Abstract
Cadmium is an exceptionally toxic industrial and environmental pollutant classified as a human carcinogen. In order to provide insight into how we can keep our environment safe from cadmium contamination and prevent the accumulation of it in the food chain, we aim to elucidate how Aspergillus nidulans, one of the most abundant fungi in soil, survives and handles cadmium stress. As AtfA is the main transcription factor governing stress responses in A. nidulans, we examined genome-wide expression responses of wild-type and the atfA null mutant exposed to CdCl2. Both strains showed up-regulation of the crpA Cu2+/Cd2+ pump gene and AN7729 predicted to encode a putative bis(glutathionato)-cadmium transporter, and transcriptional changes associated with elevated intracellular Cys availability leading to the efficient adaptation to Cd2+. Although the deletion of atfA did not alter the cadmium tolerance of the fungus, the cadmium stress response of the mutant differed from that of a reference strain. Promoter and transcriptional analyses of the “Phospho-relay response regulator” genes suggest that the AtfA-dependent regulation of these genes can be relevant in this phenomenon. We concluded that the regulatory network of A. nidulans has a high flexibility allowing the fungus to adapt efficiently to stress both in the presence and absence of this important transcription factor.
1. Introduction
Cadmium is a highly toxic, nonessential heavy metal. Although it is regularly found in ores of zinc, lead, and copper, the elevated cadmium levels detected in the environment are of anthropogenic origin in most cases [1,2,3,4]. Atmospheric deposition, water irrigation, and the use of phosphate fertilizers with high cadmium content can all increase cadmium concentrations in agricultural fields and lead to accumulation of this toxic heavy metal in the food chain [1,4]. Deeper understanding of cadmium stress response may help in improving Cd2+ biosorption techniques based on microbial biomass [5,6,7,8,9,10] as well as developing new strategies to prevent the accumulation of Cd2+ in plants [10,11].
AtfA is a bZIP-type transcription factor of Aspergillus nidulans [12,13]. AtfA and its orthologs regulate sexual and asexual development, different stress responses, and secondary metabolism in filamentous fungi [14,15,16,17,18,19]. Not surprisingly, it contributes substantially to the virulence of both the human and plant pathogenic fungi [16,17,18,20,21,22]. Previously we found that the ΔatfA strain was more sensitive to oxidative stress induced by H2O2, menadione sodium bisulfite, terc-butylhydroperoxide, or diamide than the reference strain [23]. Transcriptomic studies also demonstrated that AtfA plays an important role in the regulation of oxidative stress: Deletion of its gene affected the transcription of a large number of genes during stress treatments [23,24,25]. In addition to the large number of genes affected, the stressor specificity of the AtfA-dependent genes and similar numbers from up- and down-regulated genes suggest that most affected genes are indirectly regulated by AtfA [24]. Elements of the phosphorelay response regulator systems (e.g., fphA, nikA, phkA, srrB, srrC, sskA, or tcsB) are among the few genes that showed consequent down-regulation in the ΔatfA mutant in comparison to the reference strain [23,24,25]. The strong AtfA-dependence of these regulatory genes raises the possibility that they are under direct control of AtfA. This would explain why deletion of atfA had a strong effect on the transcriptome.
Here we used a transcriptomics-based approach using RNAseq to elucidate the CdCl2-induced stress response in A. nidulans as a model organism. In addition to the reference strain, we also recorded the cadmium stress response of a ΔatfA mutant. Investigating two different strains helped us to better identify key genes and processes of Cd2+ tolerance and also allowed us to better understand the role of AtfA in the regulation of stress responses.
2. Materials and Methods
2.1. Strains and Culture Conditions
The A. nidulans THS30 (pyrG89; AfupyrG+; pyroA4; pyroA+) as reference strain and TNJ92 (pyrG89; AfupyrG+; pyroA4; ΔatfA::pyroA) as a ΔatfA gene deletion mutant [23] were used in this study. Strains were maintained on Barratt’s minimal medium agar plates at 37 °C [26]. Conidia freshly collected from 6 d cultures were used for inoculation in all experiments.
For submerged cultivation, 100 mL Barratt’s minimal broth (in 500 mL Erlenmeyer flasks) was inoculated with 100 × 106 conidia and was incubated at 37 °C and at 3.7 Hz shaking frequency for 16.5–26 h. Before stress treatment (0.2 mM CdCl2, at 16 h) the biomass of cultures were set to equal levels as described earlier [23].
2.2. Recording Growth, Measuring GSH and GSSG Contents, 2′,7′-Dichlorofluorescein-Assay (DCF-Test)
Mycelia from 10 mL fermentation broth were filtered and dried at room temperature. The increase in the dry cell mass (DCM) values between 16 h and 26 h were calculated and used to characterize the growth of CdCl2-treated and untreated cultures.
Mycelia samples taken at 18.5 h and treated with 5′-sulfosalicylic acid [27,28] were used for the determination of (reduced) glutathione (GSH) and oxidized glutathione (GSSG) content with the DTNB-glutathione reductase assay [29] as well as for DCF-test to record changes in the redox homeostasis of the cultures [30].
2.3. Measuring Specific Enzyme Activities
Specific catalase and superoxide dismutase (SOD) activities were measured by rate assays [31,32] with cell-free extracts prepared by X-pressing [33]. The same cell free extracts were used to detect Cys synthase activities by the colorimetric method of Warrilow and Hawkesford [34] with O-acetyl-serine and Na2S as substrates. Protein content of the samples was measured with Bradford reagent. Chitinase, N-acetyl-glucosaminidase, and β-glucosidase activities were determined from fermentation broth with 4-nitrophenyl-β-D-N,N′,N′′-triacetylchitotriose,4-nitrophenyl-N-acetyl-β-D-glucosaminide and 4-nitrophenyl-β-D-glucoside as substrates, respectively, according to Pusztahelyi et al. [35]. Due to low enzyme activity values, the fermentation broth was concentrated tenfold before the hydrolase activity measurements using Amicon Ultra (Merck, Budapest, Hungary) centrifugal filters.
2.4. Reverse-Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR) Assays
Lyophilized mycelia were used for isolation of total RNA as described previously by Chomczynski [36]. RT-qPCR assays were carried out as detailed in Kovács et al. [37], with the primer pairs listed in Table S1 and using the actA gene as reference. Relative transcription was characterized with either ΔCP or ΔΔCP. ΔCP = CPr − CPt where r, t, and CP stand for the reference gene, target gene, and the crossing point value detected in the RT-qPCR assays, respectively. ΔΔCPtreatment = ΔCPtreated culture − ΔCPuntreated culture and ΔΔCPmutation = ΔCPmutant − ΔCPreference strain.
2.5. High-Throughput RNA Sequencing
Four different cultures with three biological replicates were studied. For “untreated cultures” of the reference strain (THS30) and the ΔatfA mutant (TNJ92), Barrat’s minimal broths were inoculated with conidia of the THS30 or the TNJ92 strains as described above and were incubated for 16.5 h at 37 °C and at 3.7 Hz shaking frequency. For “CdCl2-treated cultures” of the same strains, Barrat’s minimal broths were inoculated with conidia of the THS30 or the TNJ92 strains and were incubated for 16.5 h at 37 °C and at 3.7 Hz shaking frequency. These cultures were treated with 0.2 mM CdCl2 at 16 h.
Total RNA were isolated from cultures independent to those used for RT-qPCR assays. RNA sequencing (from library preparation to generation of fastq.gz files) were carried out at the Genomic Medicine and Bioinformatics Core Facility, Department of Biochemistry and Molecular Biology, Faculty of Medicine, University of Debrecen, Debrecen, Hungary. RNAseq libraries were prepared with TruSeq RNA Sample preparation kit (Illumina, Praha, Czech Republic) according to the manufacturer’s protocol. Each library pool was sequenced (single-read 75 bp sequencing on an Illumina HiScan SQ instrument; Illumina, San Diego, CA, USA) in one lane of a sequencing flow cell, and 11–16 million reads per sample were obtained. Reads were aligned to the genome of A. nidulans FGSC A4 with hisat2 (version 2.1.0) [38] using the following genome and genome features files (GFF) http://www.aspergillusgenome.org/download/sequence/A_nidulans_FGSC_A4/archive/A_nidulans_FGSC_A4_version_s10-m04-r12_chromosomes.fasta.gz (accessed on 2 July 2021) and http://www.aspergillusgenome.org/download/gff/A_nidulans_FGSC_A4/archive/A_nidulans_FGSC_A4_version_s10-m04-r12_features_with_chromosome_sequences.gff.gz (accessed on 2 July 2021), respectively. The percentage of the successfully aligned reads varied between 80–91%. Differentially expressed genes were determined with DESeq2 (version 1.24.0) [39]. RPKM values (reads per kilo base per million mapped reads) used only to visualize transcription activities of selected genes were calculated with the “rpkm” function of the edgeR package [40].
2.6. Evaluation of RNAseq Data
“Up-regulated and down-regulated genes were defined as differentially expressed genes (DESeq2; p < 0.05) where the log2FC > 0 and log2FC < 0, respectively. Log2FC stands for the number calculated by the DESeq2 software using THS30 as reference strain (TNJ92 untreated cultures vs. THS30 untreated cultures or TNJ92 Cd treated cultures vs. THS30 Cd treated cultures) or untreated cultures as reference conditions (THS30 Cd treated cultures vs. THS30 untreated cultures or TNJ92 Cd treated cultures vs. TNJ92 untreated cultures). Up-regulated and down-regulated genes identified in the CdCl2-treated vs. untreated and in the ΔatfA mutant vs. reference strain comparisons were regarded as stress responsive and AtfA-dependent genes, respectively”.
Gene groups containing functionally related genes were defined as follows:
“Antioxidative enzyme” genes: Genes of known or putative catalases, peroxidases, superoxide dismutases, or of the glutathione/glutaredoxin/thioredoxin system collected from the Aspergillus Genome Database (AspGD; http://www.aspergillusgenome.org; accessed on 2 July 2021).
“Autophagy” genes, “DNA repair” genes, and “P-type ATPase” genes: Genes with “autophagy”, “role in DNA repair”, or “P-type ATPase” feature, respectively, in their general description according to the AspGD.
“Cd2+ efflux pump” genes: Cation transporter genes of “cadmium ion import into vacuole”, “cadmium ion transport”, and “cellular detoxification of cadmium ion” GO terms (AspGD) as well as ygA (AN3624, [41]), an ortholog of A. fumigatus pcaA (Afu1g16130) putative cadmium transporter [8,10].
“Cell wall biosynthesis and degradation genes as well as cell wall integrity pathway” genes: Genes encoding enzymes of cell wall polysaccharide biosynthesis and degradation as well as of the cell wall integrity (protein kinase C) MAPK pathway according to De Groot et al. [42].
“Cys, Met and GSH metabolism” genes: Genes of GSH synthesis and degradation, Cys and Met biosynthesis, transsulfuration pathway, Met cycle., and Met salvage pathway according to the KEGG Pathway Database (https://www.genome.jp/kegg/pathway.html, accessed on 2 July 2021).
“Cys rich protein” genes: Gene encoding proteins with more than 10% Cys content according to the AspGD.
“ER to Golgi vesicle-mediated transport” genes: Genes of the “endoplasmic reticulum to Golgi vesicle-mediated transport” GO term according to the AspGD.
“Fe-S cluster binding protein” genes: Genes of the “iron-sulfur cluster binding” GO term according to the AspGD.
“Folate cycle” genes and “Glyoxylate pathway”genes: Genes of the folate cycle as well as of the glyoxalate cycle according to the KEGG Pathway Database.
“Glycolysis genes”: Genes described by Flipphi et al. [43].
“Phosphorelay response regulator” genes: Genes of the “phosphorelay response regulator activity” and “osmosensory signaling via phosphorelay pathway” GO terms in the AspGD.
“Ribosome biosynthesis” genes: Genes of the “ribosome biogenesis” Aspergillus GO-slim process category (http://www.aspergillusgenome.org/cgi-bin/GO/goTermMapper, accessed on 2 July 2021).
“Secondary metabolism cluster” genes: Manually or experimentally identified secondary metabolite cluster genes according to Inglis et al. [44].
“Squalene-ergosterol pathway” genes: Orthologs of A. fumigatus squalene–ergosterol pathway genes [45].
“Zinc transporter” genes: Genes of the “zinc ion transmembrane transporter activity”, “zinc ion transport”, and “zinc ion transmembrane transport” GO terms in the AspGD.
The enrichment of genes from the above defined genes groups within the up-regulated, down-regulated, stress responsive, and AtfA-dependent gene sets was tested with the “fisher.test” function (Fisher’s exact test; p < 0.05) of the R project (www.R-project.org, accessed on 2 July 2021).
Gene set enrichment analyses with FunCat, GO, and KEGG pathway terms were carried out with the FungiFun2 package (https://elbe.hki-jena.de/fungifun/fungifun.php, accessed on 2 July 2021) using default settings. Hits with adjusted p < 0.05 were taken into consideration during the evaluation process.
2.7. Promoter Analysis
Putative AtfA binding sites were identified according to Szabó et al. [19] in the promoter of each “Phosphorelay response regulator”, known and putative catalase, “Glycolysis”, “Squalene-ergosterol pathway”, “Folate cycle”, and Asperfuranone cluster genes of A. nidulans. In the case of A. fumigatus, A. niger, and A. oryzae, only the orthologs or best hit(s) for the A. nidulans catA, catB, and the “Phosphorelay response regulator” genes (according to AspGD) were studied. Briefly: The 5′-upstream intergenic sequences were downloaded from the AspGD and transcription start sites (TSS) were determined with the FGENESH tool (Softberry; http://www.softberry.com, accessed on 2 July 2021) in these sequences. ATF/CREB family transcription factor binding sites were searched with the PROMO (version 3.0.2) tool (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3, accessed on 2 July 2021) using 5% maximum matrix dissimilarity rate. Sites in the regions from TSS-1000 to TSS+50 and from TSS+50 to ATG were counted separately. In the case when identification of TSS failed, only the region from ATG-1500 to ATG was searched for binding motifs.
3. Results
3.1. Deletion of atfA Did Not Alter Significantly the Physiology of CdCl2-Treated Cultures
Cadmium treatment inhibited the growth in submerged cultures as expected (Figure 1A). Significant difference between the TNJ92 ΔatfA mutant and the THS30 reference strain was not detected (Figure 1A).
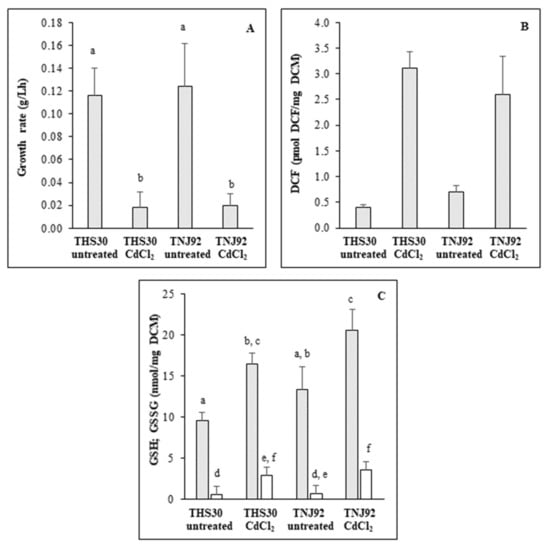
Figure 1.
Physiological consequences of cadmium treatment on A. nidulans. The ΔatfA mutant (TNJ92) and the reference (THS30) strains were treated with 0.2 mM CdCl2. Stress tolerance was characterized with the growth rate of the cultures (A). Redox imbalance caused by CdCl2 treatment was characterized with DCF-test (B). GSH (grey) and GSSG (white) content of treated and untreated cultures were recorded with the DTNB-GR assay (C). Mean ± SD. Values (n = 3) are presented. According to the two-way-ANOVA the CdCl2 treatment had a significant effect on DCM formation (p = 0.0000867), DCF production (p = 0.000003148) as well as on the GSH (p = 0.00004586) and GSSG (p = 0.00117) contents of cells. The genotype of the strains had a significant effect only on GSH content (p = 0.0097778), but the genotype did not influence the effect of CdCl2 treatment (p = 0.994) even in this case. Means of the same variable marked with a common letter are not significantly different by the Tukey post-hoc test at the p = 0.05 level.
Similar results were obtained on agar plates earlier where the relative growth values (ratio of colony diameters of treated and of untreated cultures) of the same two strains were very similar [25]. Cadmium stress significantly disturbed the redox balance in both strains, but there was no significant difference between their behaviors (Figure 1B). In addition, cadmium treatment increased GSH and GSSG contents of mycelia, and the specific SOD (but not catalase) activities displayed a very similar pattern in both strains (Figure 1C, Table 1).

Table 1.
Specific activities of selected enzymes in CdCl2-treated and untreated cultures of A. nidulans THS30 (reference) and TNJ92 (ΔatfA) strains.
3.2. Deletion of atfA Resulted in Marked Changes in Transcriptomes Observed under CdCl2 Stress
The transcriptomes of four different cultures (untreated and CdCl2-treated cultures of the TNJ92 ΔatfA mutant and the THS30 reference strain) were compared using RNA sequencing (RNAseq; Table S2). Transcriptional changes of 24 genes were also determined by RT-qPCR (Table S3) and they showed good correlation with the RNAseq data (Figure S2). According to the principal component analysis (PCA) of the RNAseq data, both the stress treatment and the genotype of the strains markedly affected the transcriptomes (Figure S1).
Composition of the differentially expressed gene sets showed that the stress responses of the TNJ92 ΔatfA mutant and the THS30 reference strain were different: 62% (685 + 1292 = 1977) of the 3167 up-regulated and 58% (663 + 1192 = 1855) of the 3177 down-regulated stress responsive genes showed altered transcription either in the mutant or in the reference strain only (Figure 2A). The stress treatment also changed the transcriptional difference between the two strains: 56% (1216 + 863 = 2079) of the 3703 up-regulated and 50% (964 + 707 = 1671) of the 3350 down-regulated “AtfA-dependent genes” showed AtfA-dependence only in untreated or only in CdCl2-treated cultures (Figure 2B).
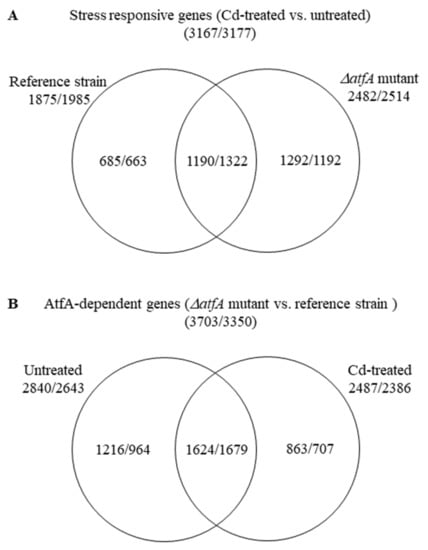
Figure 2.
Distribution of stress responsive genes between the THS30 reference strain and the TNJ92 ΔatfA mutant (A) as well as of the AtfA-dependent genes between the untreated and CdCl2-treated cultures (B). Figures represent the number of up-regulated/down-regulated genes. For definition of up-regulated, down-regulated, stress responsive, and AtfA-dependent genes. See the chapter titled “2.5 Evaluation of RNAseq data”.
3.3. CdCl2 Stress Regulates Vegetative Growth, Cd2+ Efflux System, and Sulfur Metabolism in the Reference Strain
Gene set enrichment analyses were applied to elucidate how the THS30 reference strain adapted to the CdCl2 treatment-induced stress. According to them, the stress down-regulated several processes involved in the fast vegetative growth in A. nidulans. These included glycolysis, tricarboxylic acid (TCA) cycle, respiration, and sterol biosynthesis (Table 2, Table 3, Tables S4 and S5). Interestingly “Ribosome biogenesis” genes were significantly enriched in both the up-regulated and the down-regulated gene sets (Table 3 and Table S5), suggesting a remodeling rather than bulk unidirectional changes in protein synthesis under stress.

Table 2.
Overview on the significantly enriched FunCat, GO, and KEGG pathway terms.

Table 3.
Overview on significant enrichment of the studied gene groups.
Out of the potential Cd2+ efflux pumps, crpA (AN1317) [9,46], but not ygA, was up-regulated (Tables S3 and S5). Apart from crpA, two other putative P-type ATPase genes (AN0318 and AN10367) were also up-regulated under cadmium stress (Table S5).
Fungal cell wall can bind a huge amount of Cd2+, and chitin can be particularly important in this biosorption [47,48]. Interestingly, cadmium stress did not cause substantial changes in the transcription of genes involved in cell wall biosynthesis or degradation and this was the case with extracellular chitinase, N-acetyl-β-D-glucosaminidase, and β-glucosidase activities as well (Table 1 and Table S5).
In A. nidulans, little is known on metallothioneins (Cys-moiety rich, low molecular weight proteins); however, they can be potentially important to detoxify heavy metals including cadmium [46,49]. Out of the genes encoding proteins with more than 10% Cys content, only two genes (AN7290 and AN11757) showed up-regulation (Table S5). Moreover, “Cys rich protein” genes including crdA, encoding a putative metallothionein of A. nidulans [46], were enriched in the down-regulated gene set (Table S5). Consequently, bulk up-regulation of metallothionein genes is not an important element of cadmium-induced stress response in A. nidulans.
“Glyoxylate pathway” genes, similarly to other genes involved in mitochondrial processes like TCA cycle and respiration, were down-regulated by cadmium treatment, therefore—in contrast to A. foetidus [6]—complexation of Cd2+ with secreted oxalate is unlikely to contribute substantially to the reduction of Cd2+ toxicity (Table 3 and Table S5) under the studied culturing conditions.
Sulfur metabolism was significantly affected by the stress treatment (Table 3 and Table S5, Figure 3). Surprisingly, according to the observed transcriptional changes, cells increased Cys availability by up-regulating the transsulfuration pathway, GSH degradation, and Cys biosynthesis, and down-regulated the usage of Met via Met cycle and Met salvage pathway (one carbon transfer processes) (Table S5, Figure 3). Consistent with this, specific Cys synthase activities were also significantly increased by the CdCl2 treatment (Table 1). Rearrangement (both up-regulation and down-regulation) of folate cycle genes was also observed (Table 3 and Table S5, Figure 3).
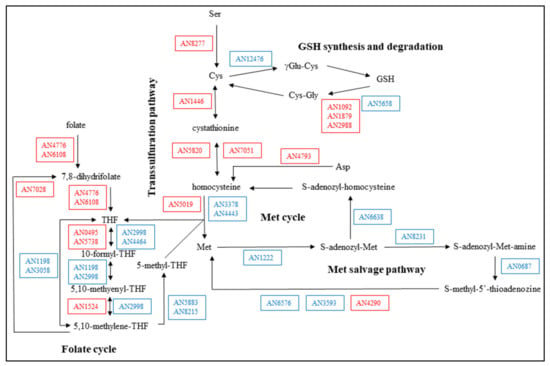
Figure 3.
Changes in the Cys and Met metabolism under CdCl2 treatment in the A. nidulans THS30 reference strain. Up-regulated genes are marked with red, down-regulated genes with blue.
Although AN7729 (ortholog of Saccharomyces cerevisiae ycf1 [50] and AN4517 (ortholog of Schizosaccharomyces pombe hmt1 [51] are both transmembrane transporter genes putatively involved in the transportation of bis(glutathionato)-cadmium complex into the vacuole, only the former was up-regulated (Tables S3 and S5).
Enrichment of “Fe-S cluster binding protein” genes within the down-regulated gene set as well as the enrichment of proteosomal degradation and autophagy genes within the up-regulated gene set were also observed (Table 3 and Table S5).
Regarding the two most important ROS producing cell organelles, mitochondrium and endoplasmic reticulum (ER), biochemical processes localized to mithochondria (e.g., TCA cycle, glyoxylate pathway, and respiration) were down-regulated (Table 2 and Table S4). In contrast, cadmium stress up-regulated the unfolded stress response together with the “ER to Golgi vesicle transport” genes but down-regulated the “Squalene-ergosterol pathway” genes (Table 2, Table 3 and Table S5). The importance of ER in cadmium stress response as well as the up-regulation of unfolded stress response under cadmium stress was also demonstrated in S. cerevisiae [52].
Interestingly the oxidative stress induced by CdCl2 treatment (Figure 1B) was not accompanied by bulk up-regulation of genes encoding antioxidative enzymes (Table 3 and Table S5). Some genes (e.g., sodB, manganese-superoxide dismutase [53]; ccp1, putative cytochrome c peroxidase [54]; catB and catC, catalases [55]) were up-regulated, while others (e.g., trxA, thioredoxin [56]; gpxA, putative glutathione peroxidase [54]; cpeA, putative catalase-peroxidase [54,55]) were down-regulated (Table S5). These transcriptional changes were accompanied with increased specific SOD activities (Table 1). In contrast, specific catalase activities remained unchanged (Table 1), which can be explained with multiple, differentially regulated catalase genes [55] affecting catalase activities [55]. Similar behavior of the “Antioxidative enzyme” genes (i.e., stress-dependent remodeling of their activity) was observed under oxidative stress induced by carbon starvation earlier [57,58].
Cd2+ can substitute Zn2+ in proteins, and elevated intracellular Zn2+ content can protect against it [59,60]. Interestingly, Zn2+ transporters showed down-regulation (Table 3 and Table S5) in our experiments. The down-regulated transport can be the consequence of reduced zinc usage caused by decreased growth (Figure 1A) under cadmium stress. It is also possible that Cd2+ enters the cells via Zn2+ transporters, therefore down-regulation of these transporters can be important to reduce the infiltration of Cd2+ into the cells.
“Excision repair” genes showed significant enrichment within the up-regulated gene set (Table 3 and Table S5) and some other “DNA repair” genes were also up-regulated (Table 3 and Table S5), which is in line with the genotoxicity of Cd2+ [61].
In contrast to oxidative, osmotic or carbon starvation stress [23,58] the applied cadmium stress had minor effect on the transcription of “Secondary metabolite cluster” genes (Table 3 and Table S5). A few clusters, however, showed up-regulation (inp cluster) or down-regulation (microperfuranone cluster, AN1242 cluster) (Table 3 and Table S5).
3.4. At the Level of Regulated Gene Groups, Deletion of atfA Caused Only Minor Changes in the Stress Response Induced by CdCl2
Cadmium stress also down-regulated several processes involved in the fast vegetative growth in the TNJ92 ΔatfA mutant. These included TCA cycle, respiration, and sterol biosynthesis (Table 2, Table 3, Tables S4 and S5). Moreover, mitotic cell cycle genes, as well as DNA synthesis and replication genes, were also significantly enriched in the down-regulated gene set of the mutant (Table 2 and Table S4). In contrast to the THS30 reference strain, “Glycolysis” genes were not down-regulated and “Ribosome biogenesis” genes showed bulk down-regulation instead of remodeling (Table 3 and Table S5).
The putative Cd2+ efflux pump gene, crpA, was up-regulated (Tables S3 and S5) and the crdA putative metallothionein gene was down-regulated (Table S5). Cadmium stress did not substantially change the transcription of cell wall homeostasis genes (Table S5); however, cell wall biosynthesis genes were significantly enriched within the stress responsive gene set, suggesting some remodeling. In line with this, minor changes in the extracellular chitinase and N-acetyl-β-D-glucosaminidase but not in β-glucosidase activities were also recorded in the ΔatfA gene deletion mutant after CdCl2 treatment (Table 1). Interestingly, β-glucosidase activities were significantly higher in the cultures of the mutant strain than in the reference strain grown in either cadmium-treated or untreated cultures (Table 1).
In the case of sulfur metabolism genes (Table 3 and Table S5, Figure S3), “Fe-S cluster protein” genes, “Autophagy” genes, and proteosomal degradation genes, changes similar to those observed with the THS30 reference strain were detected in the TNJ92 ΔatfA mutant (Table 2, Table 3, Tables S4 and S5). Up-regulation of “Folate cycle” genes, however, was not observed in the mutant (Table 3 and Table S5, Figure S3). Importantly, the specific Cys synthase activities increased after cadmium treatment (Table 1).
Mitochondrium- and ER-related processes behaved similarly in the TNJ92 mutant and THS30 reference strain: In both strains, cadmium stress down-regulated mitochondrium-related processes like TCA cycle, glyoxylate pathway, and respiration (Table 2 and Table S4), and up-regulated genes in the “Protein processing in endoplasmic reticulum” and “ER to Golgi vesicle transport” groups but down-regulated “Squalene-ergosterol pathway” genes (Table 2, Table 3, Tables S4 and S5). Rearrangements in the transcription of “Antioxidative enzyme” genes were also found (Table 3 and Table S5): In the case of the ΔatfA mutant, the up-regulated genes included sodA [62] and sodB superoxide dismutases, ccp1 putative cytochrome c peroxidase, and catA catalase [55], while the down-regulated genes included trxA thioredoxin and trxB thioredoxin reductase [56] (Table S5). Similarly to the reference strain, CdCl2 treatment increased the specific SOD but not the specific catalase activities (Table 1). Consistent with the transcriptional values of most catalase genes (Table S5), the specific catalase activities were significantly lower in the AtfA-mutant than in the reference strain (Table 1).
Zn2+ transporter genes also showed down-regulation in the TNJ92 mutant (Table 3 and Table S5). Interestingly, unlike in THS30, “DNA repair” genes were down-regulated and even the “Excision repair” genes did not show significant enrichment in the up-regulated gene set (Table 3 and Table S5).
3.5. Background of the Differences Observed between the Stress Responses of the Two Strains
In order to understand the nature of the differences between the CdCl2 stress responses of the TNJ92 ΔatfA mutant and the THS30 reference strain, we investigated the behavior of individual genes in selected gene groups.
3.5.1. Diverging Gene Sets Regulate Same Biological Processes under Cadmium Stress
During cadmium stress, the transcriptional changes of the “Cys, Met, and GSH metabolism” genes suggested a very similar conclusion, i.e., cells tried to increase Cys availability, in both strains (Figure 3 and Figure S3, Table S5). The overlap between the stress responsive genes of the two strains in this gene group, however, was only 49% (21 out of 43) (Figure 4). Similarly, the “ER to Golgi vesicle-mediated transport” genes were significantly enriched in the up-regulated gene set in both strains (Table 3 and Table S5), yet their overlap was only 45% (10 out of 22) (Figure S4). Even in the case of the “Cell wall biosynthesis and degradation” genes where bulk up-regulations or down-regulations were not observed in either strain (Table S5), the overlap between the stress responsive genes of the ΔatfA mutant and the reference strain was only 39% (24 out of 61) (Figure S5). These data suggest that the two strains initiated similar physiological changes under cadmium stress but they did it with partly different gene sets. It concurs with our previous observations when the effects of two different stressors were investigated on the same strain: We found that both menadione and diamide inhibited the growth of A. nidulans THS30 and down-regulated several “Ribosome biogenesis” genes (36 and 30 genes for menadione and diamide stress treatment, respectively), but the overlap (1 gene) was negligible [24].
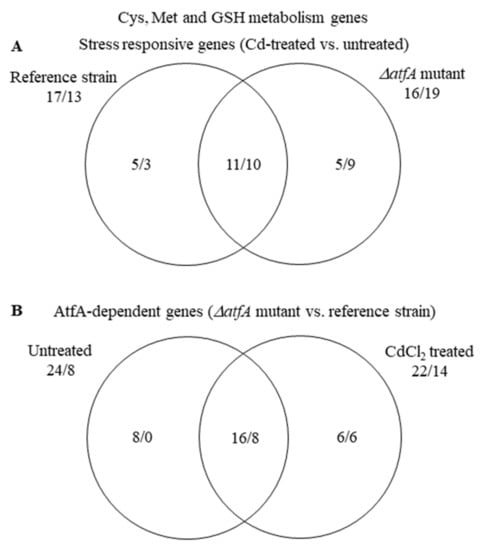
Figure 4.
Distribution of stress responsive “Cys, Met and GSH metabolism” genes between the THS30 reference strain and the TNJ92 ΔatfA mutant (A) as well as of the AtfA-dependent “Cys, Met and GSH metabolism” genes between the untreated and CdCl2-treated cultures (B). Figures represent the number of up-regulated/down-regulated genes. Further data on this gene group are available in Table S5.
3.5.2. Differences between the Transcriptomes of Untreated Cultures Led to Different Stress Responses
In the THS30 reference strain, the “Ribosome biogenesis” genes were significantly enriched in both the up-regulated and the down-regulated gene sets, in the TNJ92 ΔatfA mutant, however, only in the down-regulated gene set (Table 3 and Table S5). As Figure 5A shows, 72 and 1 genes were up-regulated exclusively in the reference strain and the ΔatfA mutant, respectively, while the numbers of exclusively down-regulated genes were 2 and 89.
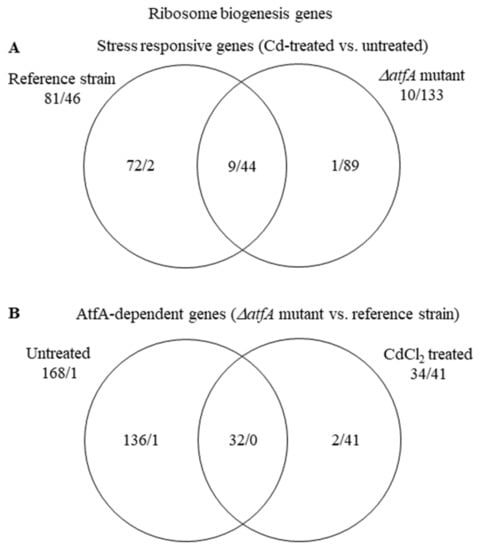
Figure 5.
Distribution of stress responsive “Ribosome biogenesis” genes between the THS30 reference strain and the TNJ92 ΔatfA mutant (A) as well as of the AtfA-dependent “Ribosome biogenesis” genes between the untreated and CdCl2-treated cultures (B). Figures represent the number of up-regulated/down-regulated genes. Further data on the gene group are available in Table S5.
This difference between the strains may reflect the difference between the two untreated cultures: Before stress treatment, 168 genes showed higher and only 1 lower transcription value in the mutant than in the reference strain. After stress treatment, only 34 genes had higher and 43 had lower transcription activity in the atfA gene deletion mutant than in the reference strain (Figure 5B). The divergent response found in the two strains may be the consequence of both strains aiming to set similar transcription profile of the “Ribosome biogenesis” genes under stress but they started from very different transcription levels (Figure 5; Table S5).
The behavior of “Folate cycle” genes can also be explained in a similar manner. Cadmium stress up-regulated six genes (AN0495, AN1524, AN4776, AN5738, AN6108, AN7028) in the reference strain and only one (AN0495) in the ΔatfA mutant (Table 3 and Table S5), resulting in significant enrichment of up-regulated “Folate-cycle” genes in THS30, but not in TNJ92. All six genes showed lower transcriptional activity in the reference strain than in the ΔatfA mutant before stress treatment, which may explain the divergent response of the two strains (Figure S6 and Table S5).
3.5.3. Transcriptional Activities of Certain Genes Are Not Properly Regulated in the Absence of AtfA
Among the 20 “Phosphorelay response regulator” genes, in untreated cultures, the majority (18) of the genes showed significantly smaller transcription in the TNJ92 mutant than in the THS30 reference strain (Table S5, Figure 6). Under stress, the mutant up-regulated 10 genes and the reference strain down-regulated 6 (Figure 6A), yet the difference between the two strains remained substantial (Figure 6B). The stress condition only reduced, but did not eliminate, the difference between the activities of “Phosphorelay response regulator” genes in the two strains.
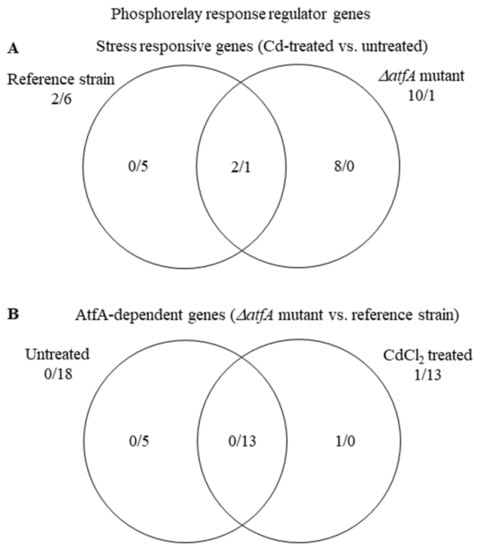
Figure 6.
Distribution of stress responsive “Phosphorelay response regulator” genes between the THS30 reference strain and the TNJ92 ΔatfA mutant (A) as well as of the AtfA-dependent “Phosphorelay response regulator” genes between the untreated and CdCl2-treated cultures (B). Figures represent the number of up-regulated/down-regulated genes. Further data on the gene group are available in Table S5.
In the above case, the mutant—in the absence of AtfA—appeared unable to appropriately regulate the studied gene group. Therefore, we investigated the presence of putative AtfA binding sites in the promoters of these genes. As it was, according to what our previous studies [23,24,25] expected, the promoters of the “Phosphorelay response regulator” genes contained significantly more putative AtfA binding sites than the reference gene groups did (Table 4 and Table S6). The AtfA dependence of catalase gene expressions also seems likely (Table 4 and Table S6), which concurs with previous studies [12,15,20,63] and the transcription of data of catA and catB (Table S5, Figure S7). Promoter analyses of the orthologs of A. nidulans “Phosphorelay response regulator” genes as well as catA and catB were also conducted in three other Aspergillus species (A. fumigatus, A. niger, and A. oryzae). In the case of the catalase genes, the numbers of the found putative binding sites in each species were similar to those recorded with A. nidulans (Table S6). Moreover, an evolutionary conserved binding site was also found in the promoter of catA orthologs in A. fumigatus, A. niger, and A. oryzae (Figure 7). In the case of the “Phosphorelay response regulator” genes, the orthologs of A. niger had significantly less putative binding sites than those of A. nidulans (Table S6). In the case of this gene group, no evolutionary conserved binding sites were recorded. It can be explained by the rather different biology of the four Aspergillus species, which increases the number of species-specific regulatory sequences and eradicates the evolutionary conserved ones. The weak conservation of the AtfA binding sites in the promoter of A. nidulans catA also supports this view (Figure 7).

Table 4.
Abundance of putative AtfA binding sites in selected gene groups.

Figure 7.
Evolutionary conserved putative AtfA binding site in the promoters of the catA ortholog genes.
Promoter regions were aligned with the MAFFT software (http://www.ebi.ac.uk/Tools/msa/mafft, accessed on 2 July 2021). Blue, purple, light green, and yellow highlight putative binding sites characterized with less than 5% maximum matrix dissimilarity rate (Promo tool). The dark green color highlights a putative AtfA binding site of A. nidulans where the dissimilarity rate for CRE-BP2 [T01017] (Promo tool) binding site is 13.5%. The following sequences, downloaded from AspGD, are presented: A. fumigatus Af293 catA promoter region between ATG-221 and ATG-109, A. niger catA promoter region between ATG-242 and ATG-144, A. oryzae RIB40 catA promoter region between ATG-236 and ATG-140, and A. nidulans FGSC A4 catA promoter region between ATG-197 and ATG-108.
3.5.4. Alternative Stress Response Elements Can Increase the Difference between the Stress Responses of the Two Strains
Both up-regulation and down-regulation of “Glycolysis” genes could have adaptive value during stress. Up-regulation of this biochemical pathway can support the protection against Cd2+ with increased ATP production. On the other hand, its down-regulation can adapt to the reduced growth rate under stress. “Glycolysis” genes were enriched in the down-regulated gene set of the THS30 reference strain and many of them were up-regulated in the TNJ92 mutant; however, their enrichment was not significant (Table S5, Figure 8A). Due to these changes, the ratio of genes showing higher/lower transcriptional activity in the TNJ92 mutant in comparison to the THS30 reference strain turned from 4/10 (untreated cultures) to 9/4 (stress treated cultures) (Table S5, Figure 8B).
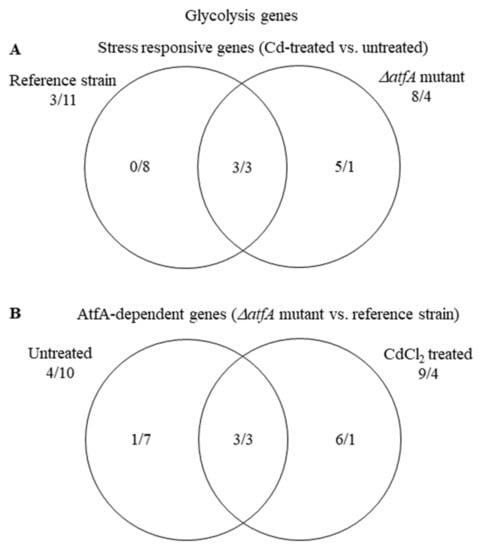
Figure 8.
Distribution of stress responsive “Glycolysis” genes between the THS30 reference strain and the TNJ92 ΔatfA mutant (A) as well as of the AtfA-dependent “Glycolysis” genes between the untreated and CdCl2-treated cultures (B). Figures represent the number of up-regulated/down-regulated genes. Further data on the gene group are available in Table S5.
In addition to its genotoxicity [61] cadmium can also disturb the activity of repair enzymes [64]. Therefore, similarly to “Glycolysis” genes, either up-regulation or down-regulation of “DNA repair” genes could be an adaptive response under cadmium stress. The ratio of genes with higher/lower transcriptional activity in the ΔatfA mutant than in the reference strain also turned to the opposite under stress, from 22/12 (untreated cultures) to 14/24 (cadmium-treated cultures) (Table S5).
In the case of these gene groups, the two strains seem to apply two different strategies to adapt to the stress.
4. Discussion
4.1. Increased Cys Availability Is an Important Element of the CdCl2-Induced Stress Response
Genome-wide transcriptional changes induced by CdCl2 were studied in an A. nidulans model organism in order to gain deeper insight into the reasons of Cd2+ toxicity and the biochemical processes necessary to the detoxification of this heavy metal. Entering the cells, Cd2+ interacts with susceptible proteins (e.g., different metallo- or thiol-containing proteins) perturbing their activity and functions. It disturbs metal ion (e.g., iron, zinc, copper) homeostasis, protein folding, DNS repair processes, and leads to oxidative stress, aggregation of nascent proteins, or increased mutation rate, which all affect severely the viability of cells [65,66,67,68]. Many of the observed transcriptional changes concur well with these consequences of cadmium treatment. Significant alterations in the transcription of DNA repair genes and up-regulation of the unfolded stress response genes together with several ER specific genes were observed (Table 2, Table 3 and Table S5). Down-regulation of Fe-S cluster biosynthesis genes as well as other mitochondrium specific genes encoding iron-dependent proteins (e.g., respiration and TCA cycle genes) (Table 2, Table 3 and Table S5) can be the consequence of disturbed iron homeostasis caused by Cd2+. Besides the altered transcription of the antioxidative enzyme genes (Table S5), the increased specific SOD activities and GSSG concentrations as well as the disturbed redox homeostasis (Figure 1, Table 1) demonstrate developing oxidative stress during the CdCl2 treatment. Up-regulation of specific SOD, catalase, and glutathione reductase activities induced by cadmium stress were also recorded previously in A. nidulans by Guelfi et al. [69]. Since cadmium is not a Fenton metal, it can induce oxidative stress only indirectly. Both the disturbed iron/mitochondrium homeostasis and the changes in the activity of ER as another important ROS-generating organelle [70,71] can be vital in this process.
The cell wall of A. nidulans, similarly to those of many other fungi, can bind a huge amount of Cd2+ [9,10,72,73], which prevents this toxic metal from entering the cells. This property of fungal biomass can be used both for bioleaching and biomining purposes in the industry [72,74,75]. Although cadmium stress affected the transcription of some genes involved in cell wall biosynthesis or degradation, significant enrichment of the up-regulated or down-regulated cell wall homeostasis genes was not observed (Table S5). This supports the view that the high Cd2+ (metal ion) binding capacity is an innate property of the fungal cell wall and no substantial cell wall remodeling is needed for the efficient binding. It is an important feature that increases the applicability of fungal biomass in biosorption based processes.
There are multiple ways in fungi to detoxify Cd2+ entering their cells. These include secreting Cd2+ by efflux pumps, binding Cd2+ with metallothioneins, and the formation and vacuolar accumulation of the bis(glutathionato)-cadmium complex [76]. Up-regulation of crpA (Tables S3 and S5) encoding a P-type ATPase by cadmium stress in our experiments concurs with the observation that deletion of this gene was accompanied with increased CdCl2 sensitivity of A. nidulans [9,46]. Transcriptome data did not provide clear evidence on the working of Cd2+ pumps other than CrpA in A. nidulans: Apart from crpA, only two other putative P-type ATPase genes (AN0318 and AN10367) were up-regulated by cadmium stress (Table S5). Of these, the AN10367 gene, like crpA, was also up-regulated in the TNJ92 mutant (Table S5). The AN0318 protein is an ortholog of Neurospora crassa Pma-1 and S. cerevisiae Pma1p H+-ATPase [77,78], while AN10367 is an ortholog of S. cerevisiae Spf1p and S. pombe Cta4p Ca2+ pumps, which are responsible for the calcium homeostasis of the ER [79,80]. The function of the two A. nidulans genes is waiting for experimental justification; however, according to the abovementioned data, CrpA seems to be the sole Cd2+ pump of A. nidulans under the tested conditions.
Although crdA, like many other genes encoding “Cys rich proteins”, was down-regulated by the cadmium stress (Table S5), two genes from this group (AN7290 and AN11757) showed up-regulation. AN7290, which was also up-regulated in TNJ92 mutant under cadmium stress (Table S5), is an ortholog of S. cerevisiae Rds3p and S. pombe Ini1p zinc cluster proteins involved in mRNA splicing [81,82]. The AN7290 protein is 12.3 kDa and consists of 108 amino acids including 12 Cys (AspGD). The AN11757 protein has no orthologs with known function. It is 11.4 kDa and consists of 100 amino acids including 13 Cys (AspGD). Further studies are needed to test whether these proteins function as metallothioneins under cadmium stress in A. nidulans.
Ycf1p of S. cerevisiae is an ABC transporter involved in the transportation of bis(glutathionato)–cadmium complex into the vacuole [50]. Up-regulation of the ycf1 ortholog AN7727 under cadmium stress (Table S5) supports the view that GSH dependent detoxification of this heavy metal is an important element of stress response under Cd2+ treatment in A. nidulans, similarly to other organisms from bacteria to human [51].
Fe-S cluster binding proteins and proteins with high Cys content are particularly sensitive to the presence of Cd2+ and/or the oxidative stress caused by Cd2+ [66,83]. To manage this problem, cells can down-regulate genes encoding nonessential Cys rich and Fe-S cluster binding proteins as well as they can eliminate the destroyed proteins and can re-synthetize those that are important for surviving this stress. Enrichment of the “Cys-rich protein” and “Fe-S cluster binding protein” genes within the down-regulated gene set as well as the enrichment of “Proteosomal degradation” and “Autophagy” genes within the up-regulated gene set were all observed (Table 3 and Table S5). Moreover, the up-regulation of the transsulfuration pathway, GSH degradation, Cys biosynthesis genes and Cys synthase activities, and down-regulation of Met cycle and Met salvage pathway genes (Table S5, Figure 3) suggest that cells increased Cys availability during cadmium stress. It concurs with the high Cys demand of the continuous re-synthesis of Cys rich and/or Fe-S cluster binding proteins. These data demonstrate that GSH is involved not only directly in Cd2+ detoxification, but also indirectly as an important intracellular Cys storage. During oxidative stress, the thiol-moiety of proteins are continuously oxidized to disulfide bridges and cells continuously reduce them back. This process substantially contributes to ROS elimination [84]. It is possible that a similar mechanism (i.e., replacing Cd2+-bound proteins with new ones) can help cells to collect and detoxify Cd2+.
4.2. Adaptation to the Missing AtfA under Unstressed Conditions Influences Substantially the CdCl2 Induced Stress Response
In addition to the THS30 reference strain, the CdCl2-induced stress response was also studied in the TNJ92 ΔatfA mutant to better understand how this transcription factor is involved in the regulation of stress responses. Previously, we found that deletion of atfA increased the stress sensitivity of A. nidulans against various oxidative agents [23,24,25], which concur with its reduced specific catalase activity recorded in this study (Table 1). Accordingly, the stress response of the ΔatfA mutant substantially differed from that of the reference strain [23,24,25]. In contrast, AtfA was not essential for the efficient stress response under CdCl2 stress: The growth reduction induced by the stress treatment in the THS30 reference strain did not differ significantly to that of the TNJ92 ΔatfA mutant either on the surface [25] or in submerged cultures (Figure 1A). There were no significant differences between the two strains when the physiological consequences (i.e., alterations in the GSH and GSSG concentrations, in the specific SOD, catalases and Cys synthase activities, or in the redox homeostasis) of the cadmium treatment were studied either (Figure 1B,C, Table 1), while in the case of the extracellular chitinase or N-acetyl-β-D-glucosaminidase activities, only mild differences were detected (Table 1). Moreover, the transcription of atfA was even down-regulated under CdCl2 stress in the reference strain (Table S3), suggesting that this transcription factor was not a key player in the regulation of stress responses in these experiments. Surprisingly, the genome-wide transcriptional changes induced by the cadmium stress treatment in the THS30 and TNJ92 strains were very different (Figure 2A). It demonstrates that AtfA is involved in the cadmium stress response, but cells can efficiently compensate the consequences of its absence. It is very unlikely that A. nidulans has “learnt” during its evolution how to survive CdCl2 stress without AtfA. Therefore, we should assume that stress responses cannot be rigid programs worked out in detail for lots of possible stress conditions. Instead, cells should have flexible regulatory mechanisms that can find a suitable stress response even under brand new conditions.
The differences between the cadmium stress responses of the two strains are linked to the (missing) AtfA and can help to better understand the function of this transcription factor. In our previous oxidative stress treatment experiments, the two strains showed different sensitivities to the applied stressors [23,24,25]. In spite of the two strains receiving the same treatments, the effect of the treatments was not the same because the decreased stress tolerance of the ΔatfA mutant increased the difference between the observed stress responses, making identification of the direct effects of AtfA deletion more difficult. In this respect, cadmium stress experiments are more favorable since the cadmium tolerance of the THS30 and TNJ92 strains are very similar. Even so, identification of potential targets of AtfA is still challenging. There are at least two possible reasons of the altered cadmium stress responses of the two strains:
(1) The transcriptome, and as a consequence, the physiology of the THS30 and TNJ92 strains, were different under unstressed conditions (Figure 2B). Cells with dissimilar physiology necessarily respond differently to the same stress treatment. As a consequence, gene sets with dissimilar transcription profiles at unstressed conditions have to respond to the stress in different ways to reach the same transcription profile (Figure 5 and Figure S6). The dissimilar physiological status of the untreated cultures can also lead to alternative stress responses (Figure 8). The small physiological/transcriptomic differences between the untreated cultures may also explain why gene sets respond in the same overall manner to the stress but by the regulation of partly different genes (Figure 3; Figure 4 and Figures S3–S5).
(2) Due to the missing AtfA, cells could not regulate properly certain genes. The improper behavior of these genes can also lead to the development of alternative stress response elements or to the development of the same stress response element in an alternative manner. Moreover, the improper regulation of AtfA-dependent genes is necessarily the primary source of the observed differences between the two strains under unstressed conditions. Unfortunately, based on only transcription data, due to strong indirect effects of the missing AtfA, it is difficult to identify genes directly regulated by this transcription factor. The “Phosphorelay response regulator” genes and the “Catalase” genes are our best candidates for this (Figure 6 and Figure S7). In addition to the data gained in previous investigations [12,15,20,23,24,25,63] and the observed transcriptional behavior of these genes (Table S5), the presence of putative AtfA binding sites in their promoters (Table 4 and Table S6) supports this view. Although the number of putative binding sites was large, none but one was evolutionary conserved for the four Aspergillus species tested (Table S6, Figure 7). Therefore, detailed molecular biological studies are needed to ascertain which species-specific putative binding sites have regulatory function in A. nidulans.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9071433/s1, Figure S1: Principal component analysis (PCA) of the transcriptomes of CdCl2-treated and untreated A. nidulans THS30 (reference strain) and TNJ92 (ΔatfA mutant) cultures, Figure S2: Correlation between RNAseq and RT-qPCR data, Figure S3: Changes in the Cys and Met metabolism under CdCl2 treatment in the A. nidulans TNJ92 ΔatfA mutant, Figure S4: Distribution of stress responsive “ER to Golgi vesicle-mediated transport” genes between the THS30 reference strain and the TNJ92 ΔatfA mutant (A) as well as of the AtfA-dependent “ER to Golgi vesicle-mediated transport” genes between the untreated and CdCl2-treated cultures (B), Figure S5: Distribution of stress responsive “Cell wall biosynthesis and degradation” genes between the THS30 reference strain and the TNJ92 ΔatfA mutant (A) as well as of the AtfA-dependent “Cell wall biosynthesis and degradation” genes between the untreated and CdCl2-treated cultures (B), Figure S6: Changes in the transcription of selected “Folate cycle” genes, Figure S7: Changes in the transcription of genes encoding known and putative catalases, Table S1: RT-qPCR primer pairs used in the study, Table S2: Read mapping statistics, Table S3: RT-qPCR data of the selected genes, Table S4: Significantly enriched FunCat, GO, and KEGG pathway terms. Table S5: RNAseq data and the results of enrichment analyses of the studied gene groups, Table S6: Results of the promoter analyses.
Author Contributions
Conceptualization, I.P., J.-H.Y. and T.E.; methodology, H.M., K.A. and T.E.; investigation, B.G., F.F. and T.E.; validation, K.A. and T.E.; formal analysis, B.G., H.M., I.P., J.-H.Y., K.A. and T.E.; data curation, K.A. and T.E.; writing—original draft preparation, H.M., I.P., J.-H.Y., K.A. and T.E.; writing—review and editing, H.M., I.P., J.-H.Y., K.A. and T.E.; visualization, B.G., K.A. and T.E.; funding acquisition, I.P. and T.E. All authors have read and agreed to the published version of the manuscript.
Funding
The research was financed by the European Union and the European Social Fund through the project EFOP-3.6.1–16–2016–00022, by the National Research, Development, and Innovation Office (Hungary) projects K112181, K119494, NN125671 and K131767, by the Thematic Excellence Programme (TKP2020-IKA-04) and by the New National Excellence Program (ÚNKP-20-3) of the Ministry for Innovation and Technology in Hungary.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw and processed transcriptome datasets were uploaded to the Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo/, accessed on 2 July 2021) with the following accession number: GSE166128.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Järup, L.; Akesson, A. Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol. 2009, 238, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Godt, J.; Scheidig, F.; Grosse-Siestrup, C.; Esche, V.; Brandenburg, P.; Reich, A.; Groneberg, D.A. The toxicity of cadmium and resulting hazards for human health. J. Occup. Med. Toxicol. 2006, 1, 22. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar] [PubMed]
- Tang, X.; Li, Q.; Wu, M.; Lin, L.; Scholz, M. Review of remediation practices regarding cadmium-enriched farmland soil with particular reference to China. J. Environ. Manag. 2016, 181, 646–662. [Google Scholar] [CrossRef]
- Abdel-Aty, A.M.; Ammar, N.S.; Abdel Ghafar, H.H.; Ali, R.K. Biosorption of cadmium and lead from aqueous solution by fresh water alga Anabaena sphaerica biomass. J. Adv. Res. 2013, 4, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Mukherjee, A.; Khuda-Bukhsh, A.R.; Das, T.K. Cadmium-induced ox idative stress tolerance in cadmium resistant Aspergillus foetidus: Its possible role in cadmium bioremediation. Ecotoxicol. Environ. Saf. 2014, 106, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Bano, A.; Hussain, J.; Akbar, A.; Mehmood, K.; Anwar, M.; Hasni, M.S.; Ullah, S.; Sajid, S.; Ali, I. Biosorption of heavy metals by obligate halophilic fungi. Chemosphere 2018, 199, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Bakti, F.; Sasse, C.; Heinekamp, T.; Pócsi, I.; Braus, G.H. Heavy Metal-Induced Expression of PcaA Provides Cadmium Tolerance to Aspergillus fumigatus and Supports Its Virulence in the Galleria mellonella Model. Front. Microbiol. 2018, 9, 744. [Google Scholar] [CrossRef]
- Boczonádi, I.; Török, Z.; Jakab, Á.; Kónya, G.; Gyurcsó, K.; Baranyai, E.; Szoboszlai, Z.; Döncző, B.; Fábián, I.; Leiter, É.; et al. Increased Cd2+ biosorption capability of Aspergillus nidulans elicited by crpA deletion. J. Basic Microbiol. 2020, 60, 574–584. [Google Scholar] [CrossRef]
- Kurucz, V.; Kiss, B.; Szigeti, Z.M.; Nagy, G.; Orosz, E.; Hargitai, Z.; Harangi, S.; Wiebenga, A.; de Vries, R.P.; Pócsi, I.; et al. Physiological background of the remarkably high Cd2+ tolerance of the Aspergillus fumigatus Af293 strain. J. Basic Microbiol. 2018, 58, 957–967. [Google Scholar] [CrossRef]
- Banakar, R.; Alvarez Fernández, Á.; Abadía, J.; Capell, T.; Christou, P. The expression of heterologous Fe(III) phytosiderophore transporter HvYS1 in rice increases Fe uptake, translocation and seed loading and excludes heavy metals by selective Fe transport. Plant Biotechnol. J. 2016, 15, 423–432. [Google Scholar] [CrossRef]
- Hagiwara, D.; Asano, Y.; Yamashino, T.; Mizuno, T. Characterization of bZip-type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2008, 72, 2756–2760. [Google Scholar] [CrossRef]
- Balázs, A.; Pócsi, I.; Hamari, Z.; Leiter, É.; Emri, T.; Miskei, M.; Oláh, J.; Tóth, V.; Hegedűs, N.; Prade, R.A.; et al. AtfA BZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol. Genet. Genom. 2010, 283, 289–303. [Google Scholar] [CrossRef]
- Sakamoto, K.; Iwashita, K.; Yamada, O.; Kobayashi, K.; Mizuno, A.; Akita, O.; Mikami, S.; Shimoi, H.; Gomi, K. Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet. Biol. 2009, 46, 887–897. [Google Scholar] [CrossRef]
- Lara-Rojas, F.; Sánchez, O.; Kawasaki, L.; Aguirre, J. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 2011, 80, 436–454. [Google Scholar] [CrossRef]
- Temme, N.; Oeser, B.; Massaroli, M.; Heller, J.; Simon, A.; Collado, I.G.; Viaud, M.; Tudzynski, P. BcAtf1, a global regulator, controls various differentiation processes and phytotoxin production in Botrytis cinerea. Mol. Plant Pathol. 2012, 13, 704–718. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, T.; Kroger, C.; Bonnighausen, J.; Schäfer, W.; Bormann, J. The ATF/CREB transcription factor Atf1 is essential for full virulence, deoxynivalenol production, and stress tolerance in the cereal pathogen Fusarium graminearum. Mol. Plant. Microbe Interact. 2013, 26, 1378–1394. [Google Scholar] [CrossRef]
- Hagiwara, D.; Takahashi, H.; Kusuya, Y.; Kawamoto, S.; Kamei, K.; Gonoi, T. Comparative transcriptome analysis revealing dormant conidia and germination associated genes in Aspergillus species: An essential role for AtfA in conidial dormancy. BMC Genom. 2016, 17, 358. [Google Scholar] [CrossRef] [PubMed]
- Szabó, Z.; Pákozdi, K.; Murvai, K.; Pusztahelyi, T.; Kecskeméti, Á.; Gáspár, A.; Logrieco, A.F.; Emri, T.; Ádám, A.L.; Leiter, É.; et al. FvatfA regulates growth, stress tolerance as well as mycotoxin and pigment productions in Fusarium verticillioides. Appl. Microbiol. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Nathues, E.; Joshi, S.; Tenberge, K.B.; von den Driesch, M.; Oeser, B.; Bäumer, N.; Mihlan, M.; Tudzynski, P. CPTF1, a CREB-like transcription factor, is involved in the oxidative stress response in the phytopathogen Claviceps purpurea and modulates ROS level in its host Secale cereale. Mol. Plant Microbe Interact. 2004, 17, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Guo, W.; Chen, Y.; Dong, S.; Zhang, X.; Zhang, H.; Song, W.; Wang, W.; Wang, Q.; Lv, R.; et al. The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact. 2010, 23, 1053–1068. [Google Scholar] [CrossRef]
- Pereira Silva, L.; Alves de Castro, P.; Dos Reis, T.F.; Paziani, M.H.; Von Zeska Kress, M.R.; Riaño-Pachón, D.M.; Hagiwara, D.; Ries, L.N.; Brown, N.A.; Goldman, G.H. Genome-wide transcriptome analysis of Aspergillus fumigatus exposed to osmotic stress reveals regulators of osmotic and cell wall stresses that are SakAHOG1 and MpkC dependent. Cell Microbiol. 2017, 19, e12681. [Google Scholar] [CrossRef]
- Emri, T.; Szarvas, V.; Orosz, E.; Antal, K.; Park, H.; Han, K.H.; Yu, J.H.; Pócsi, I. Core oxidative stress response in Aspergillus nidulans. BMC Genom. 2015, 16, 478. [Google Scholar] [CrossRef]
- Orosz, E.; Antal, K.; Gazdag, Z.; Szabó, Z.; Han, K.H.; Yu, J.H.; Pócsi, I.; Emri, T. Transcriptome-based modeling reveals that oxidative stress induces modulation of the AtfA-dependent signaling networks in Aspergillus nidulans. Int. J. Genomics. 2017, 2017, 6923849. [Google Scholar] [CrossRef] [PubMed]
- Antal, K.; Gila, B.C.; Pócsi, I.; Emri, T. General stress response or adaptation to rapid growth in Aspergillus nidulans? Fungal Biol. 2020, 124, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Barratt, R.W.; Johnson, G.B.; Ogata, W.N. Wild-type and mutant stocks of Aspergillus nidulans. Genetics 1965, 52, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Emri, T.; Pócsi, I.; Szentirmai, A. Glutathione metabolism and protection against oxidative stress caused by peroxides in Penicillium chrysogenum. Free. Radic. Biol. Med. 1997, 23, 809–814. [Google Scholar] [CrossRef]
- Emri, T.; Pócsi, I.; Szentirmai, A. Analysis of the oxidative stress response of Penicillium chrysogenum to menadione. Free. Radic. Res. 1999, 30, 125–132. [Google Scholar] [CrossRef]
- Anderson, M.E. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzym. 1985, 113, 548–555. [Google Scholar]
- Sámi, L.; Emri, T.; Pócsi, I. Autolysis and aging of Penicillium chrysogenum cultures under carbon starvation: III: Glutathione metabolism and formation of reactive oxygen species. Mycol. Res. 2001, 105, 1246–1250. [Google Scholar] [CrossRef]
- Roggenkamp, R.; Sahm, H.; Wagner, F. Microbial assimilation of methanol induction and function of catalase in Candida boidinii. FEBS Lett. 1974, 41, 283–286. [Google Scholar] [CrossRef]
- Oberley, L.W.; Spitz, D.R. Assay of superoxide dismutase activity in tumor tissue. Methods Enzymol. 1984, 105, 457–464. [Google Scholar]
- Emri, T.; Bartók, G.; Szentirmai, A. Regulation of specific activity of glucose-6-phosphate-dehydrogenase and 6-phosphogluconate dehydrogenase in Penicillium chrysogenum. FEMS Microbiol. Lett. 1994, 117, 67–70. [Google Scholar] [CrossRef][Green Version]
- Warrilow, A.G.; Hawkesford, M.J. Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J. Exp. Bot. 2000, 51, 985–993. [Google Scholar] [CrossRef]
- Pusztahelyi, T.; Pócsi, I.; Szentirmai, A. Ageing of Penicillium chrysogenum cultures under carbon starvation. II. Protease and N-acetyl—D-hexosaminidase production. Biotechnol. Appl. Biochem. 1997, 25, 87–93. [Google Scholar] [CrossRef]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Bio. Techniques. 1993, 15, 536–537. [Google Scholar]
- Kovács, Z.; Szarka, M.; Kovács, S.; Boczonádi, I.; Emri, T.; Abe, K.; Pócsi, I.; Pusztahelyi, T. Effect of cell wall integrity stress and RlmA transcription factor on asexual development and autolysis in Aspergillus nidulans. Fungal Genet. Biol. 2013, 54, 1–14. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Clutterbuck, A.J. The genetics of conidiophore pigmentation in Aspergillus nidulans. J. Gen. Microbiol. 1990, 136, 1731–1738. [Google Scholar] [CrossRef]
- De Groot, P.W.; Brandt, B.W.; Horiuchi, H.; Ram, A.F.; de Koster, C.G.; Klis, F.M. Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet. Biol. 2009, 46, S72–S81. [Google Scholar] [CrossRef]
- Flipphi, M.; Sun, J.; Robellet, X.; Karaffa, L.; Fekete, E.; Zeng, A.P.; Kubicek, C.P. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol. 2009, 46, S19–S44. [Google Scholar] [CrossRef]
- Inglis, D.O.; Binkley, J.; Skrzypek, M.S.; Arnaud, M.B.; Cerqueira, G.C.; Shah, P.; Wymore, F.; Wortman, J.R.; Sherlock, G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013, 13, 91. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Fuoli, L.; Mellado, E. Ergosterol biosynthesis in Aspergillus fumigatus: Its relevance as an antifungal target and role in antifungal drug resistance. Front Microbiol. 2013, 3, 439. [Google Scholar] [CrossRef]
- Antsotegi-Uskola, M.; Markina-Iñarrairaegui, A.; Ugalde, U. Copper Resistance in Aspergillus nidulans Relies on the PI-Type ATPase CrpA, Regulated by the Transcription Factor AceA. Front. Microbiol. 2017, 8, 912. [Google Scholar] [CrossRef]
- Benguella, B.; Benaissa, H. Cadmium removal from aqueous solutions by chitin: Kinetic and equilibrium studies. Water Res. 2002, 36, 2463–2474. [Google Scholar] [CrossRef]
- Bhanoori, M.; Venkateswerlu, G. In vivo chitin-cadmium complexation in cell wall of Neurospora crassa. Biochim. Biophys. Acta. 2000, 1523, 21–28. [Google Scholar] [CrossRef]
- Chatterjee, S.; Kumari, S.; Rath, S.; Priyadarshanee, M.; Das, S. Diversity, structure and regulation of microbial metallothionein: Metal resistance and possible applications in sequestration of toxic metals. Metallomics 2020, 12, 1637–1655. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.S.; Lu, Y.P.; Zhen, R.G.; Szczypka, M.; Thiele, D.J.; Rea, P.A. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. USA 1997, 94, 42–47. [Google Scholar] [CrossRef]
- Prévéral, S.; Gayet, L.; Moldes, C.; Hoffmann, J.; Mounicou, S.; Gruet, A.; Reynaud, F.; Lobinski, R.; Verbavatz, J.M.; Vavasseur, A.; et al. A common highly conserved cadmium detoxification mechanism from bacteria to humans: Heavy metal tolerance conferred by the ATP-binding cassette (ABC) transporter SpHMT1 requires glutathione but not metal-chelating phytochelatin peptides. J. Biol. Chem. 2009, 284, 4936–4943. [Google Scholar] [CrossRef]
- Le, Q.G.; Ishiwata-Kimata, Y.; Kohno, K.; Kimata, Y. Cadmium impairs protein folding in the endoplasmic reticulum and induces the unfolded protein response. FEMS Yeast Res. 2016, 16, fow049. [Google Scholar] [CrossRef]
- Oberegger, H.; Schoeser, M.; Zadra, I.; Abt, B.; Haas, H. SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 2001, 41, 1077–1089. [Google Scholar] [CrossRef]
- Sato, I.; Shimizu, M.; Hoshino, T.; Takaya, N. The glutathione system of Aspergillus nidulans involves a fungus-specific glutathione S-transferase. J. Biol. Chem. 2009, 284, 8042–8053. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, L.; Aguirre, J. Multiple catalase genes are differentially regulated in Aspergillus nidulans. J. Bacteriol. 2001, 183, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Thön, M.; Al-Abdallah, Q.; Hortschansky, P.; Brakhage, A.A. The thioredoxin system of the filamentous fungus Aspergillus nidulans: Impact on development and oxidative stress response. J. Biol. Chem. 2007, 282, 27259–27269. [Google Scholar] [CrossRef] [PubMed]
- Szilágyi, M.; Miskei, M.; Karányi, Z.; Lenkey, B.; Pócsi, I.; Emri, T. Transcriptome changes initiated by carbon starvation in Aspergillus nidulans. Microbiology 2013, 159, 176–190. [Google Scholar] [CrossRef]
- Gila CsB Moon, H.; Antal, K.; Hajdu, M.; Kovács, R.; Jónás, P.A.; Pusztahelyi, T.; Yu, J.H.; Pócsi, I.; Emri, T. DUG pathway governs degradation of intracellular glutathione in Aspergillus nidulans. Appl. Environ. Microbiol. 2021, 87, e01321-20. [Google Scholar] [CrossRef]
- Brzóska, M.M.; Moniuszko-Jakoniuk, J. Interactions between cadmium and zinc in the organism. Food Chem. Toxicol. 2001, 39, 967–980. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, J.; Gao, J.; Shahzad, M.; Han, Z.; Wang, Z.; Li, J.; Sjölinder, H. Zinc supplementation protects against cadmium accumulation and cytotoxicity in Madin-Darby bovine kidney cells. PLoS ONE. 2014, 9, e103427. [Google Scholar] [CrossRef]
- Rossman, T.G.; Roy, N.K.; Lin, W.C. Is cadmium genotoxic? IARC Sci. Publ. 1992, 118, 367–375. [Google Scholar]
- Oberegger, H.; Zadra, I.; Schoeser, M.; Haas, H. Iron starvation leads to increased expression of Cu/Zn-superoxide dismutase in Aspergillus. FEBS Lett. 2000, 485, 113–116. [Google Scholar] [CrossRef]
- Nakagawa, C.W.; Yamada, K.; Mutoh, N. Role of Atf1 and Pap1 in the induction of the catalase gene of fission yeast Schizosaccharomyces pombe. J. Biochem. 2000, 127, 233–238. [Google Scholar] [CrossRef]
- Giaginis, C.; Gatzidou, E.; Theocharis, S. DNA repair systems as targets of cadmium toxicity. Toxicol. Appl. Pharmacol. 2006, 213, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Beyersmann, D.; Hartwig, A. Carcinogenic metal compounds: Recent insight into molecular and cellular mechanisms. Arch. Toxicol. 2008, 82, 493–512. [Google Scholar] [CrossRef] [PubMed]
- Maret, W.; Moulis, J.M. The bioinorganic chemistry of cadmium in the context of its toxicity. Met. Ions Life Sci. 2013, 11, 1–29. [Google Scholar]
- Tamás, M.J.; Sharma, S.K.; Ibstedt, S.; Jacobson, T.; Christen, P. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 2014, 4, 252–267. [Google Scholar] [CrossRef]
- Tamás, M.J.; Fauvet, B.; Christen, P.; Goloubinoff, P. Misfolding and aggregation of nascent proteins: A novel mode of toxic cadmium action in vivo. Curr. Genet. 2018, 64, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Guelfi, A.; Azevedo, R.A.; Lea, P.J.; Molina, S.M. Growth inhibition of the filamentous fungus Aspergillus nidulans by cadmium: An antioxidant enzyme approach. J. Gen. Appl. Microbiol. 2003, 49, 63–73. [Google Scholar] [CrossRef]
- Tu, B.P.; Weissman, J.S. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J. Cell Biol. 2004, 164, 341–346. [Google Scholar] [CrossRef]
- Rinnerthaler, M.; Büttner, S.; Laun, P.; Heeren, G.; Felder, T.K.; Klinger, H.; Weinberger, M.; Stolze, K.; Grousl, T.; Hasek, J.; et al. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc. Natl. Acad. Sci. USA 2012, 109, 8658–8663. [Google Scholar] [CrossRef]
- Todorova, D.; Nedeva, D.; Abrashev, R.; Tsekova, K. Cd (II) stress response during the growth of Aspergillus niger B 77. J. Appl. Microbiol. 2008, 104, 178–184. [Google Scholar] [CrossRef]
- Boczonádi, I.; Jakab, Á.; Baranyai, E.; Tóth, C.N.; Daróczi, L.; Csernoch, L.; Kis, G.; Antal, M.; Pusztahelyi, T.; Grawunder, A.; et al. Rare earth element sequestration by Aspergillus oryzae biomass. Environ. Technol. 2020, 16, 1–11. [Google Scholar] [CrossRef]
- Kapahi, M.; Sachdeva, S. Bioremediation options for heavy metal pollution. J. Health Pollut. 2019, 9, 191203. [Google Scholar] [CrossRef]
- Dusengemungu, L.; Kasali, G.; Gwanama, C.; Ouma, K.O. Recent advances in biosorption of copper and cobalt by filamentous fungi. Front Microbiol. 2020, 11, 582016. [Google Scholar] [CrossRef]
- Wysocki, R.; Tamás, M.J. How Saccharomyces cerevisiae copes with toxic metals and metalloids. FEMS Microbiol. Rev. 2010, 34, 925–951. [Google Scholar] [CrossRef] [PubMed]
- Fajardo-Somera, R.A.; Bowman, B.; Riquelme, M. The Plasma Membrane Proton Pump PMA-1 Is Incorporated into Distal Parts of the Hyphae Independently of the Spitzenkörper in Neurospora crassa. Eukaryot Cell 2013, 12, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Lecchi, S.; Allen, K.E.; Pardo, J.P.; Mason, A.B.; Slayman, C.W. Conformational changes of yeast plasma membrane H(+)-ATPase during activation by glucose: Role of threonine-912 in the carboxy-terminal tail. Biochemistry 2005, 44, 16624–16632. [Google Scholar] [CrossRef] [PubMed]
- Façanha, A.L.; Appelgren, H.; Tabish, M.; Okorokov, L.; Ekwall, K. The endoplasmic reticulum cation P-type ATPase Cta4p is required for control of cell shape and microtubule dynamics. J. Cell Biol. 2002, 157, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Cronin, S.R.; Rao, R.; Hampton, R.Y. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 2002, 157, 1017–1028. [Google Scholar] [CrossRef]
- Vincent, K.; Wang, Q.; Jay, S.; Hobbs, K.; Rymond, B.C. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics 2003, 164, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Oltra, E.; Verde, F.; Werner, R.; D’Urso, G. A novel RING-finger-like protein Ini1 is essential for cell cycle progression in fission yeast. J. Cell Sci. 2004, 117, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Vernis, L.; El Banna, N.; Baïlle, D.; Hatem, E.; Heneman, A.; Huang, M.E. Fe-S Clusters Emerging as Targets of Therapeutic Drugs. Oxid. Med. Cell. Longev. 2017, 2017, 3647657. [Google Scholar] [CrossRef] [PubMed]
- Aung-Htut, M.T.; Ayer, A.; Breitenbach, M.; Dawes, I.W. Oxidative stresses and ageing. Subcell Biochem. 2012, 57, 13–54. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).