Colonization of Clostridium butyricum in Rats and Its Effect on Intestinal Microbial Composition
Abstract
:1. Introduction
2. Materials and Methods
2.1. C. butyricum Cultivation
2.2. Primers and PCR Amplification
2.3. Real-Time qPCR
2.4. Animals and Experimental Groups
2.5. Genomic DNA Extraction from Pure Culture Strains and Fecal Samples of Rat Intestinal
2.6. 16S rDNA V3-V4 Region Sequencing and Bioinformatics Analysis
2.7. Statistical Analysis
3. Results
3.1. Specificity of Primer
3.2. Quantitation of C. Butyricum Populations in Rat Intestinal
3.3. C. butyricum Changes the Composition of Rat Intestinal Flora
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Engevik, M.A.; Versalovic, J. Biochemical features of beneficial microbes: Foundations for therapeutic microbiology. Bugs Drugs 2018, 5, 1–47. [Google Scholar]
- Liu, M.; Xie, W.; Wan, X.; Deng, T. Clostridium butyricum modulates gut microbiota and reduces colitis associated colon cancer in mice. Int. Immunopharmacol. 2020, 88, 106862. [Google Scholar] [CrossRef]
- Hooper, L.V.; Falk, P.G.; Gordon, J.I. Analyzing the molecular foundations of commensalism in the mouse intestine. Curr. Opin. Microbiol. 2000, 3, 79–85. [Google Scholar] [CrossRef]
- Guarner, F.; Malagelada, J.-R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Seo, M.; Inoue, I.; Tanaka, M.; Matsuda, N.; Nakano, T.; Awata, T.; Katayama, S.; Alpers, D.H.; Komoda, T. Clostridium butyricum MIYAIRI 588 improves high-fat diet-induced non-alcoholic fatty liver disease in rats. Digest. Dis. Sci. 2013, 58, 3534–3544. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, X.; Liu, P.; Zhao, J.; Sun, J.; Guan, W.; Johnston, L.J.; Levesque, C.L.; Fan, P.; He, T.; et al. Dietary Clostridium butyricum Induces a phased shift in fecal microbiota structure and increases the acetic acid-producing bacteria in a weaned piglet model. J. Agric. Food Chem. 2018, 66, 5157–5166. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Hold, G.L.; Barcenilla, A.; Stewart, C.S.; Flint, H.J. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 2002, 52, 1615–1620. [Google Scholar] [CrossRef] [Green Version]
- Shang, H.; Sun, J.; Chen, Y.Q. Clostridium butyricum CGMCC0313.1 modulates lipid profile, insulin resistance and colon homeostasis in obese mice. PLoS ONE 2016, 11, e0154373. [Google Scholar] [CrossRef] [PubMed]
- de Vries, M.C.; Vaughan, E.E.; Kleerebezem, M.; de Vos, W.M. Lactobacillus plantarum survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 2006, 16, 1018–1028. [Google Scholar] [CrossRef]
- Li, X.; Ringø, E.; Hoseinifar, S.H.; Lauzon, H.L.; Birkbeck, H.; Yang, D. The adherence and colonization of microorganisms in fish gastrointestinal tract. Rev. Aquacult. 2019, 11, 603–618. [Google Scholar] [CrossRef]
- Han, S.; Lu, Y.; Xie, J.; Fei, Y.; Zheng, G.; Wang, Z.; Liu, J.; Lv, L.; Ling, Z.; Berglund, B.; et al. Probiotic gastrointestinal transit and colonization after oral administration: A long journey. Front. Cell. Infect. Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Grimm, V.; Radulovic, K.; Riedel, C.U. Colonization of C57BL/6 Mice by a potential probiotic Bifidobacterium bifidum strain under germ-free and specific pathogen-free conditions and during experimental colitis. PLoS ONE 2015, 10, e0139935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, J. Ecological role of Lactobacilli in the gastrointestinal tract: Implications for fundamental and biomedical research. Appl. Environ. Microbiol. 2008, 74, 4985–4996. [Google Scholar] [CrossRef] [Green Version]
- Frese, S.A.; Hutkins, R.W.; Walter, J. Comparison of the colonization ability of autochthonous and allochthonous strains of Lactobacilli in the human gastrointestinal Tract. Adv. Microbiol. 2012, 2, 11. [Google Scholar] [CrossRef] [Green Version]
- Sato, R.; Tanaka, M. Intestinal distribution and intraluminal localization of orally administered Clostridium butyricum in rats. Microbiol. Immunol. 1997, 41, 665–671. [Google Scholar] [CrossRef]
- Bartosch, S.; Fite, A.; Macfarlane, G.T.; McMurdo, M.E. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 2004, 70, 3575–3581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, D.; Zhao, Q.; Yousaf, L.; Khan, J.; Xue, Y.; Shen, Q. Consumption of mung bean (Vigna radiata L.) attenuates obesity, ameliorates lipid metabolic disorders and modifies the gut microbiota composition in mice fed a high-fat diet. J. Funct. Foods 2019, 64, 103687. [Google Scholar] [CrossRef]
- Robles-Vera, I.; Toral, M.; de la Visitación, N.; Sánchez, M.; Gómez-Guzmán, M.; Romero, M.; Yang, T.; Izquierdo-Garcia, J.L.; Jiménez, R.; Ruiz-Cabello, J.; et al. Probiotics prevent dysbiosis and the rise in blood pressure in genetic hypertension: Role of short-chain fatty acids. Mol. Nutr. Food Res. 2020, 64, e1900616. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Canfora, E.E.; Meex, R.C.R.; Venema, K.; Blaak, E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019, 15, 261–273. [Google Scholar] [CrossRef]
- Kamada, N.; Seo, S.-U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Araki, Y.; Andoh, A.; Takizawa, J.; Takizawa, W.; Fujiyama, Y. Clostridium butyricum, a probiotic derivative, suppresses dextran sulfate sodium-induced experimental colitis in rats. Int. J. Mol. Med. 2004, 13, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Itoh, H. Hypertension as a metabolic disorder and the novel role of the gut. Curr. Hypertens. Rep. 2019, 21, 63. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Fu, Y.; Zhang, H.; Wang, J.; Zhu, J.; Wang, Y.; Guo, Y.; Wang, G.; Xu, T.; Chu, M.; et al. The hepatoprotective effect of the probiotic Clostridium butyricum against carbon tetrachloride-induced acute liver damage in mice. Food Funct. 2017, 8, 4042–4052. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, J.; Yang, B.; Chen, K.; Kong, Y.; Fang, N.; Gong, T.; Wang, F.; Ling, Z.; Liu, J. Effect of Clostridium butyricum against microglia-mediated neuroinflammation in alzheimer’s disease via regulating gut microbiota and metabolites butyrate. Mol. Nutr. Food Res. 2020, 64, e1900636. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, F.; Ling, Z.; Yu, X.; Chen, W.; Li, H.; Jin, J.; Pang, M.; Zhang, H.; Yu, J.; et al. Clostridium butyricum attenuates cerebral ischemia/reperfusion injury in diabetic mice via modulation of gut microbiota. Brain Res. 2016, 1642, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Kanai, T.; Mikami, Y.; Hayashi, A. A breakthrough in probiotics: Clostridium butyricum regulates gut homeostasis and anti-inflammatory response in inflammatory bowel disease. J. Gastroenterol. 2015, 50, 928–939. [Google Scholar] [CrossRef] [Green Version]
- Casula, G.; Cutting, S.M. Probiotics: Spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachisuka, Y.; Suzuki, I.; Morikawa, K.; Maeda, S. The effect of oxidation-reduction potential on spore germination, outgrowth, and vegetative growth of Clostridium tetani, Clostridium butyricum, and Bacillus subtilis. Microbiol. Immunol. 1982, 26, 803–811. [Google Scholar] [CrossRef]
- Yue, Y.; Xu, X.; Yang, B.; Lu, J.; Zhang, S.; Liu, L.; Nassar, K.; Zhang, C.; Zhang, M.; Pang, X.; et al. Stable colonization of orally administered Lactobacillus casei SY13 alters the gut microbiota. Biomed Res. Int. 2020, 2020, 5281639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunne, C.; Murphy, L.; Flynn, S.; O’Mahony, L.; O’Halloran, S.; Feeney, M.; Morrissey, D.; Thornton, G.; Fitzgerald, G.; Daly, C.; et al. Probiotics: From myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. In Lactic Acid Bacteria: Genetics, Metabolism and Applications, Proceedings of the Sixth Symposium on Lactic Acid Bacteria: Genetics, Metabolism and Applications, Veldhoven, The Netherlands, 19–23 September 1999; Konings, W.N., Kuipers, O.P., In’t Veld, J.H.J.H., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 279–292. [Google Scholar]
- Finnie, I.A.; Dwarakanath, A.D.; Taylor, B.A.; Rhodes, J.M. Colonic mucin synthesis is increased by sodium butyrate. Gut 1995, 36, 93–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sicard, J.-F.; Le Bihan, G.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of intestinal bacteria with components of the Intestinal mucus. Front. Cell. Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef]
- Tu, Q.V.; McGuckin, M.A.; Mendz, G.L. Campylobacter jejuni response to human mucin muc2: Modulation of colonization and pathogenicity determinants. J. Med. Microbiol. 2008, 57, 795–802. [Google Scholar] [CrossRef] [Green Version]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? a changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Zhao, Q.; Yousaf, L.; Xue, Y.; Shen, Q. Whole mung bean (Vigna radiata L.) supplementation prevents high-fat diet-induced obesity and disorders in a lipid profile and modulates gut microbiota in mice. Eur. J. Nutr. 2020, 59, 3617–3634. [Google Scholar] [CrossRef]
- Zhang, J.; Song, L.; Wang, Y.; Liu, C.; Zhang, L.; Zhu, S.; Liu, S.; Duan, L. Beneficial effect of butyrate-producing Lachnospiraceae on stress-induced visceral hypersensitivity in rats. J. Gastroen. Hepatol. 2019, 34, 1368–1376. [Google Scholar] [CrossRef] [Green Version]
- Qu, W.; Yuan, X.; Zhao, J.; Zhang, Y.; Hu, J.; Wang, J.; Li, J. Dietary advanced glycation end products modify gut microbial composition and partially increase colon permeability in rats. Mol. Nutr. Food Res. 2017, 61, 1700118. [Google Scholar] [CrossRef]
- Hu, S.; Wang, J.; Xu, Y.; Yang, H.; Wang, J.; Xue, C.; Yan, X.; Su, L. Anti-inflammation effects of fucosylated chondroitin sulphate from acaudina molpadioides by altering gut microbiota in obese mice. Food Funct. 2019, 10, 1736–1746. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.-X.; Hu, Y.-N.; Li, J.-W.; Yuan, K. Hypoglycemic mechanism of the berberine organic acid salt under the synergistic effect of intestinal flora and oxidative stress. Oxid. Med. Cell. Longev. 2018, 2018, 8930374. [Google Scholar] [CrossRef] [PubMed]
- Ruppin, H.; Bar-Meir, S.; Soergel, K.H.; Wood, C.M.; Schmitt, M.G., Jr. Absorption of short-chain fatty acids by the colon. Gastroenterology 1980, 78, 1500–1507. [Google Scholar] [CrossRef]
- Rechkemmer, G.; Rönnau, K.; von Engelhardt, W. Fermentation of polysaccharides and absorption of short chain fatty acids in the mammalian hindgut. Comp. Biochem. Phys. A 1988, 90, 563–568. [Google Scholar] [CrossRef]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef] [Green Version]
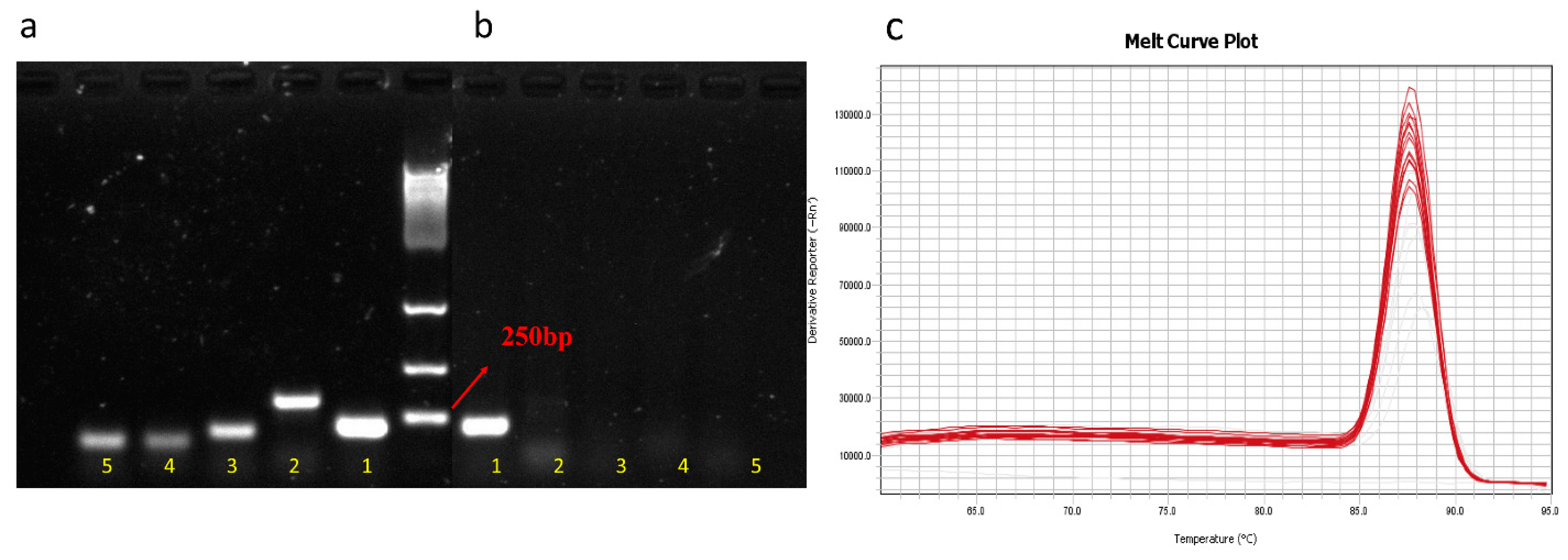

| Primer Set | Primers Sequences (5′–3′) | Annealing Temp (°C) | Approximate Amplicon Size (bp) | Reference or Source | |
|---|---|---|---|---|---|
| 1 | Cbtu-1F | GTGCCGCCGCTAACGCATTAAGTAT | 72 | 213 | [17] |
| Cbtu-1R | ACCATGCACCACCTGTCTTCCTGCC | ||||
| 2 | Cbtu-2F | GCATCCAGCAGACTTAGCAG | 63 | 275 | This study |
| Cbtu-2R | CTTCCGACTGTCTCATCTTC | ||||
| 3 | Cbtu-3F | CAATGGTTGTGAATGCTGAGG | 60 | 142 | This study |
| Cbtu-3R | CCAACTATGCTCATTTCGCCC | ||||
| 4 | Cbtu-4F | CCTCAAATCCGCCTTCTGG | 58 | 102 | This study |
| Cbtu-4R | GGATCTTGTTATCGTTCCG | ||||
| 5 | Cbtu-5F | GAGTCTGATTCGGTTGTGGC | 60 | 135 | This study |
| Cbtu-5R | CCTCTCTTCCTTGAACTGGATG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Kong, Q.; Wang, Y.; Duan, X.; Wang, P.; Li, C.; Huan, Y. Colonization of Clostridium butyricum in Rats and Its Effect on Intestinal Microbial Composition. Microorganisms 2021, 9, 1573. https://doi.org/10.3390/microorganisms9081573
Luo X, Kong Q, Wang Y, Duan X, Wang P, Li C, Huan Y. Colonization of Clostridium butyricum in Rats and Its Effect on Intestinal Microbial Composition. Microorganisms. 2021; 9(8):1573. https://doi.org/10.3390/microorganisms9081573
Chicago/Turabian StyleLuo, Xianshu, Qing Kong, Yuming Wang, Xuefeng Duan, Peng Wang, Chenman Li, and Yuchen Huan. 2021. "Colonization of Clostridium butyricum in Rats and Its Effect on Intestinal Microbial Composition" Microorganisms 9, no. 8: 1573. https://doi.org/10.3390/microorganisms9081573
APA StyleLuo, X., Kong, Q., Wang, Y., Duan, X., Wang, P., Li, C., & Huan, Y. (2021). Colonization of Clostridium butyricum in Rats and Its Effect on Intestinal Microbial Composition. Microorganisms, 9(8), 1573. https://doi.org/10.3390/microorganisms9081573






