Chironomus ramosus Larval Microbiome Composition Provides Evidence for the Presence of Detoxifying Enzymes
Abstract
:1. Introduction
2. Materials and Methods
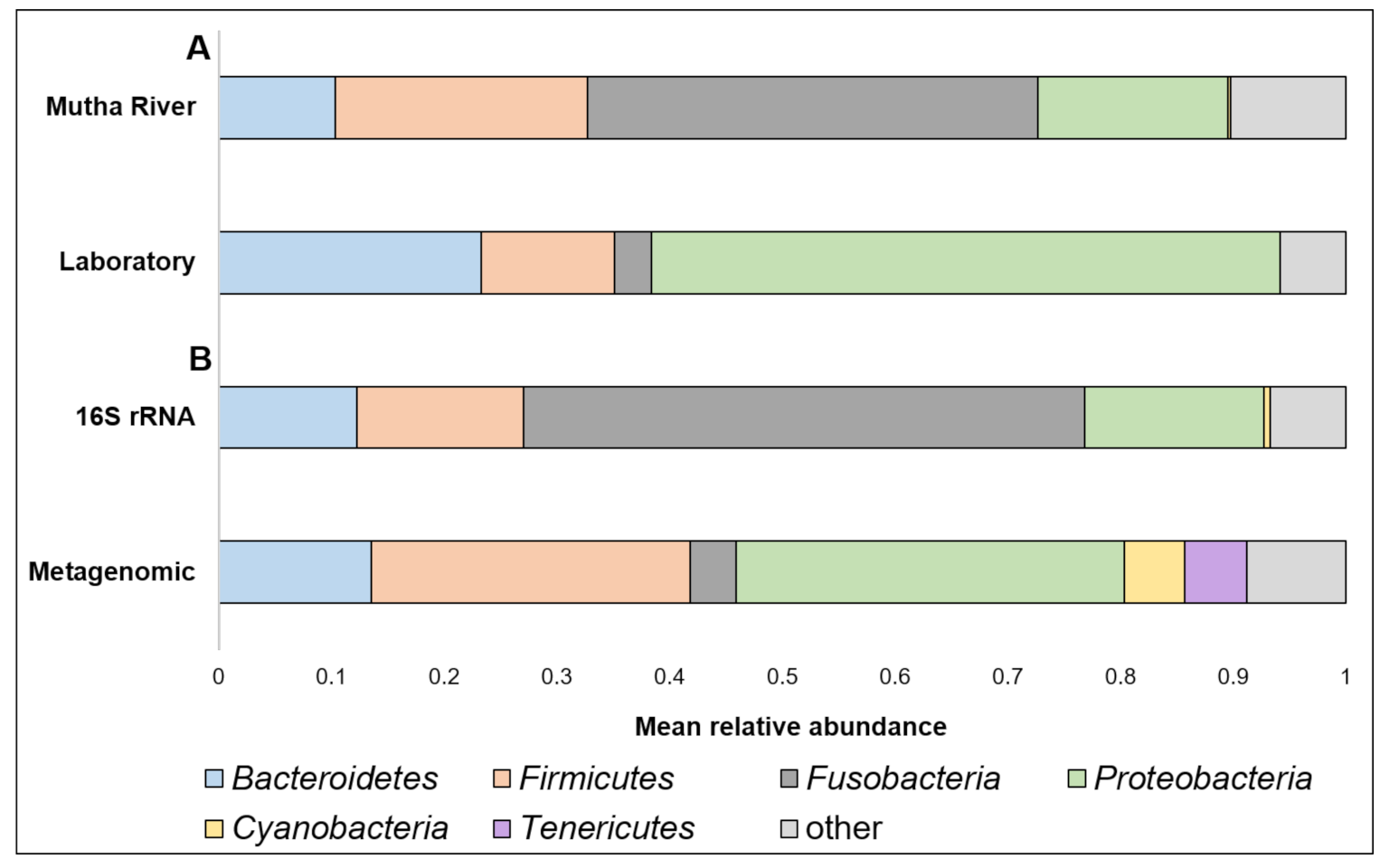
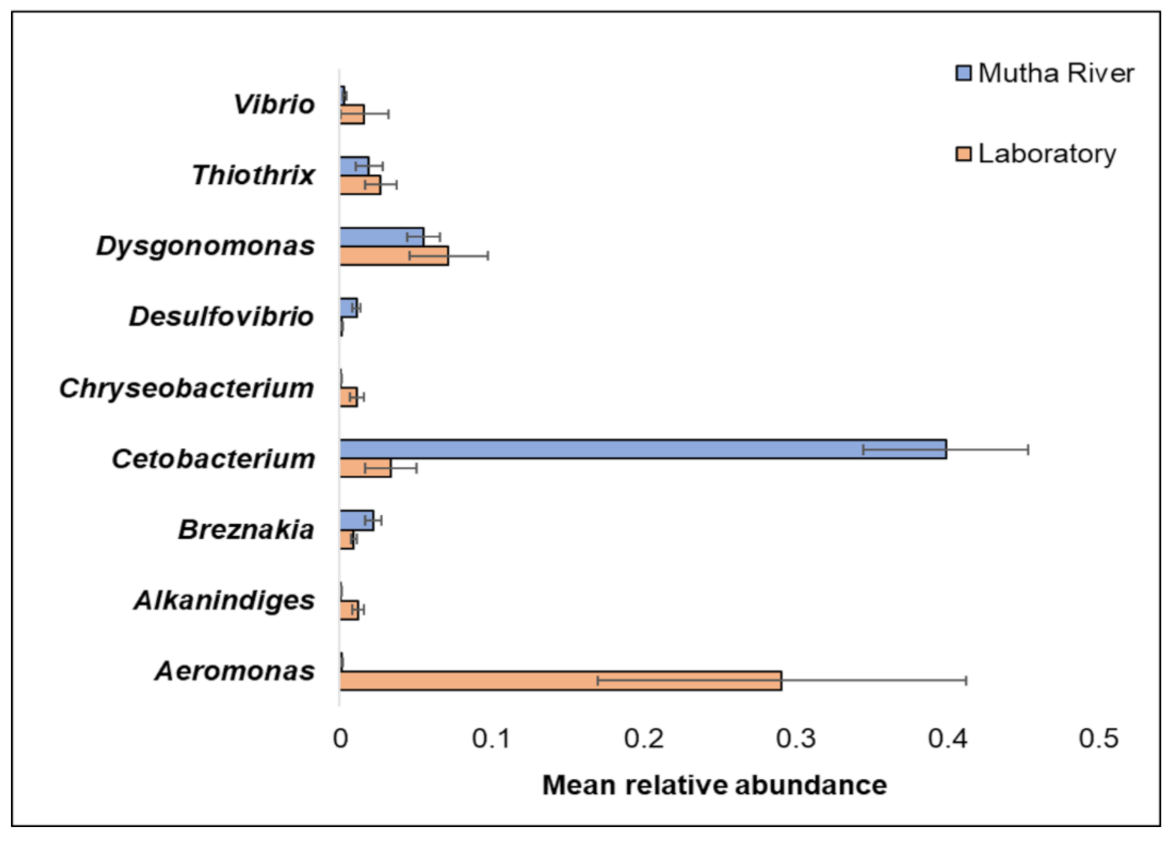
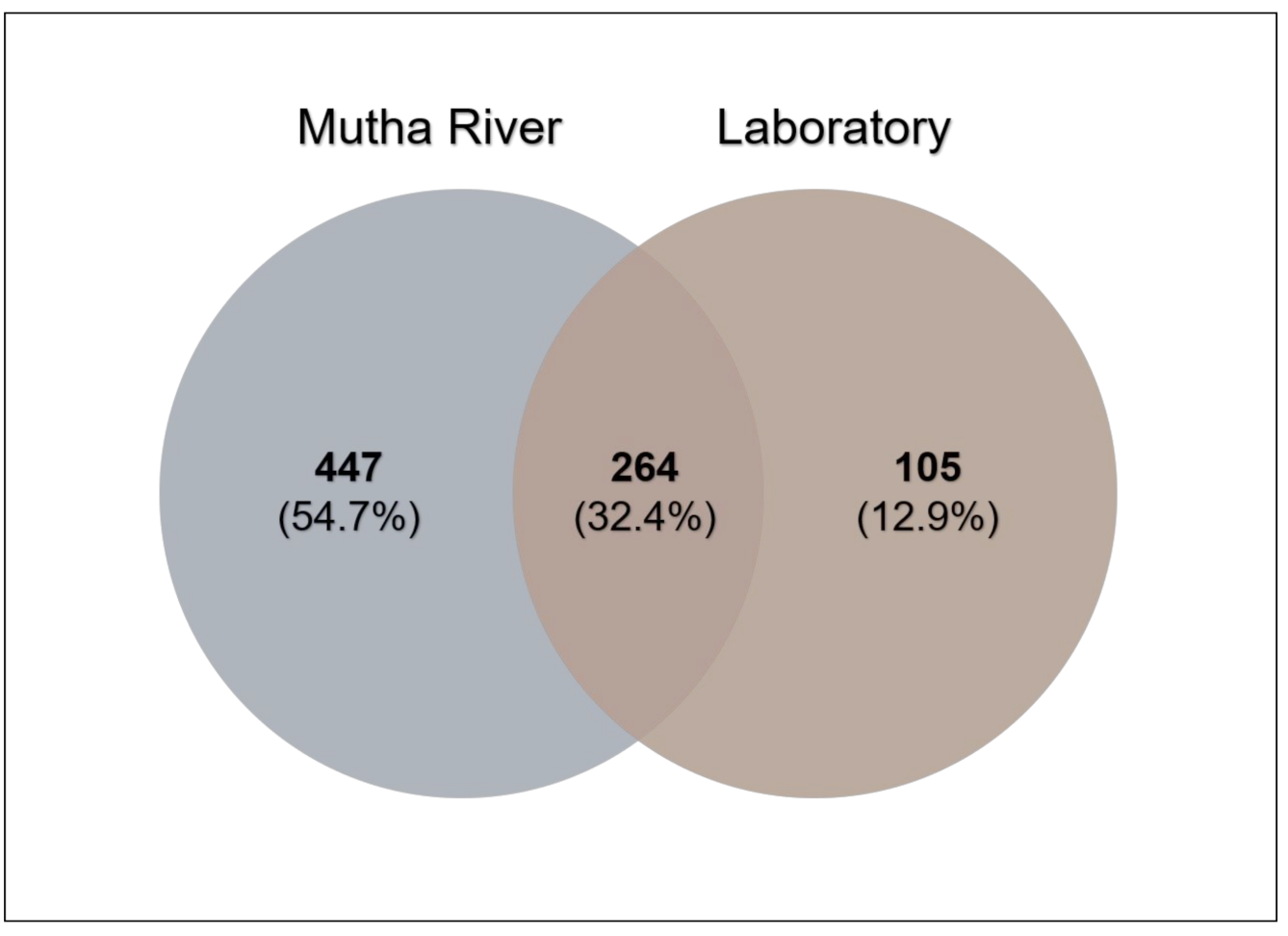
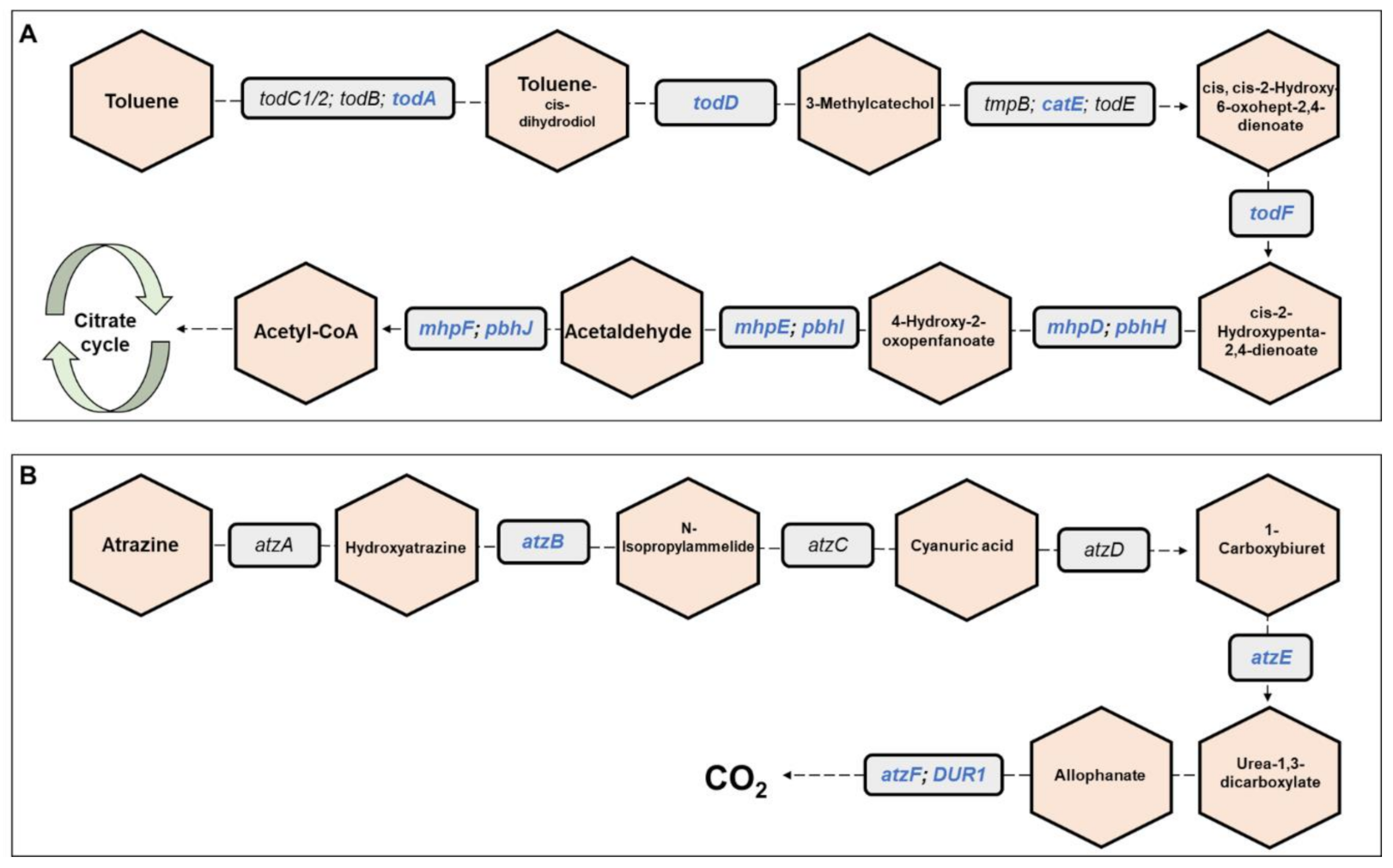
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Halpern, M.; Senderovich, Y. Chironomid Microbiome. Microb. Ecol. 2015, 70, 1–8. [Google Scholar] [CrossRef]
- Thorat, L.; Joseph, E.; Nisal, A.; Shukla, E.; Ravikumar, A.; Nath, B.B. Structural and physical analysis of underwater silk from housing nest composites of a tropical chironomid midge. Int. J. Biol. Macromol. 2020, 163, 934–942. [Google Scholar] [CrossRef]
- Hare, L.; Shooner, F. Do aquatic insects avoid cadmium-contaminated sediments? Environ. Toxicol. Chem. 1995, 14, 1071–1077. [Google Scholar] [CrossRef]
- Nath, B.B. Extracellular hemoglobin and environmental stress tolerance in Chironomus larvae. J. Limnol. 2018, 77. [Google Scholar] [CrossRef] [Green Version]
- Akbulut, M.; Çelik, E.; Odabaşi, D.; Kaya, H.; Selvi, K.; Arslan, N.; Odabaşı, S. Seasonal Distribution and Composition of Benthic Macroinvertebrate Communities in Menderes Creek, Çanakkale, Turkey. Fresenius Environ. Bull. 2009, 18, 2136–2145. [Google Scholar]
- Senderovich, Y.; Halpern, M. The protective role of endogenous bacterial communities in chironomid egg masses and larvae. ISME J. 2013, 7, 2147–2158. [Google Scholar] [CrossRef] [Green Version]
- Armitage, P.; Cranston, P.S.; Pinder, L.C.V.; Berg, M.B. The Chironomidae: The Biology and Ecology of Non-Biting Midges; Chapman & Hall: London, UK, 1995; ISBN 978-94-011-0715-0. [Google Scholar]
- Laviad-Shitrit, S.; Sharaby, Y.; Sela, R.; Thorat, L.; Nath, B.B.; Halpern, M. Copper and chromium exposure affect chironomid larval microbiota composition. Sci. Total. Environ. 2021, 771, 145330. [Google Scholar] [CrossRef] [PubMed]
- Ali, A. Perspectives on management of pestiferous Chironomidae (Diptera), an emerging global problem. J. Am. Mosq. Control. Assoc. 1991, 7, 260–281. [Google Scholar]
- Serra, S.R.Q.; Cobo, F.; Graça, M.A.S.; Dolédec, S.; Feio, M.J. Synthesising the trait information of European Chironomidae (Insecta: Diptera): Towards a new database. Ecol. Indic. 2016, 61, 282–292. [Google Scholar] [CrossRef] [Green Version]
- Sela, R.; Halpern, M. Seasonal dynamics of Chironomus transvaalensis populations and the microbial community composition of their egg masses. FEMS Microbiol. Lett. 2019, 366. [Google Scholar] [CrossRef]
- Sela, R.; Hammer, B.K.; Halpern, M. Quorum-sensing signaling by chironomid egg masses’ microbiota, affects haemagglutinin/protease (HAP) production by Vibrio cholerae. Mol. Ecol. 2021, 30, 1736–1746. [Google Scholar] [CrossRef] [PubMed]
- Boush, M.G.; Matsumura, F. Insecticidal Degradation by Pseudomonas melophthora, the Bacterial Symbiote of the Apple Maggot. J. Econ. Èntomol. 1967, 60, 918–920. [Google Scholar] [CrossRef]
- Pietri, J.E.; Liang, D. The Links Between Insect Symbionts and Insecticide Resistance: Causal Relationships and Physiological Tradeoffs. Ann. Èntomol. Soc. Am. 2018, 111, 92–97. [Google Scholar] [CrossRef]
- Sela, R.; Laviad-Shitrit, S.; Halpern, M. Changes in Microbiota Composition Along the Metamorphosis Developmental Stages of Chironomus transvaalensis. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Laviad-Shitrit, S.; Sela, R.; Thorat, L.; Sharaby, Y.; Izhaki, I.; Nath, B.B.; Halpern, M. Identification of chironomid species as natural reservoirs of toxigenic Vibrio cholerae strains with pandemic potential. PLOS Negl. Trop. Dis. 2020, 14, e0008959. [Google Scholar] [CrossRef]
- Halpern, M.; Landsberg, O.; Raats, D.; Rosenberg, E. Culturable and VBNC Vibrio cholerae: Interactions with Chironomid Egg Masses and Their Bacterial Population. Microb. Ecol. 2007, 53, 285–293. [Google Scholar] [CrossRef]
- Laviad-Shitrit, S.; Lev-Ari, T.; Katzir, G.; Sharaby, Y.; Izhaki, I.; Halpern, M. Great cormorants (Phalacrocorax carbo) as potential vectors for the dispersal of Vibrio cholerae. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Naqib, A.; Poggi, S.; Wang, W.; Hyde, M.; Kunstman, K.; Green, S.J. Making and Sequencing Heavily Multiplexed, High-Throughput 16S Ribosomal RNA Gene Amplicon Libraries Using a Flexible, Two-Stage PCR Protocol. In Gene Expression Analysis: Methods and Protocols; Raghavachari, N., Garcia-Reyero, N., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 149–169. ISBN 978-1-4939-7834-2. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Clarke, K.R.; Gorley, R.N. PRIMER v7: User Manual/Tutorial; Primer-E Ltd: Plymouth, UK, 2015. [Google Scholar]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, R.T.; Sanchez, L.G.; Fierer, N. A Cross-Taxon Analysis of Insect-Associated Bacterial Diversity. PLoS ONE 2013, 8, e61218. [Google Scholar] [CrossRef]
- Pechal, J.L.; Benbow, M.E. Microbial ecology of the salmon necrobiome: Evidence salmon carrion decomposition influences aquatic and terrestrial insect microbiomes. Environ. Microbiol. 2016, 18, 1511–1522. [Google Scholar] [CrossRef]
- Ramírez, C.; Coronado, J.; Silva, A.; Romero, J. Cetobacterium Is a Major Component of the Microbiome of Giant Amazonian Fish (Arapaima gigas) in Ecuador. Animals 2018, 8, 189. [Google Scholar] [CrossRef] [Green Version]
- Halpern, M.; Izhaki, I. Fish as Hosts of Vibrio cholerae. Front. Microbiol. 2017, 8, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugita, H.; Miyajima, C.; Deguchi, Y. The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture 1991, 92, 267–276. [Google Scholar] [CrossRef]
- Mostes Vidal, D.; Von Rymon-Lipinski, A.-L.; Ravella, S.; Groenhagen, U.; Herrmann, J.; Zaburannyi, N.; Zarbin, P.H.G.; Varadarajan, A.R.; Ahrens, C.H.; Weisskopf, L.; et al. Long-Chain Alkyl Cyanides: Unprecedented Volatile Compounds Released by Pseudomonas and Micromonospora Bacteria. Angew. Chem. Int. Ed. 2017, 56, 4342–4346. [Google Scholar] [CrossRef] [PubMed]
- Özdal, M.; Ozdal, O.G.; Algur, O.F. Isolation and Characterization of α-Endosulfan Degrading Bacteria from the Microflora of Cockroaches. Pol. J. Microbiol. 2016, 65, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Heydarnezhad, F.; Hoodaji, M.; Shahriarinour, M.; Tahmourespour, A.; Shariati, S. Optimizing Toluene Degradation by Bacterial Strain Isolated from Oil-Polluted Soils. Pol. J. Environ. Stud. 2018, 27, 655–663. [Google Scholar] [CrossRef]
- Beller, H.; Grbić-Galić, D.; Reinhard, M. Microbial degradation of toluene under sulfate-reducing conditions and the influence of iron on the process. Appl. Environ. Microbiol. 1992, 58, 786–793. [Google Scholar] [CrossRef] [Green Version]
- Hamzah, A.; Tavakoli, A.; Rabu, A. Detection of Toluene Degradation in Bacteria Isolated from Oil Contaminated Soils. Sains Malays 2011, 40, 1231–1235. [Google Scholar]
- Graymore, M.; Stagnitti, F.; Allinson, G. Impacts of atrazine in aquatic ecosystems. Environ. Int. 2001, 26, 483–495. [Google Scholar] [CrossRef]
- Steinberg, C.E.W.; Lorenz, R.; Spieser, O.H. Effects of atrazine on swimming behavior of zebrafish, Brachydanio rerio. Water Res. 1995, 29, 981–985. [Google Scholar] [CrossRef]
- Anderson, T.D. Toxicological, Biochemical, and Molecular Effects of Atrazine to the Aquatic Midge Chironomus tentans (Diptera: Chironomidae). Ph.D. Thesis, Kansas State University, Manhattan, KS, USA, 2006. [Google Scholar]
- Mandelbaum, R.T.; Allan, D.L.; Wackett, L.P. Isolation and Characterization of a Pseudomonas sp. That Mineralizes the s-Triazine Herbicide Atrazine. Appl. Environ. Microbiol. 1995, 61, 1451–1457. [Google Scholar] [CrossRef] [Green Version]
- Behki, R.; Topp, E.; Dick, W.; Germon, P. Metabolism of the herbicide atrazine by Rhodococcus strains. Appl. Environ. Microbiol. 1993, 59, 1955–1959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirgain, I.; Green, G.; Monteil, H. Degradation of atrazine in laboratory microcosms: Isolation and identification of the biodegrading bacteria. Environ. Toxicol. Chem. 1993, 12, 1627–1634. [Google Scholar] [CrossRef]
- Cai, B.; Han, Y.; Liu, B.; Ren, Y.; Jiang, S. Isolation and characterization of an atrazine-degrading bacterium from industrial wastewater in China. Lett. Appl. Microbiol. 2003, 36, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Salmelin, J.; Karjalainen, A.K.; Hämäläinen, H.; Leppänen, M.T.; Kiviranta, H.; Kukkonen, J.V.K.; Vuori, K.M. Biological responses of midge (Chironomus riparius) and lamprey (Lampetra fluviatilis) larvae in ecotoxicity assessment of PCDD/F-, PCB- and Hg-contaminated river sediments. Environ. Sci. Pollut. Res. 2016, 23, 18379–18393. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Category | Id (Detoxifying Pathways) |
|---|---|
| ko00633 | Nitrotoluene degradation |
| ko00623 | Toluene degradation |
| ko00622 | Xylene degradation |
| ko00642 | Ethylbenzene degradation |
| ko00362 | Benzoate degradation |
| ko00627 | Aminobenzoate degradation |
| ko00364 | Fluorobenzoate degradation |
| ko00945 | Stilbenoid, diarylheptanoid, and gingerol biosynthesis |
| ko00626 | Naphthalene degradation |
| ko00627 | Aminobenzoate degradation |
| ko00364 | Fluorobenzoate degradation |
| ko00625 | Chloroalkane and chloroalkene degradation |
| ko00361 | Chlorocyclohexane and chlorobenzene degradation |
| ko00642 | Ethylbenzene degradation |
| ko00643 | Styrene degradation |
| ko00791 | Atrazine degradation |
| ko00930 | Caprolactam degradation |
| ko00363 | Bisphenol degradation |
| ko00621 | Dioxin degradation |
| ko00624 | Polycyclic aromatic hydrocarbon degradation |
| ko00365 | Furfural degradation |
| ko00984 | Steroid degradation |
| ko00980 | Metabolism of xenobiotics by cytochrome P450 |
| ko00982 | Drug metabolism—cytochrome P450 |
| ko00983 | Drug metabolism—other enzymes |
| Category | Gene | Function (Resistance) |
|---|---|---|
| K07665 | cusR, copR, silR | two-component system, OmpR family, copper resistance phosphate regulon response regulator CusR |
| K03327 | TC.MATE, SLC47A, norM, mdtK, dinF | multidrug resistance protein, MATE family |
| K03297 | emrE, qac, mmr, smr | small multidrug resistance pump |
| K08163 | mdtL | MFS transporter, DHA1 family, multidrug resistance protein |
| K18924 | ykkC | paired small multidrug resistance pump |
| K11741 | sugE | quaternary ammonium compound resistance protein SugE |
| K03712 | marR | MarR family transcriptional regulator, multiple antibiotic resistance protein MarR |
| K11811 | arsH | arsenical resistance protein ArsH |
| K07245 | pcoD | copper resistance protein D |
| K07156 | copC, pcoC | copper resistance protein C |
| K21740 | rclB | reactive chlorine resistance protein B |
| K02547 | mecR1 | methicillin resistance protein |
| K21252 | fosX | fosfomycin resistance protein FosX |
| K07233 | pcoB, copB | copper resistance protein B |
| K02617 | paaY | phenylacetic acid degradation protein |
| K22011 | cfbA | sirohydrochlorin cobalto/nickelchelatase [EC:4.99.1.3 4.99.1.11] |
| K07803 | zraP | zinc resistance-associated protein |
| K15726 | czcA | cobalt-zinc-cadmium resistance protein CzcA |
| K16264 | czcD, zitB | cobalt-zinc-cadmium efflux system protein |
| K21903 | cadC, smtB | ArsR family transcriptional regulator, lead/cadmium/zinc/bismuth-responsive transcriptional repressor |
| K00520 | merA | mercuric reductase [EC:1.16.1.1] |
| K21903 | cadC, smtB | ArsR family transcriptional regulator, lead/cadmium/zinc/bismuth-responsive transcriptional repressor |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sela, R.; Laviad-Shitrit, S.; Thorat, L.; Nath, B.B.; Halpern, M. Chironomus ramosus Larval Microbiome Composition Provides Evidence for the Presence of Detoxifying Enzymes. Microorganisms 2021, 9, 1571. https://doi.org/10.3390/microorganisms9081571
Sela R, Laviad-Shitrit S, Thorat L, Nath BB, Halpern M. Chironomus ramosus Larval Microbiome Composition Provides Evidence for the Presence of Detoxifying Enzymes. Microorganisms. 2021; 9(8):1571. https://doi.org/10.3390/microorganisms9081571
Chicago/Turabian StyleSela, Rotem, Sivan Laviad-Shitrit, Leena Thorat, Bimalendu B. Nath, and Malka Halpern. 2021. "Chironomus ramosus Larval Microbiome Composition Provides Evidence for the Presence of Detoxifying Enzymes" Microorganisms 9, no. 8: 1571. https://doi.org/10.3390/microorganisms9081571
APA StyleSela, R., Laviad-Shitrit, S., Thorat, L., Nath, B. B., & Halpern, M. (2021). Chironomus ramosus Larval Microbiome Composition Provides Evidence for the Presence of Detoxifying Enzymes. Microorganisms, 9(8), 1571. https://doi.org/10.3390/microorganisms9081571





