Effects of Aspergillus fumigatus Conidia on Apoptosis and Proliferation in an In Vitro Model of the Lung Microenvironment
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Suspension of AF293 Conidia
2.3. A Two-Dimensional (2D) Proliferation Assay
2.4. Three-Dimensional Floating (3DF) Cell Culture
2.5. Detection of Apoptotic Cells in Spheroids
2.6. Western Blotting
2.7. RNA Sequencing Analysis (RNA-Seq)
2.8. Real-Time Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
2.9. Ontology Analyses
2.10. Statistical Analyses in Cell Culture Experiments
3. Results
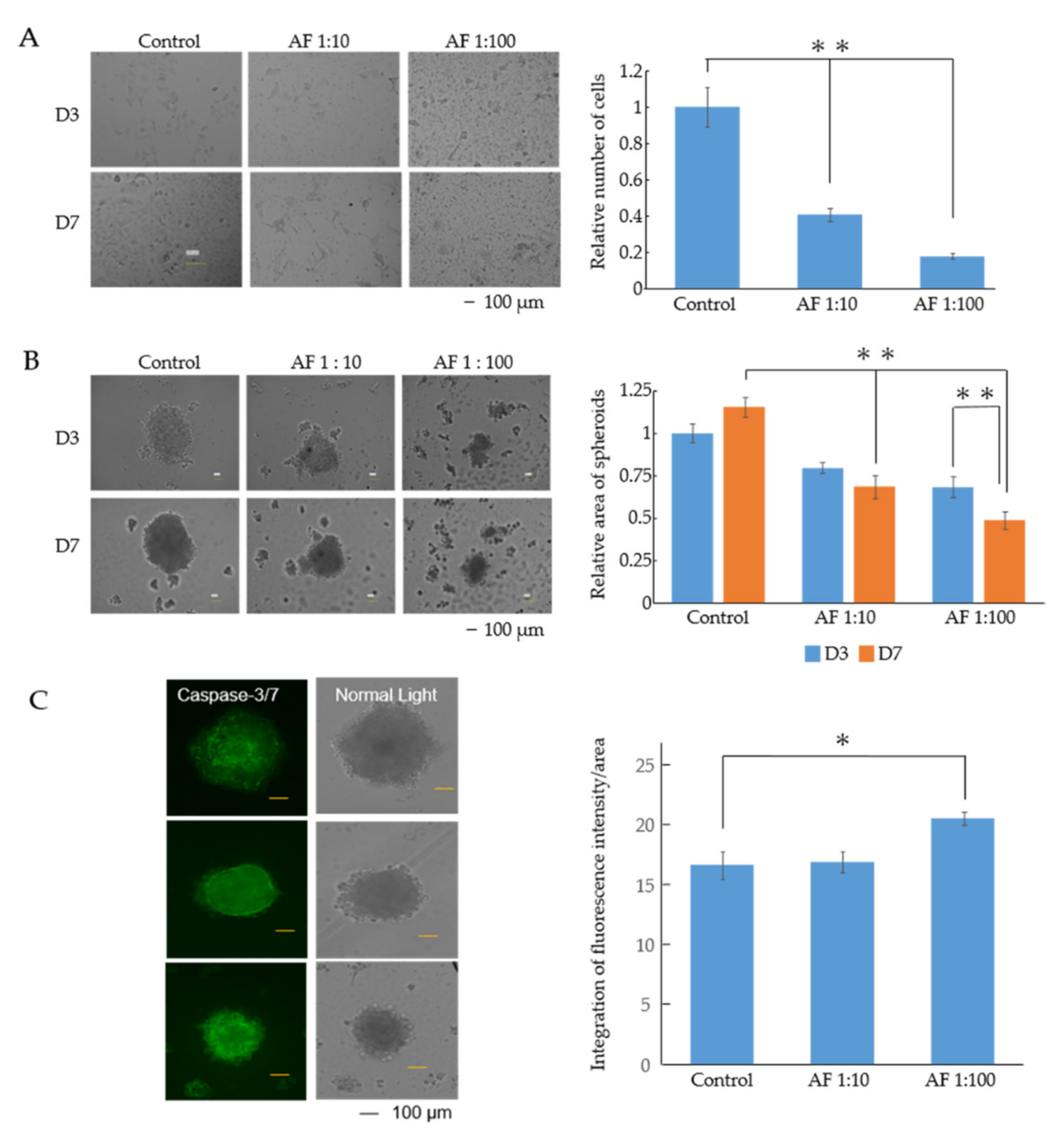
3.1. AF293 Conidia Suppress the Growth of A549 Cells
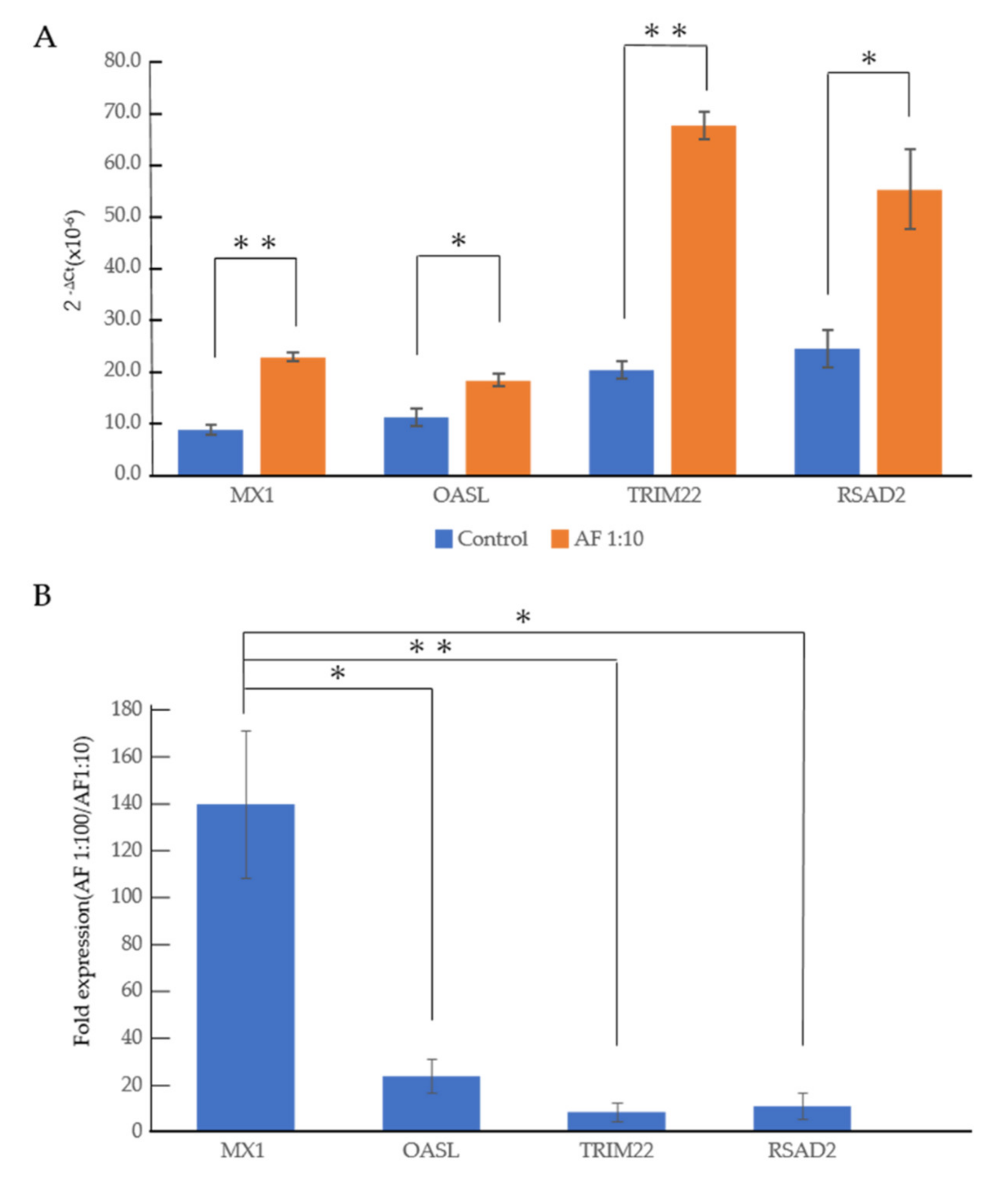
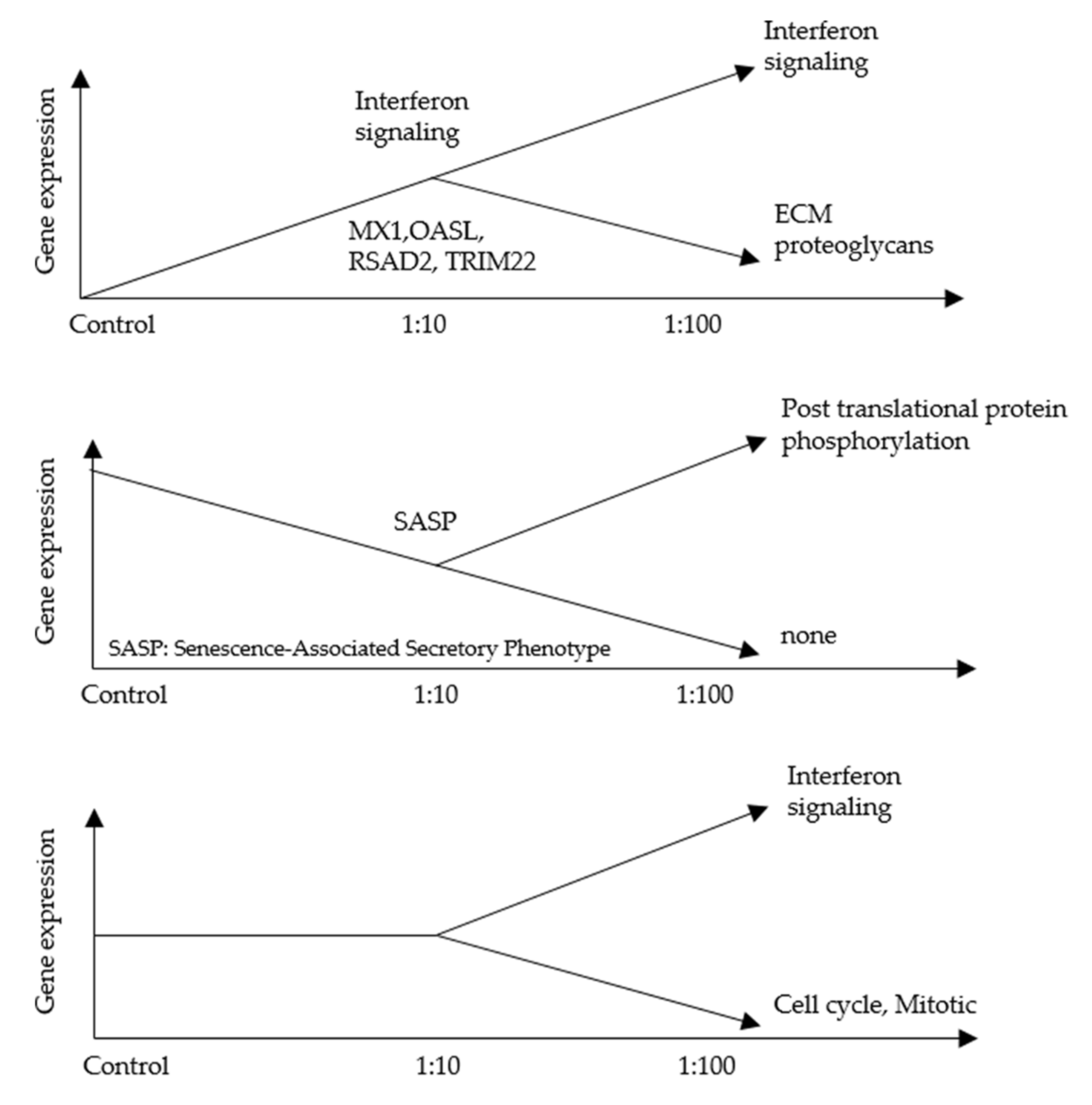
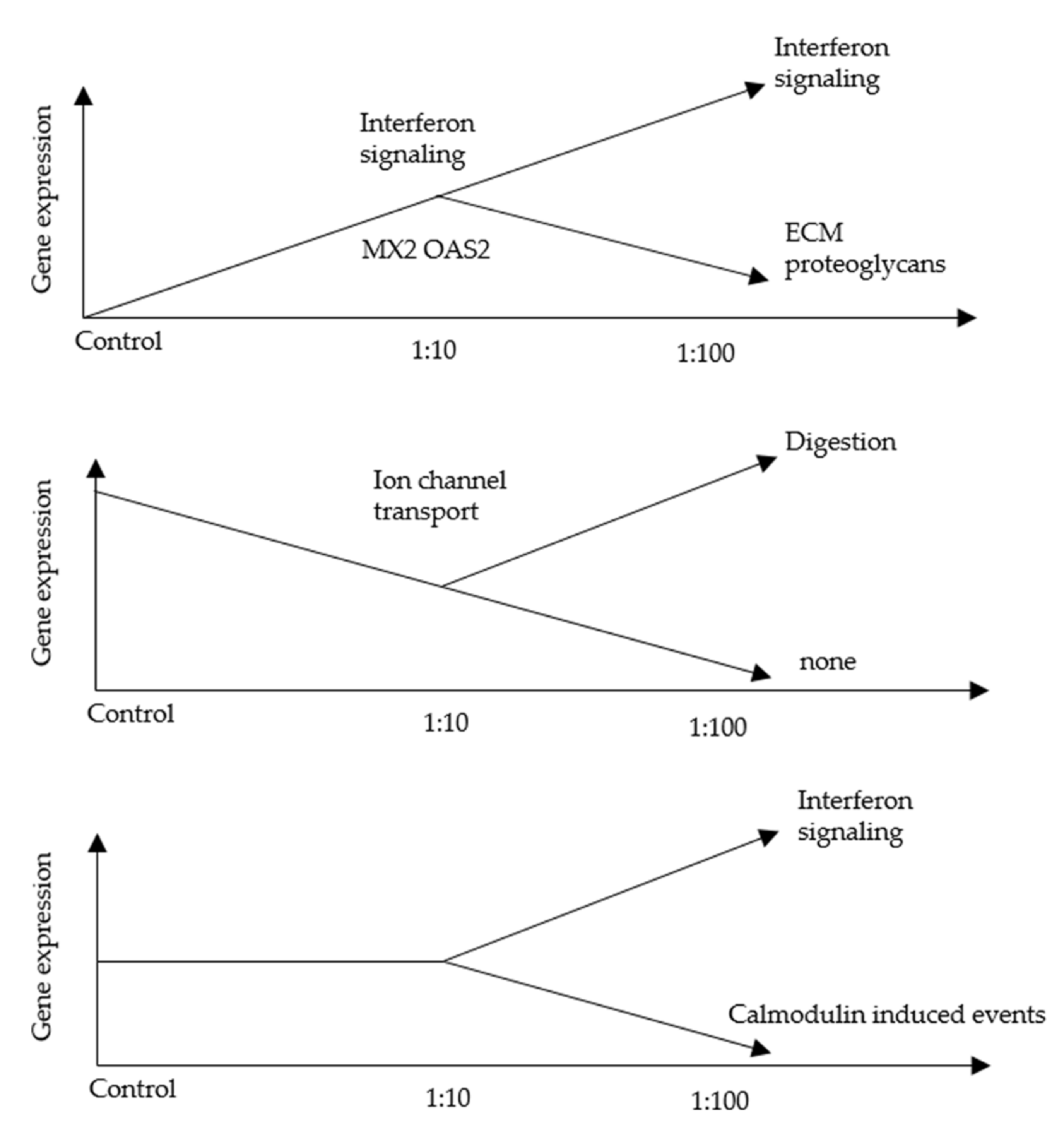
3.2. An RNA-Seq Analysis of Genes Induced by AF293 Conidia in A549 Cells
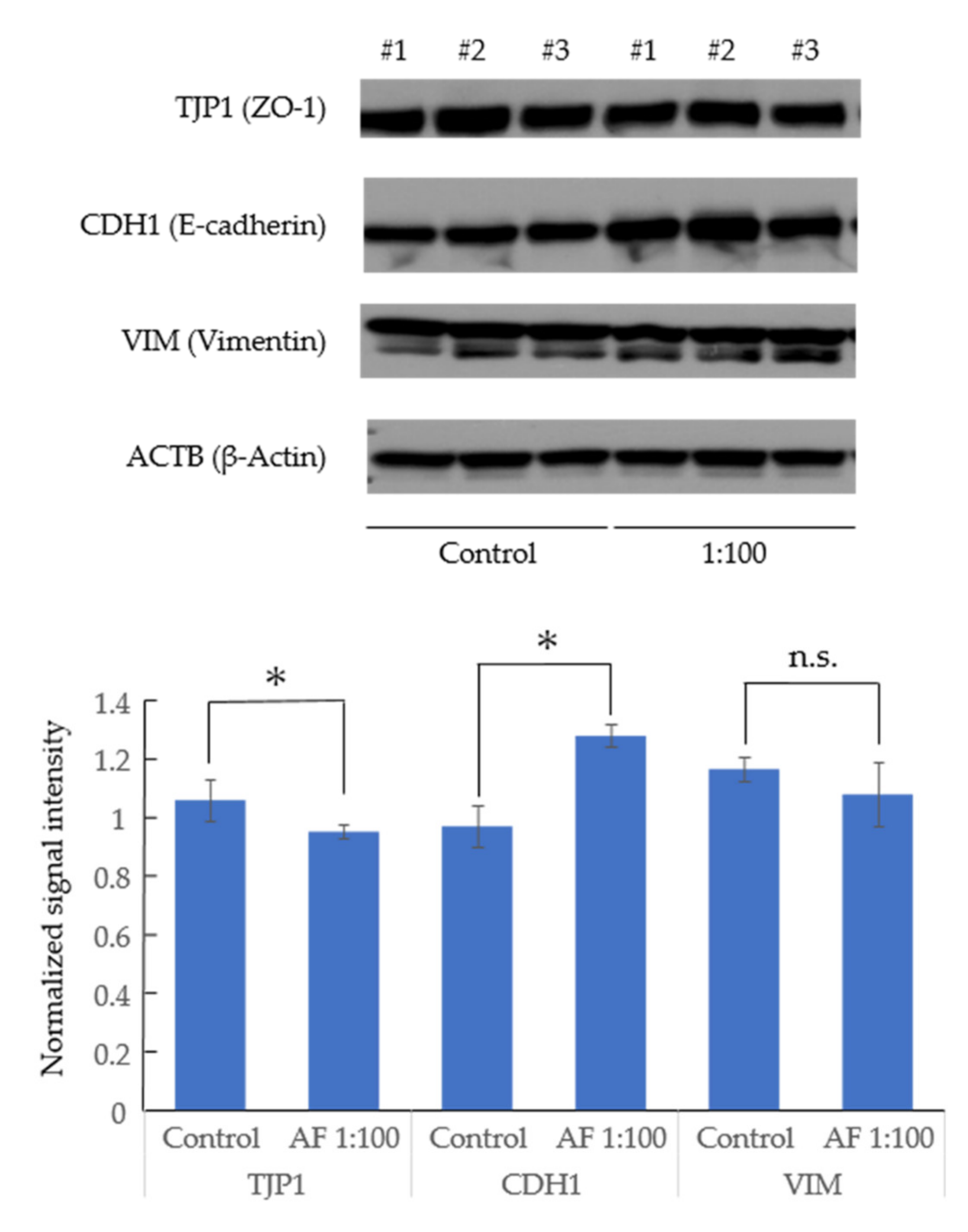
3.3. AF293 Conidia Did Not Increase the Expression of Epithelial-Mesenchymal Transition (EMT)-Related Genes in A549 Cells
3.4. AF293 Conidia Promoted the Growth of Mouse NIH/3T3 Cells
3.5. An RNA-Seq Analysis of Genes Induced by AF293 Conidia in NIH/3T3 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sabino, R.; Veríssimo, C.; Viegas, C.; Viegas, S.; Brandão, J.; Alves-Correia, M.; Borrego, L.M.; Clemons, K.V.; Stevens, D.A.; Richardson, M. The role of occupational aspergillus exposure in the development of diseases. Med. Mycol. 2019, 57 (Suppl. 2), S196–S205. [Google Scholar] [CrossRef]
- Tashiro, T.; Izumikawa, K.; Tashiro, M.; Takazono, T.; Morinaga, Y.; Yamamoto, K.; Imamura, Y.; Miyazaki, T.; Seki, M.; Kakeya, H.; et al. Diagnostic significance of Aspergillus species isolated from respiratory samples in an adult pneumology ward. Med. Mycol. 2011, 49, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.J.; Anaissie, E.J.; Denning, D.W.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Segal, B.H.; Steinbach, W.J.; Stevens, D.A.; et al. Treatment of aspergillosis: Clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect Dis. 2008, 46, 327–360. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Chen, S.Y.; Yeh, W.S.; Maroni, B.; Li, Q.; Lee, Y.C.; Collard, H.R. Idiopathic pulmonary fibrosis in US Medicare beneficiaries aged 65 years and older: Incidence, prevalence, and survival, 2001–2011. Lancet Respir. Med. 2014, 2, 566–572. [Google Scholar] [CrossRef]
- Strongman, H.; Kausar, I.; Maher, T.M. Incidence, Prevalence, and Survival of Patients with Idiopathic Pulmonary Fibrosis in the UK. Adv. Ther. 2018, 35, 724–736. [Google Scholar] [CrossRef]
- Yokohori, N.; Aoshiba, K.; Nagai, A.; Respiratory Failure Research Group in Japan. Increased levels of cell death and proliferation in alveolar wall cells in patients with pulmonary emphysema. Chest 2004, 125, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Kuwano, K.; Kunitake, R.; Kawasaki, M.; Nomoto, Y.; Hagimoto, N.; Nakanishi, Y.; Hara, N. P21Waf1/Cip1/Sdi1 and p53 expression in association with DNA strand breaks in idiopathic pulmonary fibrosis. Am. J. Respir Crit. Care Med. 1996, 154, 477–483. [Google Scholar] [CrossRef]
- Cottin, V.; Nunes, H.; Brillet, P.Y.; Delaval, P.; Devouassoux, G.; Tillie-Leblond, I.; Israel-Biet, D.; Court-Fortune, I.; Valeyre, D.; Cordier, J.F.; et al. Combined pulmonary fibrosis and emphysema: A distinct underrecognised entity. Eur. Respir J. 2005, 26, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Yoshimi, M.; Maeyama, T.; Hagimoto, N.; Kuwano, K.; Hara, N. Resistance to Fas-mediated apoptosis in human lung fibroblast. Eur. Respir J. 2002, 20, 359–368. [Google Scholar] [CrossRef]
- Kuwano, K.; Hagimoto, N.; Nakanishi, Y. The role of apoptosis in pulmonary fibrosis. Histol. Histopathol. 2004, 19, 867–881. [Google Scholar] [CrossRef]
- Yamada, M.; Kuwano, K.; Maeyama, T.; Hamada, N.; Yoshimi, M.; Nakanishi, Y.; Kasper, M. Dual-immunohistochemistry provides little evidence for epithelial-mesenchymal transition in pulmonary fibrosis. Histochem. Cell Biol. 2008, 129, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Homma, T.; Suzuki, S.; Takuma, T.; Tanaka, A.; Yokoe, T.; Ohnishi, T.; Niki, Y.; Sagara, H. High burden of Aspergillus fumigatus infection among chronic respiratory diseases. Chron. Respir Dis. 2018, 15, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Beisswenger, C.; Hess, C.; Bals, R. Aspergillus fumigatus conidia induce interferon-β signalling in respiratory epithelial cells. Eur. Respir J. 2012, 39, 411–418. [Google Scholar] [CrossRef]
- Berkova, N.; Lair-Fulleringer, S.; Féménia, F.; Huet, D.; Wagner, M.C.; Gorna, K.; Tournier, F.; Ibrahim-Granet, O.; Guillot, J.; Chermette, R.; et al. Aspergillus fumigatus conidia inhibit tumour necrosis factor- or staurosporine-induced apoptosis in epithelial cells. Int. Immunol. 2006, 18, 139–150. [Google Scholar] [CrossRef][Green Version]
- Tsunoda, T.; Takashima, Y.; Fujimoto, T.; Koyanagi, M.; Yoshida, Y.; Doi, K.; Tanaka, Y.; Kuroki, M.; Sasazuki, T.; Shirasawa, S. Three-dimensionally specific inhibition of DNA repair-related genes by activated KRAS in colon crypt model. Neoplasia 2010, 12, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.A.; Oster, C.G.; Mayer, M.M.; Avery, M.L.; Audus, K.L. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 1998, 243, 359–366. [Google Scholar] [CrossRef]
- Blasi, E.; Brozzetti, A.; Francisci, D.; Neglia, R.; Cardinali, G.; Bistoni, F.; Vidotto, V.; Baldelli, F. Evidence of microevolution in a clinical case of recurrent Cryptococcus neoformans meningoencephalitis. Eur. J. Clin. Microbiol. Infect Dis. 2001, 20, 535–543. [Google Scholar] [CrossRef]
- Tsunoda, T.; Ishikura, S.; Doi, K.; Iwaihara, Y.; Hidesima, H.; Luo, H.; Hirose, Y.; Shirasawa, S. Establishment of a Three-dimensional Floating Cell Culture System for Screening Drugs Targeting KRAS-mediated Signaling Molecules. Anticancer Res. 2015, 35, 4453–4459. [Google Scholar]
- Nishi, K.; Luo, H.; Ishikura, S.; Doi, K.; Iwaihara, Y.; Wills, L.; Baillie, G.S.; Sakata, T.; Shirasawa, S.; Tsunoda, T. Apremilast Induces Apoptosis of Human Colorectal Cancer Cells with Mutant KRAS. Anticancer Res. 2017, 37, 3833–3839. [Google Scholar] [CrossRef]
- Tsunoda, T.; Ota, T.; Fujimoto, T.; Doi, K.; Tanaka, Y.; Yoshida, Y.; Ogawa, M.; Matsuzaki, H.; Hamabashiri, M.; Tyson, D.R.; et al. Inhibition of phosphodiesterase-4 (PDE4) activity triggers luminal apoptosis and AKT dephosphorylation in a 3-D colonic-crypt model. Mol. Cancer 2012, 11, 46. [Google Scholar] [CrossRef]
- Kamburov, A.; Stelzl, U.; Lehrach, H.; Herwig, R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013, 41, D793–D800. [Google Scholar] [CrossRef]
- Hall, J.C.; Rosen, A. Type I interferons: Crucial participants in disease amplification in autoimmunity. Nat. Rev. Rheumatol. 2010, 6, 40–49. [Google Scholar] [CrossRef]
- Jiao, P.; Fan, W.; Cao, Y.; Zhang, H.; Tian, L.; Sun, L.; Luo, T.; Liu, W.; Li, J. Robust induction of interferon and interferon-stimulated gene expression by influenza B/Yamagata lineage virus infection of A549 cells. PLoS ONE 2020, 15, e0231039. [Google Scholar] [CrossRef]
- Vancheri, C. Idiopathic pulmonary fibrosis: An altered fibroblast proliferation linked to cancer biology. Proc. Am. Thorac. Soc. 2012, 9, 153–157. [Google Scholar] [CrossRef]
- Haller, O.; Staeheli, P.; Schwemmle, M.; Kochs, G. Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015, 23, 154–163. [Google Scholar] [CrossRef]
- Brown, S.G.; Knowell, A.E.; Hunt, A.; Patel, D.; Bhosle, S.; Chaudhary, J. Interferon inducible antiviral MxA is inversely associated with prostate cancer and regulates cell cycle, invasion and Docetaxel induced apoptosis. Prostate 2015, 75, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Horisberger, M.A. Interferon-induced human protein MxA is a GTPase which binds transiently to cellular proteins. J. Virol. 1992, 66, 4705–4709. [Google Scholar] [CrossRef]
- Domagalski, K.; Pawłowska, M.; Kozielewicz, D.; Dybowska, D.; Tretyn, A.; Halota, W. The Impact of IL28B Genotype and Liver Fibrosis on the Hepatic Expression of IP10, IFI27, ISG15, and MX1 and Their Association with Treatment Outcomes in Patients with Chronic Hepatitis C. PLoS ONE 2015, 10, e0130899. [Google Scholar] [CrossRef] [PubMed]
- Aljohani, A.I.; Joseph, C.; Kurozumi, S.; Mohammed, O.J.; Miligy, I.M.; Green, A.R.; Rakha, E.A. Myxovirus resistance 1 (MX1) is an independent predictor of poor outcome in invasive breast cancer. Breast Cancer Res. Treat 2020, 181, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Juraleviciute, M.; Pozniak, J.; Nsengimana, J.; Harland, M.; Randerson-Moor, J.; Wernhoff, P.; Bassarova, A.; Øy, G.F.; Trøen, G.; Flørenes, V.A.; et al. MX 2 is a novel regulator of cell cycle in melanoma cells. Pigment Cell Melanoma Res. 2020, 33, 446–457. [Google Scholar] [CrossRef]
- Walker, M.; Rizzuto, P.; Godin, M.; Pelling, A.E. Structural and mechanical remodeling of the cytoskeleton maintains tensional homeostasis in 3D microtissues under acute dynamic stretch. Sci. Rep. 2020, 10, 7696. [Google Scholar] [CrossRef] [PubMed]
- Hamano, Y.; Kida, H.; Ihara, S.; Murakami, A.; Yanagawa, M.; Ueda, K.; Honda, O.; Tripathi, L.P.; Arai, T.; Hirose, M.; et al. Classification of idiopathic interstitial pneumonias using anti-myxovirus resistance-protein 1 autoantibody. Sci. Rep. 2017, 7, 43201. [Google Scholar] [CrossRef] [PubMed]
- Selman, M.; Pardo, A. Revealing the pathogenic and aging-related mechanisms of the enigmatic idiopathic pulmonary fibrosis. an integral model. Am. J. Respir Crit. Care Med. 2014, 189, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
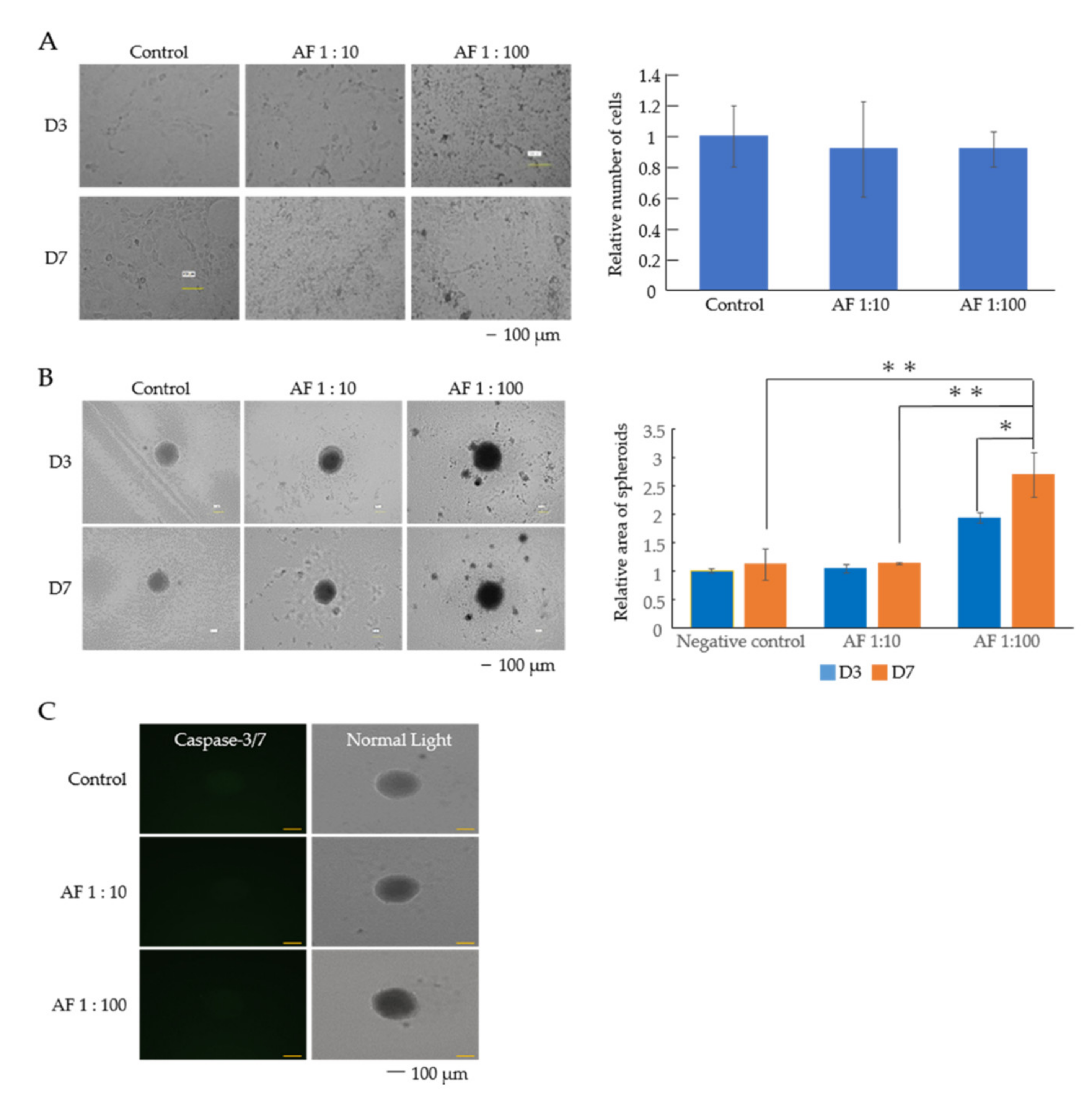
| Gene Symbol | 1:10/Control | 1:100/Control | 1:100/1:10 |
|---|---|---|---|
| MX1 | 2.53020134 | 189.2080537 | 74.77984085 |
| RSAD2 | 2.48148148 | 75.2962963 | 30.34328358 |
| OASL | 2.88034188 | 50.98290598 | 17.70029674 |
| CMPK2 | 2.19444444 | 33.41666667 | 15.2278481 |
| PARP10 | 2.03125 | 19.90625 | 9.8 |
| TRIM22 | 2.81481481 | 16.65432099 | 5.916666667 |
| RARRES3 | 3.39285714 | 14.46428571 | 4.263157895 |
| SAMD9L | 2.60608622 | 10.30092984 | 3.952643529 |
| SSPO | 2.075 | 7.4625 | 3.596385542 |
| KNDC1 | 2.56603774 | 6.132075472 | 2.389705882 |
| GOLGA8EP | 2.11111111 | 4.555555556 | 2.157894737 |
| AC068533.7 | 3.34883721 | 6.953488372 | 2.076388889 |
| Name of Pathway | Set Size | Candidates Contained | p-Value | Genes |
|---|---|---|---|---|
| Interferon signaling | 158 | 4 (2.5%) | <0.0001 | MX1, OASL, TRIM22, RSAD2 |
| Interferon alpha/beta signaling | 70 | 3 (4.3%) | <0.0001 | MX1, OASL, RSAD2 |
| Cytokine signaling in immune system | 458 | 4 (0.9%) | <0.0001 | MX1, OASL, TRIM22, RSAD2 |
| Interferon gamma signaling | 94 | 2 (2.1%) | 0.001 | OASL, TRIM22 |
| Immune system | 1840 | 4 (0.2%) | 0.001 | MX1, OASL, TRIM22, RSAD2 |
| Gene Symbol | 1:10/Control | p-Value | 1:100/Control | p-Value | 1:100/1:10 | p-Value |
|---|---|---|---|---|---|---|
| Oas2 | 17.6 | 0.01418642 | 227.0666667 | 0.000142248 | 12.90151515 | 0.000217291 |
| Oasl2 | 12.8181818 | 0.00105008 | 153.6439394 | 2.98985 × 10−8 | 11.98640662 | 1.97743 × 10−7 |
| Oas1a | 3.90163934 | 0.00115744 | 44.29508197 | 4.56841 × 10−5 | 11.35294118 | 6.24434 × 10−5 |
| Mx2 | 9.27692308 | 0.00694089 | 103.4923077 | 1.36501 × 10−6 | 11.15588723 | 4.53558 × 10−6 |
| Oas1g | 3.66666667 | 0.00878991 | 35.48888889 | 8.18609 × 10−5 | 9.678787879 | 0.000126154 |
| Usp18 | 2.55406912 | 0.00924884 | 20.79487179 | 9.33815 × 10−5 | 8.14185945 | 0.000131795 |
| Zbp1 | 2.7 | 0.08168567 | 21.74 | 0.000237643 | 8.051851852 | 0.000479164 |
| Isg15 | 4.02118003 | 0.01702177 | 30.3494705 | 9.91865 × 10−5 | 7.547404063 | 0.000208583 |
| Bst2 | 3.45379024 | 0.00064043 | 25.64382139 | 3.26371 × 10−6 | 7.424834636 | 6.17672 × 10−6 |
| Rtp4 | 3.24145786 | 0.01492393 | 21.29840547 | 9.5736 × 10−5 | 6.570625439 | 0.000206501 |
| Gbp3 | 4.82394366 | 0.004038 | 29.6971831 | 0.000166256 | 6.15620438 | 0.000345417 |
| Irgm2 | 7.52132701 | 0.005725 | 45.36966825 | 4.0298 × 10−5 | 6.032136106 | 0.000125601 |
| Rsad2 | 2.56321839 | 0.00226457 | 14.55172414 | 5.23578 × 10−5 | 5.677130045 | 8.98531 × 10−5 |
| Apol9a | 2.4 | 0.00182007 | 12.2691358 | 0.000515447 | 5.112139918 | 0.000880427 |
| Ifit3 | 3.18018018 | 0.01196764 | 16.25675676 | 3.98466 × 10−6 | 5.111898017 | 3.48212 × 10−5 |
| Xaf1 | 6.21891419 | 0.00600125 | 30.08231173 | 5.8742 × 10−6 | 4.83722895 | 6.10851 × 10−5 |
| Irf7 | 2.27311828 | 0.00105694 | 10.51397849 | 0.000500941 | 4.625354778 | 0.000872438 |
| Oas1b | 4.12820513 | 0.02932599 | 18.07692308 | 7.73289 × 10−5 | 4.378881988 | 0.000548655 |
| Ifit1 | 4.65353038 | 0.00588437 | 18.42857143 | 2.18323 × 10−7 | 3.960127029 | 4.76394 × 10−5 |
| Lgals3bp | 2.61022273 | 0.00030663 | 9.311536265 | 0.000139853 | 3.56733399 | 0.000351664 |
| Helz2 | 2.16917969 | 7.7487 × 10−5 | 6.965140407 | 8.40955 × 10−7 | 3.210955934 | 2.64734 × 10−6 |
| Ddx58 | 2.93550577 | 0.00063444 | 9.393075356 | 1.45365 × 10−5 | 3.199814986 | 7.08851 × 10−5 |
| Parp9 | 2.15489614 | 0.00044662 | 5.903264095 | 7.46111 × 10−6 | 2.739465712 | 2.5749 × 10−5 |
| Irf9 | 2.77596342 | 0.00099975 | 7.331809275 | 2.31458 × 10−5 | 2.641176471 | 0.000182173 |
| Apol8 | 2.20883534 | 0.00066116 | 4.977911647 | 0.000122632 | 2.253636364 | 0.000723058 |
| Name of Pathway | Set Size | Candidates Contained | p-Value |
|---|---|---|---|
| Interferon Signaling | 158 | 13 (8.2%) | 1.76 × 10−22 |
| Interferon alpha/beta signaling | 70 | 11 (15.7%) | 5.88 × 10−22 |
| Cytokine Signaling in Immune system | 458 | 13 (2.8%) | 2.30 × 10−16 |
| Immune System | 1840 | 14 (0.8%) | 5.06 × 10−10 |
| ISG15 antiviral mechanism | 30 | 4 (13.3%) | 5.38 × 10−8 |
| Antiviral mechanism by IFN-stimulated genes | 30 | 4 (13.3%) | 5.38 × 10−8 |
| Interferon gamma signaling | 94 | 4 (4.3%) | 5.69 × 10−6 |
| DDX58/IFIH1-mediated induction of interferon-alpha/beta | 56 | 3 (5.4%) | 4.94 × 10−5 |
| TRAF3-dependent IRF activation pathway | 13 | 2 (15.4%) | 0.000125 |
| TRAF6 mediated IRF7 activation | 17 | 2 (11.8%) | 0.000217 |
| Negative regulators of DDX58/IFIH1 signaling | 23 | 2 (8.7%) | 0.000401 |
| Cytosolic sensors of pathogen-associated DNA | 51 | 2 (3.9%) | 0.00198 |
| Innate Immune System | 1077 | 5 (0.5%) | 0.00985 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kushima, H.; Tsunoda, T.; Matsumoto, T.; Kinoshita, Y.; Izumikawa, K.; Shirasawa, S.; Fujita, M.; Ishii, H. Effects of Aspergillus fumigatus Conidia on Apoptosis and Proliferation in an In Vitro Model of the Lung Microenvironment. Microorganisms 2021, 9, 1435. https://doi.org/10.3390/microorganisms9071435
Kushima H, Tsunoda T, Matsumoto T, Kinoshita Y, Izumikawa K, Shirasawa S, Fujita M, Ishii H. Effects of Aspergillus fumigatus Conidia on Apoptosis and Proliferation in an In Vitro Model of the Lung Microenvironment. Microorganisms. 2021; 9(7):1435. https://doi.org/10.3390/microorganisms9071435
Chicago/Turabian StyleKushima, Hisako, Toshiyuki Tsunoda, Taichi Matsumoto, Yoshiaki Kinoshita, Koichi Izumikawa, Senji Shirasawa, Masaki Fujita, and Hiroshi Ishii. 2021. "Effects of Aspergillus fumigatus Conidia on Apoptosis and Proliferation in an In Vitro Model of the Lung Microenvironment" Microorganisms 9, no. 7: 1435. https://doi.org/10.3390/microorganisms9071435
APA StyleKushima, H., Tsunoda, T., Matsumoto, T., Kinoshita, Y., Izumikawa, K., Shirasawa, S., Fujita, M., & Ishii, H. (2021). Effects of Aspergillus fumigatus Conidia on Apoptosis and Proliferation in an In Vitro Model of the Lung Microenvironment. Microorganisms, 9(7), 1435. https://doi.org/10.3390/microorganisms9071435







