The Histidine Biosynthetic Genes in the Superphylum Bacteroidota-Rhodothermota-Balneolota-Chlorobiota: Insights into the Evolution of Gene Structure and Organization
Abstract
1. Introduction
1.1. Organization of the Histidine Genes
1.2. Regulation of the Histidine Biosynthesis
1.3. The Bacteroidota-Rhodothermota-Balneolota-Chlorobiota Superphylum
2. Materials and Methods
2.1. Sequence Data Source and Sequence Alignment
2.2. Phylogenetic Trees Construction
3. Results and Discussion
3.1. Retrieval of His Biosynthetic Protein Sequences from Bacteroidota, Chlorobiota, Balneolota, and Rhodothermota and from Outgroups
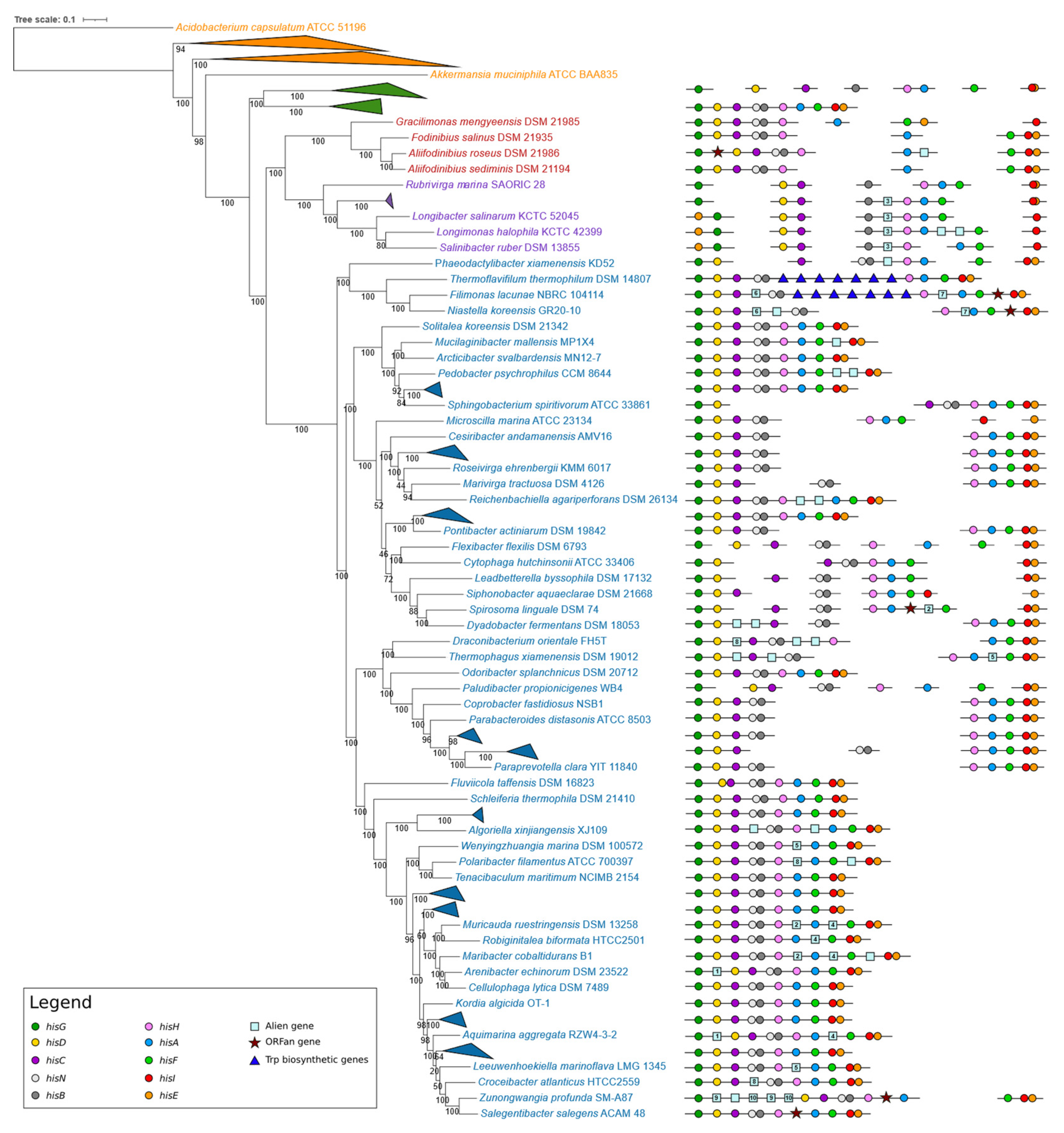
3.2. Structure of Histidine Biosynthetic Genes in Bacteroidetes, Chlorobi, Balneolota, and Rhodothermota
3.2.1. Structure of hisA/hisF
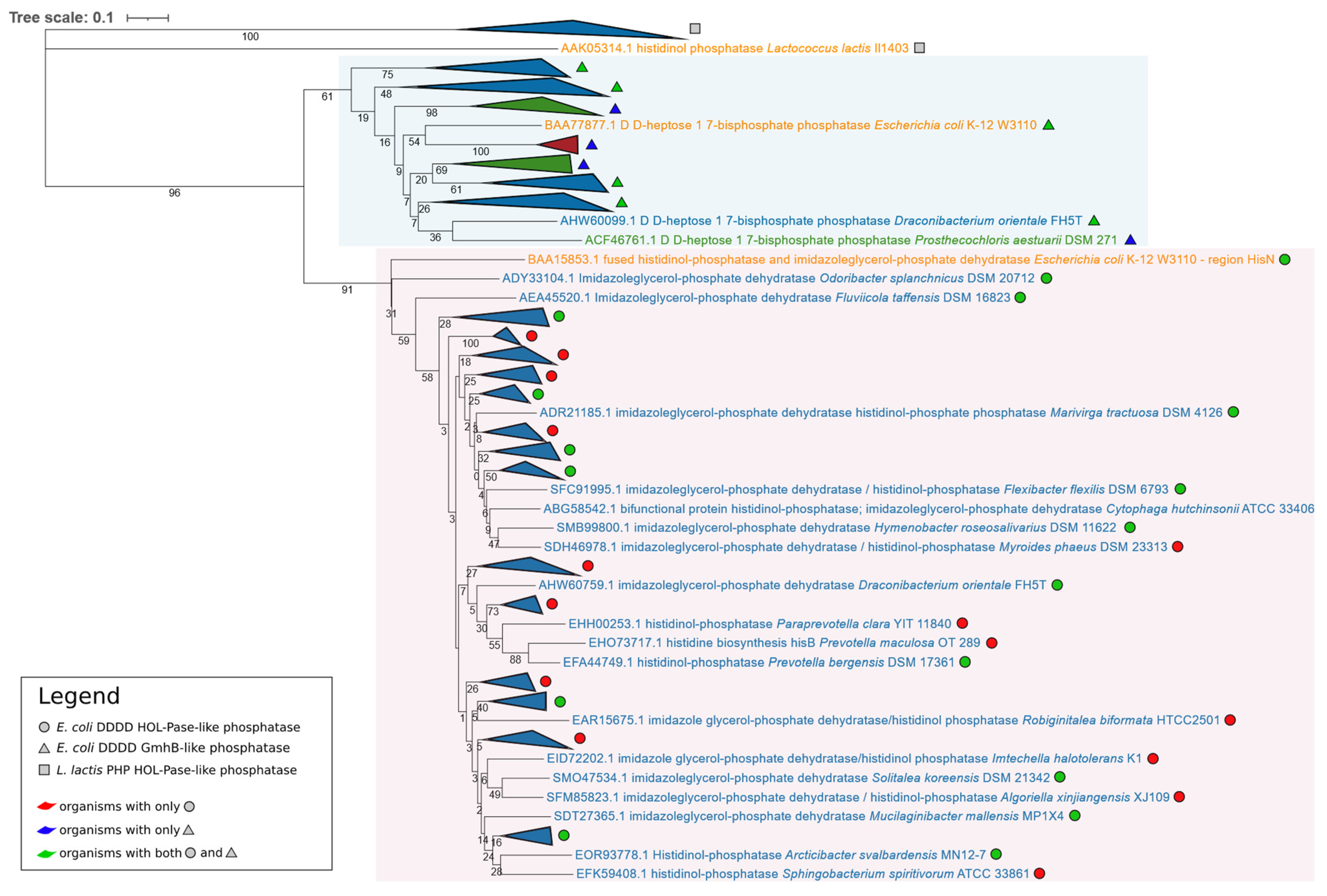
3.2.2. Structure of hisB and Identification of a hisN Homologue
- i.
- The L. lactis PHP type sequence did not retrieve any sequence at an E-value below 1.0, suggesting the absence of such phosphatases in the considered groups.
- ii.
- The E. coli DDDD HOL-Pase amino acid sequence (encoded by hisN) and GmhB amino acid sequence retrieved the same D,D-heptose 1,7-bisphosphate phosphatase from the genome of each bacterium belonging to Chlorobi. The E. coli GmhB retrieved a D,D-heptose 1,7-bisphosphate phosphatase for only two Balneolota out of four (Aliifodinibius roseus DSM 21986 and Aliifodinibius sediminis DSM 21194) and did not retrieve any sequence from Rhodothermota genomes, suggesting that bacteria belonging to these groups might use an enzyme belonging to a different family of phosphatases.
- i.
- In some Bacteroidetes, a pair gmhB/hisN was found, suggesting that in these bacteria, the two genes are the descendants of an ancestral gene coding for a bifunctional enzyme catalyzing the sixth step of histidine biosynthesis and the heptose synthesis, in agreement with Jensen’s hypothesis [45].
- ii.
- In Chlorobi, a single gmhB gene was found, and it can be speculated that it might code for a bifunctional enzyme, even though no experimental evidence in this sense is available.
- iii.
- In other cases, the eighth step of histidine biosynthesis is catalyzed by a different DDDD HOL-Pase.
3.2.3. Structure of hisIE
3.2.4. Structure of hisG and hisZ
3.2.5. Structure of hisD, hisC, and hisH
3.2.6. Organization of Histidine Biosynthetic Genes
- i.
- Genes organized in homogeneous (and more or less compact) operons; in this case, the operon embeds only genes involved in the same metabolic pathway.
- ii.
- Genes organized in heterogeneous (and more or less compact) operons; such operons include genes involved in the same metabolic pathways and one or more “alien” genes (genes apparently not involved in the same metabolic route and having homologs in other species) [2] or “ORFan” genes (lacking homologs in closely related species and probably acquired from bacteriophages) [46] responsible for other metabolic abilities.
- iii.
- Genes organized in homogeneous and/or heterogeneous (and more or less compact) suboperons.
- iv.
- Genes partially scattered and partially organized in homogeneous and/or heterogeneous suboperons.
- v.
- Genes completely scattered (regulons).
3.2.7. “Unusual” his Gene Structures and Organization
3.3. Phylogenetic Analyses
3.4. A Model for the Evolution of Histidine Biosynthetic Genes Structure and Organization in Bacteroidetes/Chlorobi/Balneolota/Rhodothermota
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fani, R.; Fondi, M. Origin and evolution of metabolic pathways. Phys. Life Rev. 2009, 6, 23–52. [Google Scholar] [CrossRef] [PubMed]
- Fondi, M.; Emiliani, G.; Fani, R. Origin and evolution of operons and metabolic pathways. Res. Microbiol. 2009, 160, 502–512. [Google Scholar] [CrossRef] [PubMed]
- Fani, R.; Brilli, M.; Fondi, M.; Lió, P. The role of gene fusions in the evolution of metabolic pathways: The histidine biosynthesis case. BMC Evol. Biol. 2007, 7, S4. [Google Scholar] [CrossRef] [PubMed]
- Alifano, P.; Fani, R.; Liò, P.; Lazcano, A.; Bazzicalupo, M.; Carlomagno, M.S.; Bruni, C.B. Histidine biosynthetic pathway and genes: Structure, regulation, and evolution. Microbiol. Rev. 1996, 60, 44–69. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Montañez, S.; Winkler, M.E. Biosynthesis of Histidine. EcoSal Plus 2009, 3. [Google Scholar] [CrossRef]
- Shen, C.; Lazcano, A.; Oró, J. The enhancement activites of histidyl-histidine in some prebiotic reactions. J. Mol. Evol. 1990, 31, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Mills, T.; Oró, J. Prebiotic synthesis of histidyl-histidine. J. Mol. Evol. 1990, 31, 175–179. [Google Scholar] [CrossRef] [PubMed]
- White, H.B. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 1976, 7, 101–104. [Google Scholar] [CrossRef]
- Fondi, M.; Emiliani, G.; Liò, P.; Gribaldo, S.; Fani, R. The evolution of histidine biosynthesis in archaea: Insights into the his genes structure and organization in luca. J. Mol. Evol. 2009, 69, 512–526. [Google Scholar] [CrossRef]
- Duca, S.D.; Chioccioli, S.; Vassallo, A.; Castronovo, L.M.; Fani, R. The role of gene elongation in the evolution of histidine biosynthetic genes. Microorganisms 2020, 8, 732. [Google Scholar] [CrossRef]
- Fani, R.; Liò, P.; Lazcano, A. Molecular evolution of the histidine biosynthetic pathway. J. Mol. Evol. 1995, 41, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Fani, R.; Brilli, M.; Liò, P. The origin and evolution of operons: The piecewise building of the proteobacterial histidine operon. J. Mol. Evol. 2005, 60, 378–390. [Google Scholar] [CrossRef]
- Fani, R.; Brilli, M.; Liò, P. Inference from proteobacterial operons shows piecewise organization: A reply to Price et al. J. Mol. Evol. 2006, 63, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Alifano, P.; Fani, R. Histidine Operon. In Brenner’s Encyclopedia of Genetics: Second Edition; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 471–476. ISBN 9780080961569. [Google Scholar]
- Sissler, M.; Delorme, C.; Bond, J.; Ehrlich, S.D.; Renault, P.; Francklyn, C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 8985–8990. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Lorenzini, E. Phylogeny and molecular signatures (conserved proteins and indels) that are specific for the Bacteroidetes and Chlorobi species. BMC Evol. Biol. 2007, 7, 71. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut Bacteroidetes: The food connection. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef]
- García-López, M.; Meier-Kolthoff, J.P.; Tindall, B.J.; Gronow, S.; Woyke, T.; Kyrpides, N.C.; Hahnke, R.L.; Göker, M. Analysis of 1000 Type-Strain Genomes Improves Taxonomic Classification of Bacteroidetes. Front. Microbiol. 2019, 10, 2083. [Google Scholar] [CrossRef]
- Eisen, J.A.; Nelson, K.E.; Paulsen, I.T.; Heidelberg, J.F.; Wu, M.; Dodson, R.J.; Deboy, R.; Gwinn, M.L.; Nelson, W.C.; Haft, D.H.; et al. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 2002, 99, 9509–9514. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.A.; Frigaard, N.U. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006, 14, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Hahnke, R.L.; Meier-Kolthoff, J.P.; García-López, M.; Mukherjee, S.; Huntemann, M.; Ivanova, N.N.; Woyke, T.; Kyrpides, N.C.; Klenk, H.P.; Göker, M. Genome-based taxonomic classification of Bacteroidetes. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Whitman, W.B.; Oren, A.; Chuvochina, M.; da Costa, M.S.; Garrity, G.M.; Rainey, F.A.; Rossello-Mora, R.; Schink, B.; Sutcliffe, I.; Trujillo, M.E.; et al. Proposal of the suffix—ota to denote phyla. Addendum to ‘proposal to include the rank of phylum in the international code of nomenclature of prokaryotes’. Int. J. Syst. Evol. Microbiol. 2018, 68, 967–969. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Khijniak, T.V.; Galinski, E.A.; Kublanov, I.V. Natronotalea proteinilytica gen. nov., sp. nov. and Longimonas haloalkaliphila sp. nov., extremely haloalkaliphilic members of the phylum Rhodothermaeota from hypersaline alkaline lakes. Int. J. Syst. Evol. Microbiol. 2017, 67, 4161–4167. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; Muntyan, M.S.; Toshchakov, S.V.; Korzhenkov, A.; Kublanov, I.V. Phenotypic and Genomic Properties of a Novel Deep-Lineage Haloalkaliphilic Member of the Phylum Balneolaeota From Soda Lakes Possessing Na+-Translocating Proteorhodopsin. Front. Microbiol. 2018, 9, 2672. [Google Scholar] [CrossRef]
- Munoz, R.; Rosselló-Móra, R.; Amann, R. Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov. Syst. Appl. Microbiol. 2016, 39, 281–296. [Google Scholar] [CrossRef]
- Johnson, E.L.; Heaver, S.L.; Walters, W.A.; Ley, R.E. Microbiome and metabolic disease: Revisiting the bacterial phylum Bacteroidetes. J. Mol. Med. 2017, 95. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Chilton, J.M.; Grüning, B.; Johnson, J.E.; Soranzo, N. NCBI BLAST+ integrated into Galaxy. Gigascience 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Proceedings of the Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Reimer, L.C.; Vetcininova, A.; Carbasse, J.S.; Söhngen, C.; Gleim, D.; Ebeling, C.; Overmann, J. BacDive in 2019: Bacterial phenotypic data for High-throughput biodiversity analysis. Nucleic Acids Res. 2019, 47, D631–D636. [Google Scholar] [CrossRef]
- Fani, R.; Liò, P.; Chiarelli, I.; Bazzicalupo, M. The evolution of the histidine biosynthetic genes in prokaryotes: A common ancestor for the hisA and hisF genes. J. Mol. Evol. 1994, 38, 489–495. [Google Scholar] [CrossRef]
- Thoma, R.; Obmolova, G.; Lang, D.A.; Schwander, M.; Jenö, P.; Sterner, R.; Wilmanns, M. Efficient expression, purification and crystallisation of two hyperthermostable enzymes of histidine biosynthesis. FEBS Lett. 1999, 454, 1–6. [Google Scholar] [CrossRef]
- Winkler, M.E. Biosynthesis of histidine. In Escherichia coli and Salmonella Typhimurium: Cellular and Molecular Biology; Neidhardt, F., Ingraham, J., Low, K., Magasanik, B., Schaechter, M., Humbarger, H., Eds.; ASM Press: Washington, DC, USA, 1987; pp. 395–411. [Google Scholar]
- Brilli, M.; Fani, R. Molecular Evolution of hisB Genes. J. Mol. Evol. 2004, 58, 225–237. [Google Scholar] [CrossRef]
- Le Coq, D.; Fillinger, S.; Aymerich, S. Histidinol phosphate phosphatase, catalyzing the penultimate step of the histidine biosynthesis pathway, is encoded by ytvp (his j) in bacillus subtilis. J. Bacteriol. 1999, 181, 3277–3280. [Google Scholar] [CrossRef] [PubMed]
- Kneidinger, B.; Marolda, C.; Graninger, M.; Zamyatina, A.; McArthur, F.; Kosma, P.; Valvano, M.A.; Messner, P. Biosynthesis pathway of ADP-L-glycero-β-D-manno-heptose in Escherichia coli. J. Bacteriol. 2002, 184, 363–369. [Google Scholar] [CrossRef]
- Shih, G.C.; Kahler, C.M.; Carlson, R.W.; Rahman, M.M.; Stephens, D.S. gmhX, a novel gene required for the incorporation of L-glycero-D-manno-heptose into lipooligosaccharide in Neisseria meningitidis. Microbiology 2001, 147, 2367–2377. [Google Scholar] [CrossRef][Green Version]
- Jensen, R.A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976, 30, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Arkin, A.P.; Alm, E.J. The life-cycle of operons. PLoS Genet. 2006, 2, 0859–0873. [Google Scholar] [CrossRef] [PubMed]
- Dabizzi, S.; Ammannato, S.; Fani, R. Expression of horizontally transferred gene clusters: Activation by promoter-generating mutations. Res. Microbiol. 2001, 152, 539–549. [Google Scholar] [CrossRef]
- Chioccioli, S.; Bogani, P.; Duca, S.D.; Castronovo, L.M.; Vassallo, A.; Puglia, A.M.; Fani, R. In vivo evaluation of the interaction between the Escherichia coli IGP synthase subunits using the bacterial two-hybrid system. FEMS Microbiol. Lett. 2020, 367, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Glansdorff, N. On the origin of operons and their possible role in evolution toward thermophily. J. Mol. Evol. 1999, 49, 432–438. [Google Scholar] [CrossRef]
| Module | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1 | - | 16.6 | 9.3 | 13.3 | 18.7 | 6.0 | 14.7 | 9.6 |
| 2 | 43.3 | - | 18.1 | 13.3 | 9.6 | 15.1 | 9.0 | 15.6 |
| 3 | 40.5 | 36.3 | - | 10.0 | 9.3 | 11.7 | 15.1 | 6.0 |
| 4 | 40.0 | 30.0 | 26.6 | - | 16.6 | 12.1 | 6.2 | 14.3 |
| 5 | 40.6 | 35.4 | 34.3 | 36.1 | - | 18.1 | 13.3 | 16.6 |
| 6 | 36.0 | 39.4 | 32.3 | 27.2 | 39.4 | - | 13.3 | 16.1 |
| 7 | 44.1 | 45.4 | 45.4 | 37.5 | 40.0 | 40.0 | - | 15.1 |
| 8 | 45.1 | 40.6 | 43.3 | 34.3 | 47.2 | 38.7 | 42.4 | - |
| Phylum | Organism | L. lactis PHP HOL-Pase | E. coli DDDD HOL-Pase | E. coli DDDD GmhB |
|---|---|---|---|---|
| Chlorobi | Chlorobaculum limnaeum DSM 1677 | - | AOS84578.1 | AOS84578.1 |
| Chlorobaculum tepidum TLS | - | AAM73229.1 | AAM73229.1 | |
| Chlorobium limicola DSM 245 | - | ACD91212.1 | ACD91212.1 | |
| Chlorobium luteolum DSM 273 | - | ABB23109.1 | ABB23109.1 | |
| Chlorobium phaeobacteroides DSM 266 | - | ABL66364.1 | ABL66364.1 | |
| Chloroherpeton thalassium ATCC 35110 | - | ACF14131.1 | ACF14131.1 | |
| Ignavibacterium album JCM 16511 | - | AFH49163.1 | AFH49163.1 | |
| Melioribacter roseus P3M-2 | - | AFN73296.1 | AFN73296.1 | |
| Prosthecochloris aestuarii DSM 271 | - | ACF46761.1 | ACF46761.1 ACF47029.1 | |
| Balneolota | Aliifodinibius roseus DSM 21986 | - | - | SHF20111.1 |
| Aliifodinibius sediminis DSM 21194 | - | - | SMO32123.1 | |
| Fodinibius salinus DSM 21935 | - | - | - | |
| Gracilimonas mengyeensis DSM 21985 | - | - | - | |
| Rhodothermota | Longibacter salinarum KCTC 52045 | - | - | - |
| Longimonas halophila KCTC 42399 | - | - | - | |
| Rhodothermus marinus DSM 4252 | - | - | - | |
| Rhodothermus profundi DSM 22212 | - | - | - | |
| Rubrivirga marina SAORIC-28 | - | - | - | |
| Salinibacter ruber DSM 13855 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Duca, S.; Riccardi, C.; Vassallo, A.; Fontana, G.; Castronovo, L.M.; Chioccioli, S.; Fani, R. The Histidine Biosynthetic Genes in the Superphylum Bacteroidota-Rhodothermota-Balneolota-Chlorobiota: Insights into the Evolution of Gene Structure and Organization. Microorganisms 2021, 9, 1439. https://doi.org/10.3390/microorganisms9071439
Del Duca S, Riccardi C, Vassallo A, Fontana G, Castronovo LM, Chioccioli S, Fani R. The Histidine Biosynthetic Genes in the Superphylum Bacteroidota-Rhodothermota-Balneolota-Chlorobiota: Insights into the Evolution of Gene Structure and Organization. Microorganisms. 2021; 9(7):1439. https://doi.org/10.3390/microorganisms9071439
Chicago/Turabian StyleDel Duca, Sara, Christopher Riccardi, Alberto Vassallo, Giulia Fontana, Lara Mitia Castronovo, Sofia Chioccioli, and Renato Fani. 2021. "The Histidine Biosynthetic Genes in the Superphylum Bacteroidota-Rhodothermota-Balneolota-Chlorobiota: Insights into the Evolution of Gene Structure and Organization" Microorganisms 9, no. 7: 1439. https://doi.org/10.3390/microorganisms9071439
APA StyleDel Duca, S., Riccardi, C., Vassallo, A., Fontana, G., Castronovo, L. M., Chioccioli, S., & Fani, R. (2021). The Histidine Biosynthetic Genes in the Superphylum Bacteroidota-Rhodothermota-Balneolota-Chlorobiota: Insights into the Evolution of Gene Structure and Organization. Microorganisms, 9(7), 1439. https://doi.org/10.3390/microorganisms9071439







