Characterization of a Novel Viral Interleukin 8 (vIL-8) Splice Variant Encoded by Marek’s Disease Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells
2.3. Viruses
2.4. Western Blotting
2.5. Indirect Immunofluorescence
2.6. Plaque Size Assays
2.7. Multi-Step Growth Kinetics
2.8. In Vivo Experiment
2.9. Virus Quantification in Blood and Feather Follicles
2.10. Statistical Analyses
3. Results
3.1. Detection of Protein Expression of the Novel vIL-8 Splice Variant
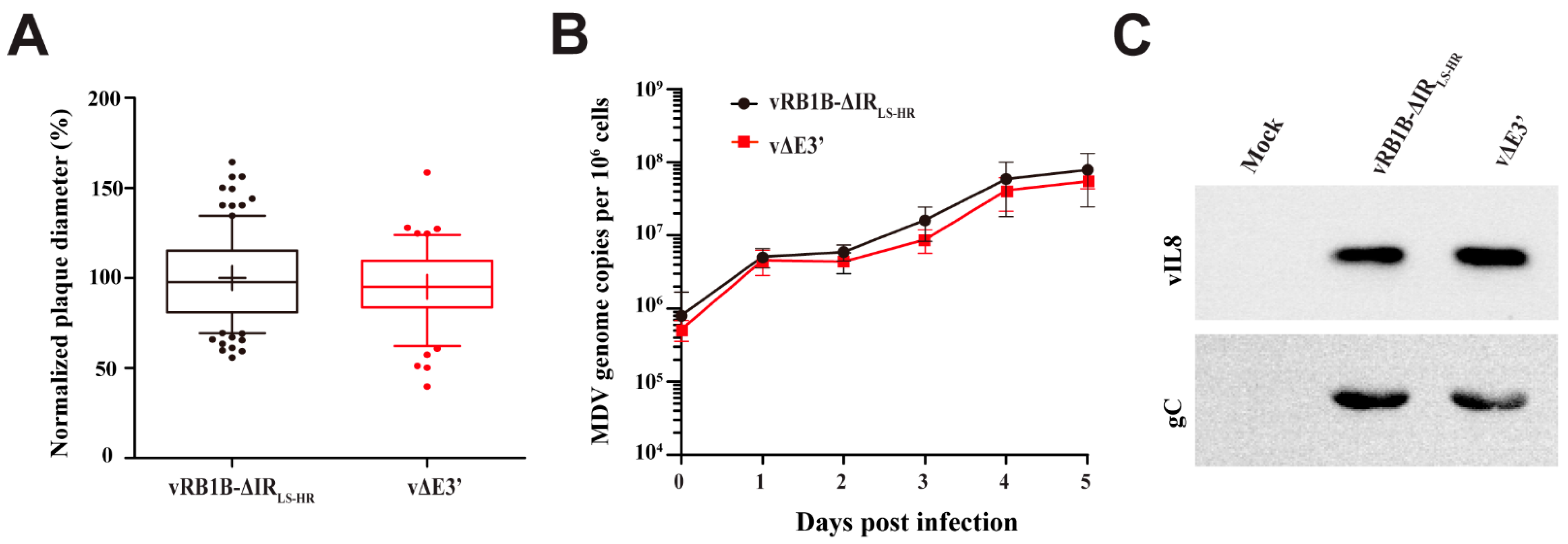
3.2. Abrogation of the Novel vIL-8 Splice Variant
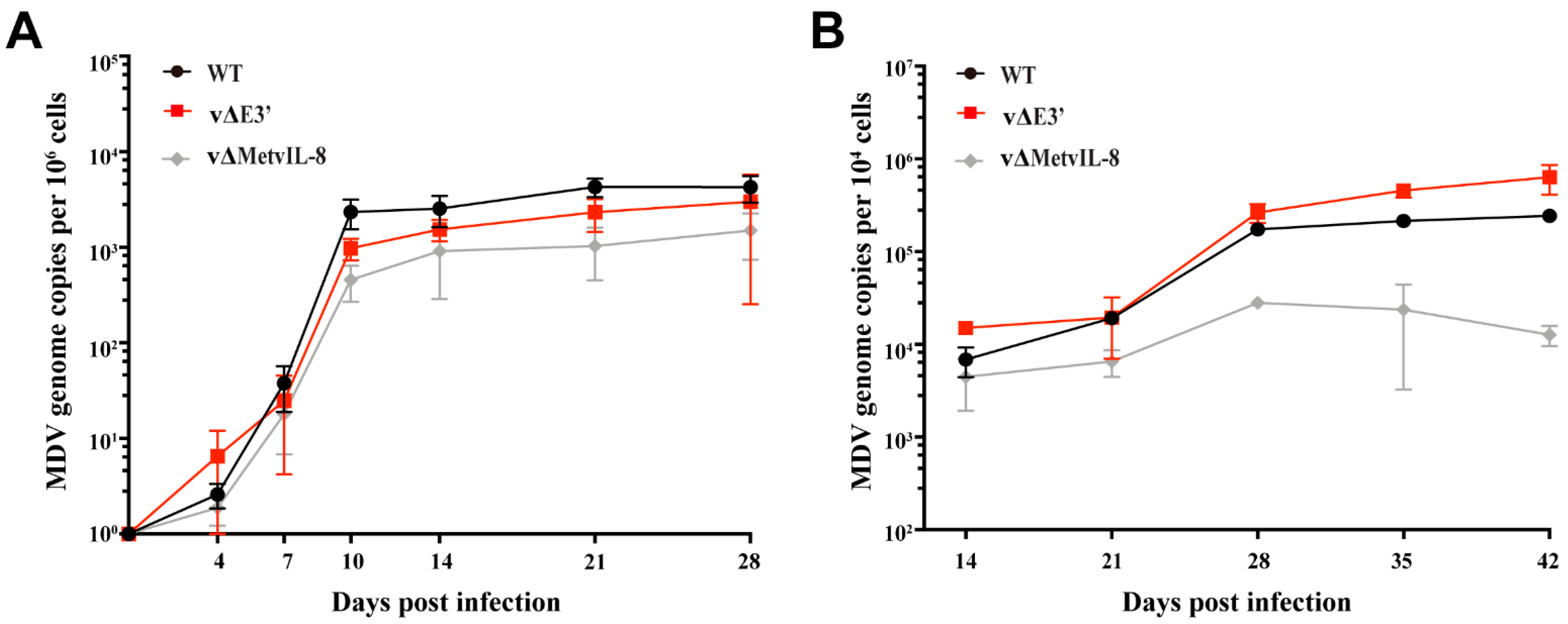
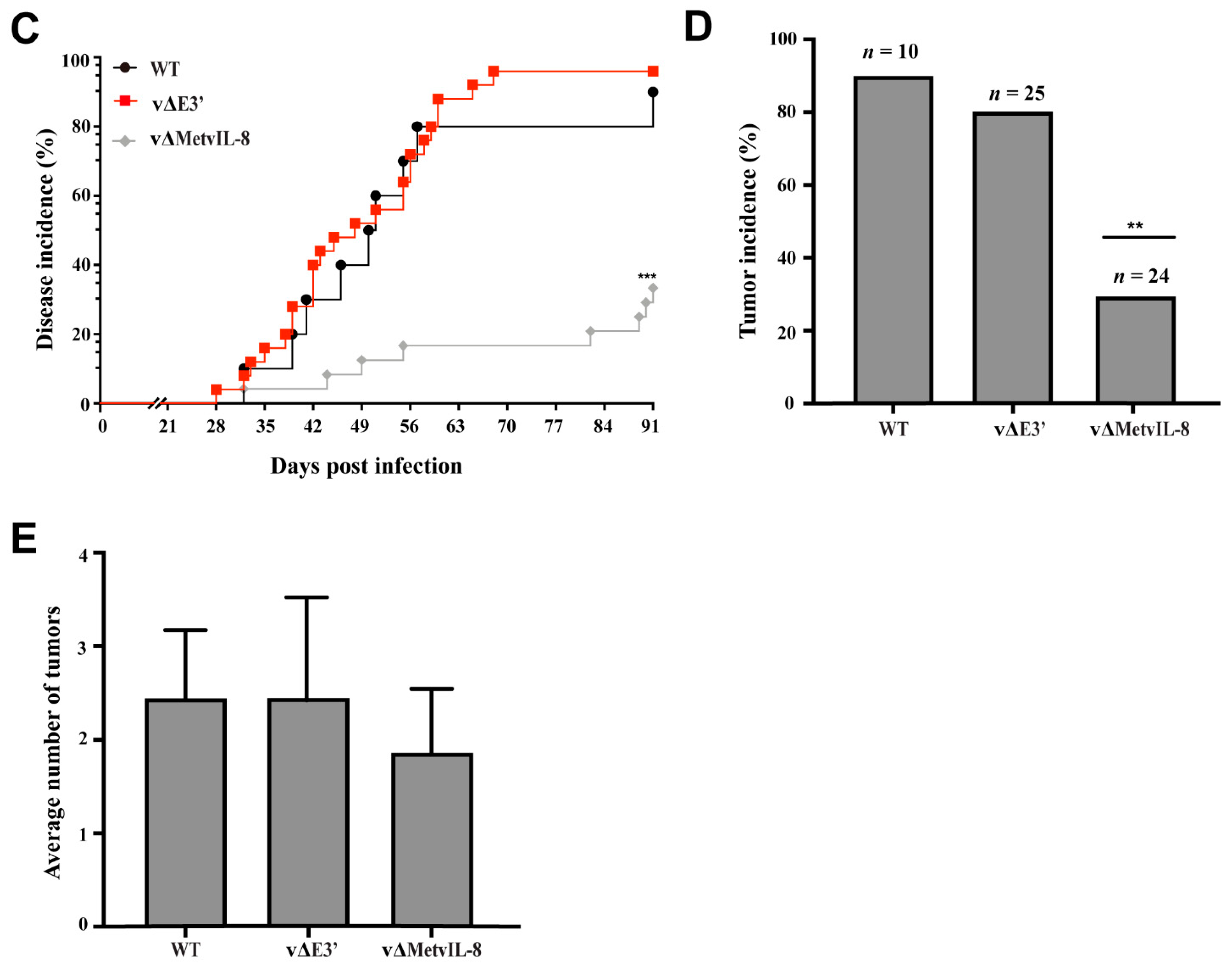
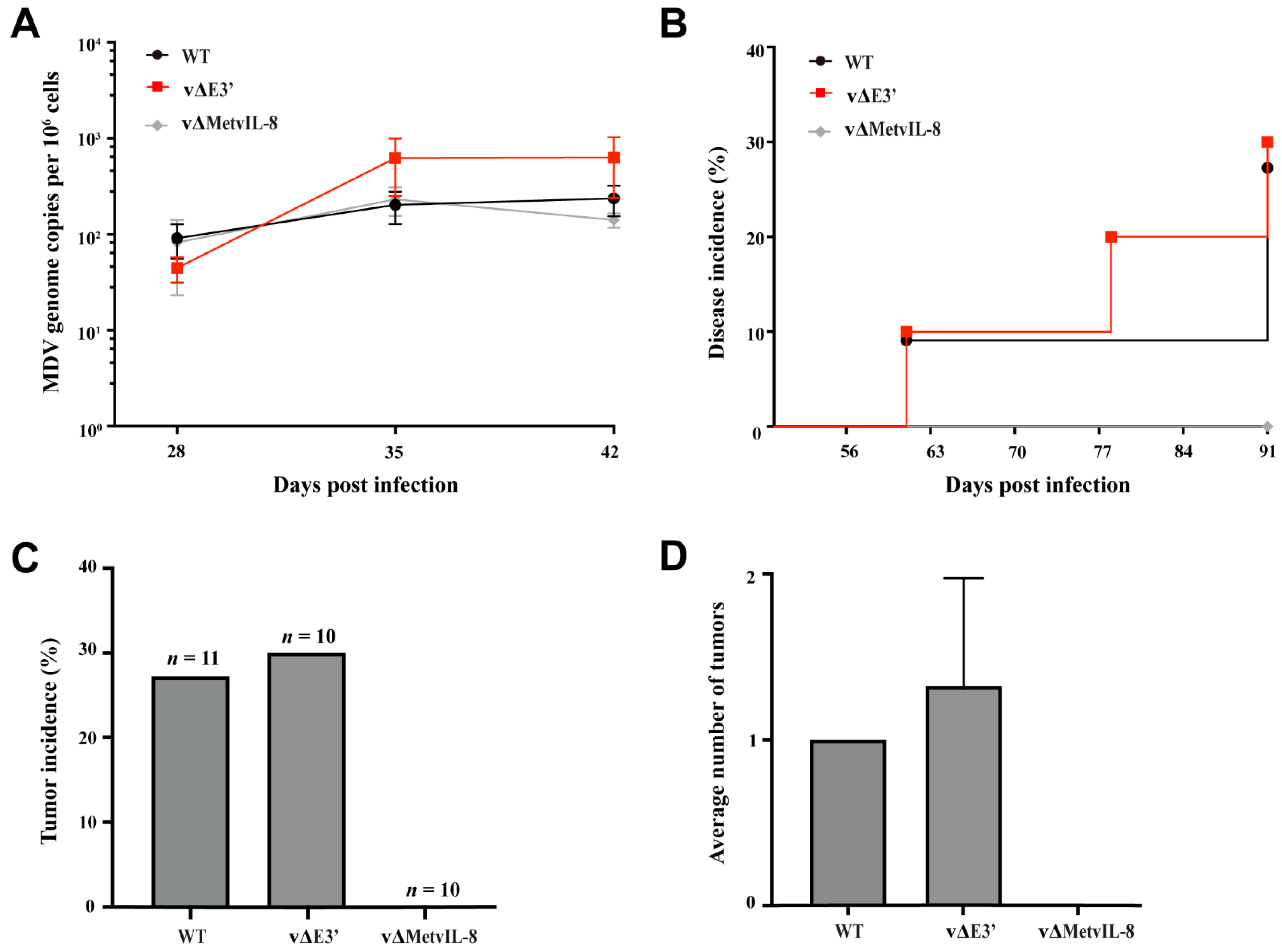
3.3. Role of the Novel vIL-8 Splice Variant in MDV Pathogenesis and Tumorigenesis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Davison, A.J. Herpesvirus systematics. Vet. Microbiol. 2010, 143, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: From miasma to model. Nat. Rev. Microbiol. 2006, 4, 283–294. [Google Scholar] [CrossRef]
- Rozins, C.; Day, T.; Greenhalgh, S. Managing Marek’s disease in the egg industry. Epidemics 2019, 27, 52–58. [Google Scholar] [CrossRef]
- Gimeno, I.; Schat, K. Virus-induced immunosuppression in chickens. Avian Dis. 2018, 62, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Bertzbach, L.D.; Conradie, A.M.; You, Y.; Kaufer, B.B. Latest insights into Marek’s disease virus pathogenesis and tumorigenesis. Cancers 2020, 12, 647. [Google Scholar] [CrossRef]
- Bertzbach, L.D.; Pfaff, F.; Pauker, V.I.; Kheimar, A.M.; Höper, D.; Härtle, S.; Karger, A.; Kaufer, B.B. The Transcriptional Landscape of Marek’s Disease Virus in Primary Chicken B Cells Reveals Novel Splice Variants and Genes. Viruses 2019, 11, 264. [Google Scholar] [CrossRef]
- Haertle, S.; Alzuheir, I.; Busalt, F.; Waters, V.; Kaiser, P.; Kaufer, B.B. Identification of the Receptor and Cellular Ortholog of the Marek’s Disease Virus (MDV) CXC Chemokine. Front. Microbiol. 2017, 8, 2543. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lee, L.F.; Reed, W.M.; Kung, H.-J.; Reddy, S.M. Marek’s Disease Virus-Encoded vIL-8 Gene Is Involved in Early Cytolytic Infection but Dispensable for Establishment of Latency. J. Virol. 2004, 78, 4753–4760. [Google Scholar] [CrossRef]
- Engel, A.T.; Selvaraj, R.K.; Kamil, J.P.; Osterrieder, N.; Kaufer, B.B. Marek’s disease viral interleukin-8 promotes lymphoma formation through targeted recruitment of B cells and CD4+ CD25+ T cells. J. Virol. 2012, 86, 8536–8545. [Google Scholar] [CrossRef] [PubMed]
- Bertzbach, L.D.; Laparidou, M.; Härtle, S.; Etches, R.J.; Kaspers, B.; Schusser, B.; Kaufer, B.B. Unraveling the role of B cells in the pathogenesis of an oncogenic avian herpesvirus. Proc. Natl. Acad. Sci. USA 2018, 115, 11603–11607. [Google Scholar] [CrossRef]
- Parcells, M.S.; Lin, S.-F.; Dienglewicz, R.L.; Majerciak, V.; Robinson, D.R.; Chen, H.-C.; Wu, Z.; Dubyak, G.R.; Brunovskis, P.; Hunt, H.D. Marek’s disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): Characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 2001, 75, 5159–5173. [Google Scholar] [CrossRef]
- Jarosinski, K.W.; Schat, K.A. Multiple alternative splicing to exons II and III of viral interleukin-8 (vIL-8) in the Marek’s disease virus genome: The importance of vIL-8 exon I. Virus Genes 2007, 34, 9–22. [Google Scholar] [CrossRef]
- Sadigh, Y.; Tahiri-Alaoui, A.; Spatz, S.; Nair, V.; Ribeca, P. Pervasive Differential Splicing in Marek’s Disease Virus can Discriminate CVI-988 Vaccine Strain from RB-1B Very Virulent Strain in Chicken Embryonic Fibroblasts. Viruses 2020, 12, 329. [Google Scholar] [CrossRef]
- Schumacher, D.; Tischer, B.K.; Fuchs, W.; Osterrieder, N. Reconstitution of Marek’s disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 2000, 74, 11088–11098. [Google Scholar] [CrossRef]
- Vychodil, T.; Conradie, A.M.; Trimpert, J.; Aswad, A.; Bertzbach, L.D.; Kaufer, B.B. Marek’s disease virus requires both copies of the inverted repeat regions for efficient in vivo replication and pathogenesis. J. Virol. 2020, 95, e01256-20. [Google Scholar] [CrossRef]
- Tischer, B.K.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 2006, 40, 191–197. [Google Scholar]
- Tischer, B.K.; Kaufer, B.B. Viral Bacterial Artificial Chromosomes: Generation, Mutagenesis, and Removal of Mini-F Sequences. J. Biomed. Biotechnol. 2012, 2012, 472537. [Google Scholar] [CrossRef]
- Bertzbach, L.D.; van Haarlem, D.A.; Härtle, S.; Kaufer, B.B.; Jansen, C.A. Marek’s Disease Virus Infection of Natural Killer Cells. Microorganisms 2019, 7, 588. [Google Scholar] [CrossRef]
- Conradie, A.M.; Bertzbach, L.D.; Trimpert, J.; Patria, J.N.; Murata, S.; Parcells, M.S.; Kaufer, B.B. Distinct polymorphisms in a single herpesvirus gene are capable of enhancing virulence and mediating vaccinal resistance. PLoS Pathog. 2020, 16, e1009104. [Google Scholar] [CrossRef]
- Schippers, T.; Jarosinski, K.; Osterrieder, N. The ORF012 gene of Marek’s disease virus type 1 produces a spliced transcript and encodes a novel nuclear phosphoprotein essential for virus growth. J. Virol. 2015, 89, 1348–1363. [Google Scholar] [CrossRef]
- Tischer, B.K.; Schumacher, D.; Chabanne-Vautherot, D.; Zelnik, V.; Vautherot, J.-F.; Osterrieder, N. High-level expression of Marek’s disease virus glycoprotein C is detrimental to virus growth in vitro. J. Virol. 2005, 79, 5889–5899. [Google Scholar] [CrossRef][Green Version]
- Previdelli, R.L.; Bertzbach, L.D.; Wight, D.J.; Vychodil, T.; You, Y.; Arndt, S.; Kaufer, B.B. The Role of Marek’s Disease Virus UL12 and UL29 in DNA Recombination and the Virus Lifecycle. Viruses 2019, 11, 111. [Google Scholar] [CrossRef]
- Conradie, A.M.; Bertzbach, L.D.; Bhandari, N.; Parcells, M.; Kaufer, B.B. A Common Live-Attenuated Avian Herpesvirus Vaccine Expresses a Very Potent Oncogene. mSphere 2019, 4, e00658-19. [Google Scholar] [CrossRef]
- Bello, N.; Francino, O.; Sánchez, A. Isolation of genomic DNA from feathers. J. Vet. Diagn. Invest. 2001, 13, 162–164. [Google Scholar] [CrossRef]
- Schommartz, T.; Loroch, S.; Alawi, M.; Grundhoff, A.; Sickmann, A.; Brune, W. Functional dissection of an alternatively spliced herpesvirus gene by splice site mutagenesis. J. Virol. 2016, 90, 4626–4636. [Google Scholar] [CrossRef]
- Anna, A.; Monika, G. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef]
- Tombácz, D.; Csabai, Z.; Szűcs, A.; Balázs, Z.; Moldován, N.; Sharon, D.; Snyder, M.; Boldogkői, Z. Long-read isoform sequencing reveals a hidden complexity of the transcriptional landscape of herpes simplex virus type 1. Front. Microbiol. 2017, 8, 1079. [Google Scholar] [CrossRef]
- O’Grady, T.; Wang, X.; Höner zu Bentrup, K.; Baddoo, M.; Concha, M.; Flemington, E.K. Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res. 2016, 44, e145. [Google Scholar] [CrossRef]
- Gatherer, D.; Seirafian, S.; Cunningham, C.; Holton, M.; Dargan, D.J.; Baluchova, K.; Hector, R.D.; Galbraith, J.; Herzyk, P.; Wilkinson, G.W. High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. USA 2011, 108, 19755–19760. [Google Scholar] [CrossRef] [PubMed]
- Anobile, J.M.; Arumugaswami, V.; Downs, D.; Czymmek, K.; Parcells, M.; Schmidt, C.J. Nuclear localization and dynamic properties of the Marek’s disease virus oncogene products Meq and Meq/vIL8. J. Virol. 2006, 80, 1160–1166. [Google Scholar] [CrossRef]
- Jarosinski, K.W.; Osterrieder, N. Marek’s disease virus expresses multiple UL44 (gC) variants through mRNA splicing that are all required for efficient horizontal transmission. J. Virol. 2012, 86, 7896–7906. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Bajwa, K.; Al-Mahmood, M.; Gimeno, I.M.; Reddy, S.M.; Lupiani, B. The role of Meq-vIL8 in regulating Marek’s disease virus pathogenesis. J. Gen. Virol. 2021, 102, 001528. [Google Scholar] [CrossRef] [PubMed]
- Arumugaswami, V.; Kumar, P.M.; Konjufca, V.; Dienglewicz, R.L.; Reddy, S.M.; Parcells, M.S. Latency of Marek’s Disease Virus (MDV) in a Reticuloendotheliosis Virus–Transformed T-Cell Line. II: Expression of the Latent MDV Genome. Avian Dis. 2009, 53, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Li, Y.; Yang, X.; Wu, Y.-P.; Lin, L.-J.; Liu, X.-M. Significance of alternative splicing in cancer cells. Chin. Med. J. 2020, 133, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Francies, F.Z.; Dlamini, Z. Aberrant Splicing Events and Epigenetics in Viral Oncogenomics: Current Therapeutic Strategies. Cells 2021, 10, 239. [Google Scholar] [CrossRef]
- Bryant, H.; Wadd, S.; Lamond, A.; Silverstein, S.; Clements, J. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 2001, 75, 4376–4385. [Google Scholar] [CrossRef]
- Tang, S.; Patel, A.; Krause, P.R. Hidden regulation of herpes simplex virus 1 pre-mRNA splicing and polyadenylation by virally encoded immediate early gene ICP27. PLoS Pathog. 2019, 15, e1007884. [Google Scholar] [CrossRef]
- Sedlackova, L.; Perkins, K.D.; Lengyel, J.; Strain, A.K.; Van Santen, V.L.; Rice, S.A. Herpes simplex virus type 1 ICP27 regulates expression of a variant, secreted form of glycoprotein C by an intron retention mechanism. J. Virol. 2008, 82, 7443–7455. [Google Scholar] [CrossRef]
- Perkins, K.D.; Gregonis, J.; Borge, S.; Rice, S.A. Transactivation of a viral target gene by herpes simplex virus ICP27 is posttranscriptional and does not require the endogenous promoter or polyadenylation site. J. Virol. 2003, 77, 9872–9884. [Google Scholar] [CrossRef]
- Amor, S.; Strassheim, S.; Dambrine, G.; Remy, S.; Rasschaert, D.; Laurent, S. ICP27 protein of Marek’s disease virus interacts with SR proteins and inhibits the splicing of cellular telomerase chTERT and viral vIL8 transcripts. J. Gen. Virol. 2011, 92, 1273–1278. [Google Scholar] [CrossRef]

| Construct | Primer or Probe Sequence (5′–3′) | |
|---|---|---|
| E3′-FLAG | For | GTAGTGTCTGGCTGTAAAGCTAATTTGGTTAAGGTTTTCCGGCAGC GATTACAAGGATGACGACGATAAGTAGGGATAACAGGGTAATCGATTT |
| Rev | ACATACCTTCCTGTTCTTCTTGAGAGCAAAGCTACAAAAGCTTAT CGTCGTCATCCTTGTAATCGCTGCCGCCAGTGTTACAACCAATTAACC | |
| vΔE3′-FLAG | For | CTTCCTGTTCTTCTTGAGAGCAAAGCTACAAAAGGGAAAACTTTA ACCAAATTAGCTTTACAGCCAGTAGGGATAACAGGGTAATCGATTT |
| Rev | CTTAGGTGTAGTGTCTGGCTGTAAAGCTAATTTGGTTAAAGTTTTCCGCCAGTGTTACAACCAATTAACC | |
| vΔE3′ | For | GCTACAAAAGCTTATCGTCGTCATCCTTGTAATCGGAAAACTTTA ACCAAATTAGCTTTACAGCCAGTAGGGATAACAGGGTAATCGATTT |
| Rev | CTTAGGTGTAGTGTCTGGCTGTAAAGCTAATTTGGTTAAAG TTTTCCGCCAGTGTTACAACCAATTAACC | |
| vIL-8 sequencing | For | CCGTATCCCTGCTCCATCCAATAGC |
| Rev | GGTCTCCAATATCACGTGTTGGTGG | |
| ICP4 | For | CGTGTTTTCCGGCATGTG |
| Rev | TCCCATACCAATCCTCATCCA | |
| Probe | FAM-CCCCCACCAGGTGCAGGCA-TAM | |
| iNOS | For | GAGTGGTTTAAGGAGTTGGATCTGA |
| Rev | TTCCAGACCTCCCACCTCAA | |
| Probe | FAM-CTCTGCCTGCTGTTGCCAACATGC-TAM | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, Y.; Hagag, I.T.; Kheimar, A.; Bertzbach, L.D.; Kaufer, B.B. Characterization of a Novel Viral Interleukin 8 (vIL-8) Splice Variant Encoded by Marek’s Disease Virus. Microorganisms 2021, 9, 1475. https://doi.org/10.3390/microorganisms9071475
You Y, Hagag IT, Kheimar A, Bertzbach LD, Kaufer BB. Characterization of a Novel Viral Interleukin 8 (vIL-8) Splice Variant Encoded by Marek’s Disease Virus. Microorganisms. 2021; 9(7):1475. https://doi.org/10.3390/microorganisms9071475
Chicago/Turabian StyleYou, Yu, Ibrahim T. Hagag, Ahmed Kheimar, Luca D. Bertzbach, and Benedikt B. Kaufer. 2021. "Characterization of a Novel Viral Interleukin 8 (vIL-8) Splice Variant Encoded by Marek’s Disease Virus" Microorganisms 9, no. 7: 1475. https://doi.org/10.3390/microorganisms9071475
APA StyleYou, Y., Hagag, I. T., Kheimar, A., Bertzbach, L. D., & Kaufer, B. B. (2021). Characterization of a Novel Viral Interleukin 8 (vIL-8) Splice Variant Encoded by Marek’s Disease Virus. Microorganisms, 9(7), 1475. https://doi.org/10.3390/microorganisms9071475






