K.A.; Johnson, S.D.; Graves, L.M.; Silk, B.J.; Mahon, B.E. Listeriosis outbreaks and associated food vehicles, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 1. [Google Scholar] [CrossRef]
- Painset, A.; Björkman, J.T.; Kiil, K.; Guillier, L.; Mariet, J.F.; Felix, B.; Amar, C.; Rotariu, O.; Roussel, S.; Perez-Reche, F.; et al. Liseq–Whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 2019. [Google Scholar] [CrossRef] [PubMed]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. [Google Scholar] [CrossRef]
- Camejo, A.; Carvalho, F.; Reis, O.; Leitão, E.; Sousa, S.; Cabanes, D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2011, 2, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Cossart, P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 2011, 108, 19484–19491. [Google Scholar] [CrossRef]
- Goulet, V.; Hebert, M.; Hedberg, C.; Laurent, E.; Vaillant, V.; De Valk, H.; Desenclos, J.C. Incidence of listeriosis and related mortality among groups at risk of acquiring listeriosis. Clin. Infect. Dis. 2012, 54, 652–660. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Wagner, M.; Scortti, M. Why are some Listeria monocytogenes genotypes more likely to cause invasive (brain, placental) infection? MBio 2020, 11, e0312. [Google Scholar] [CrossRef]
- Büla, C.J.; Bille, J.; Glauser, M.P. An epidemic of food-borne listeriosis in western switzerland: Description of 57 cases involving adults. Clin. Infect. Dis. 1995, 20, 66–72. [Google Scholar] [CrossRef]
- Heitmann, M.; Gerner-Smidt, P.; Heltberg, O. Gastroenteritis caused by Listeria monocytogenes in a private day-care facility. Pediatric Infect. Dis. J. 1997, 16, 827–828. [Google Scholar] [CrossRef]
- Dalton, C.B.; Austin, C.C.; Sobel, J.; Hayes, P.S.; Bibb, W.F.; Graves, L.M.; Swaminathan, B.; Proctor, M.E.; Griffin, P.M. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 1997, 336, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.; Zaiser, A.; Leitner, R.; Quijada, N.M.; Pracser, N.; Pietzka, A.; Ruppitsch, W.; Schmitz-Esser, S.; Wagner, M.; Rychli, K. Virulence characterization and comparative genomics of Listeria monocytogenes sequence type 155 strains. BMC Genom. 2020, 21, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple methodof estimating fifthy percent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Goetz, M.; Bubert, A.; Wang, G.; Chico-Calero, I.; Vazquez-Boland, J.A.; Beck, M.; Slaghuis, J.; Szalay, A.A.; Goebel, W. Microinjection and growth of bacteria in the cytosol of mammalian host cells. Proc. Natl. Acad. Sci. USA 2001, 98, 12221–12226. [Google Scholar] [CrossRef]
- Weinmaier, T.; Riesing, M.; Rattei, T.; Bille, J.; Arguedas-Villa, C.; Stephan, R.; Tasara, T. Complete genome sequence of Listeria monocytogenes LL195, a serotype 4b strain from the 1983-1987 listeriosis epidemic in Switzerland. Genome Announc. 2013, 1, e00152-12. [Google Scholar] [CrossRef]
- Balandyté, L.; Brodard, I.; Frey, J.; Oevermann, A.; Abril, C. Ruminant rhombencephalitis-associated Listeria monocytogenes alleles linked to a multilocus variable-number tandem-repeat analysis complex. Appl. Environ. Microbiol. 2011, 77, 8325–8335. [Google Scholar] [CrossRef]
- Cabanes, D.; Dussurget, O.; Dehoux, P.; Cossart, P. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 2004, 51, 1601–1614. [Google Scholar] [CrossRef]
- Wang, L.; Lin, M. A novel cell wall-anchored peptidoglycan hydrolase (autolysin), IspC, essential for Listeria monocytogenes virulence: Genetic and proteomic analysis. Microbiology 2008, 154, 1900–1913. [Google Scholar] [CrossRef][Green Version]
- Asano, K.; Sashinami, H.; Osanai, A.; Asano, Y.; Nakane, A. Autolysin amidase of Listeria monocytogenes promotes efficient colonization of mouse hepatocytes and enhances host immune response. Int. J. Med. Microbiol. 2011, 301, 480–487. [Google Scholar] [CrossRef]
- Milohanic, E.; Jonquières, R.; Cossart, P.; Berche, P.; Gaillard, J.L. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 2001, 39, 1212–1224. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Bowen, B.; Wiedmann, M.; Boor, K.J. Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 2012, 78, 2602–2612. [Google Scholar] [CrossRef] [PubMed]
- Travier, L.; Lecuit, M. Listeria monocytogenes ActA: A new function for a “classic” virulence factor. Curr. Opin. Microbiol. 2014, 17, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.J.; Wiedmann, M. Allelic exchange and site-directed mutagenesis probe the contribution of ActA amino-acid variability to phosphorylation and virulence-associated phenotypes among Listeria monocytogenes strains. FEMS Microbiol. Lett. 2006, 254, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Pamer, E.G. Immune responses to Listeria monocytogenes. Nat. Rev. Immunol. 2004, 4, 812–823. [Google Scholar] [CrossRef] [PubMed]
- D’Orazio, S.E.F. Innate and adaptive immune responses during Listeria monocytogenes infection. Microbiol. Spectr. 2019, 7, 12. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.; Ferrer, D.; Rocourt, J. Heterogeneity of strains of Listeria monocytogenes isolated during an outbreak of listeriosis among adults in Valencia in 1989. Enferm. Infecc. Microbiol. Clin. 1991, 9, 442–444. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).