Abstract
Tick-borne Rickettsia pathogens have become an emerging source of zoonotic infections and have a major impact on human health worldwide. In this study, the prevalence and genetic identity of Rickettsia infections in Ixodes granulatus ticks was firstly determined in Kinmen Island of Taiwan. A total of 247 I. granulatus ticks were examined for Rickettsia infection by nested-PCR assay targeting the citrate synthase (gltA) gene of Rickettsia. The Rickettsia infection was detected with a general infection rate of 4.86%, and was detected in nymph, male and female stages with an infection rate of 3.81%, 0% and 6.84%, respectively. Phylogenetic relationships were analyzed by comparing the gltA sequences obtained from four Taiwan strains and 19 other strains representing 13 genospecies of Rickettsia. Phylogenetic analyses reveal that all Taiwan strains were genetically affiliated to the genospecies of spotted fever (R. parkeri) and transitional (R. felis) groups of Rickettsia. Our findings reveal the first detection of R. parkeri-like and R. felis in I. granulatus ticks from Kinmen Island. As a tourist island between Taiwan and mainland China, these results demonstrate the epidemiological significance of diverse Rickettsia species existed in I. granulatus ticks and highlight the potential threat of geographical transmission among humans in the Taiwan area.
1. Introduction
The genus Rickettsia is composed of approximately 27 species of obligate intracellular gram-negative bacteria that can be classified into four major groups: ancestral group (AG), typhus group (TG), transitional group (TRG) and spotted fever group (SFG) [1,2,3]. The Ixodid ticks may serve as vectors and reservoirs of amplifying hosts for Rickettsia agents [4]. The SFG rickettsiae are mainly transmitted by vector ticks that can transmit the Rickettsia agents through the transovarial and transstadial pathways [5,6]. During recent decades, Rickettsia infections have become a global threat for emerging and re-emerging tick-borne diseases [7,8]. Global climate change has enhanced the geographical distribution of ticks and also expanded the spread of tick-borne pathogens [9,10]. Most SFG rickettsioses are found in a particular geographic location [10,11,12,13,14,15,16], and many validated SFG rickettsial species have been discovered in Australia, Central and South America, and Asia [17,18,19,20,21,22,23,24,25]. Thus, an epidemiological survey on tick-borne rickettsiae is important to understand the potential threat of emerging and re-emerging tick-borne Rickettsia infections in the Taiwan area.
Ticks are obligate hematophagous arthropod that may act as vectors with the ability to transmit various pathogens including bacteria, rickettsiae, protozoans and viruses [26]. The abundance and spread of Ixodes granulatus ticks had been recorded from various countries in Southeast Asia and Taiwan [27]. This tick species is found mainly on rodent hosts captured in grassy areas and vegetable gardens [28]. The medical and veterinary importance of the molecular detection of Lyme disease spirochetes (Borrelia burgdorferi sensu lato) and related spirochete (Borrelia valaisiana) has been identified in various stages of I. granulatus ticks of Taiwan [28,29]. In addition, the Rickettsia species of spotted fever group rickettsia had also been identified in I. granulatus ticks collected from Japan and offshore Orchid Island (Lanyu) of Taiwan [30,31]. Although the I. granulatus ticks have been recognized as the incriminated vector ticks for a variety of pathogens in Taiwan, there has no research demonstrating the prevalence of Rickettsia infection and confirming the genetic diversity in this tick species in Taiwan.
A DNA-based approach provides the feasibility to investigate the genetic variance at the individual base-pair level and gives a much more direct pathway for measuring the genetic diversity between and within species of Rickettsia [32]. Previous studies based on the molecular marker of citrate synthase gene (gltA) have concluded that it is sufficiently informative for the analysis of evolutionary relationships between the genetic diversity of Rickettsia species among various vectors and hosts [16,22,23,33]. Thus, molecular detection and genetic analysis based on the phylogenetic analysis of gltA gene has facilitated the identification and discrimination of Rickettsia species within ticks.
It may be that the Rickettsia infection in I. granulatus ticks of Taiwan is a genetically distinct genospecies, as compared with the existing common genospeciess of Rickettsia around the world. Thus, the objectives of this study intend to investigate the prevalence of Rickettsia infection in I. granulatus ticks collected from offshore Kinmen Island of Taiwan, and to determine the phylogenetic relationships between and within the genospecies of Rickettsia in these ticks. The genetic affiliation of Rickettsia strains detected in I. granulatus ticks of Taiwan was analyzed by comparing their differential nucleotide composition with other Rickettsia strains identified from various biological and geographical sources which have been documented in GenBank.
2. Materials and Methods
2.1. Tick Collection and Species Identification
All specimens of I. granulatus ticks used in this study were collected from rodents captured in the grassy areas and vegetable gardens, especially the peanut and sweet potato. The collection sites in four townships of Kinmen Island include Kinhu (24°41′ N, 118°43′ E; 24°43′ N, 118°46′ E), Kinsha (24°52′ N, 118°41′ E; 24°50′ N, 118°44′ E), Kinning (24°45′ N, 118°37′ E) and Kincheng (24°40′ N, 118°31′ E) (Figure 1).
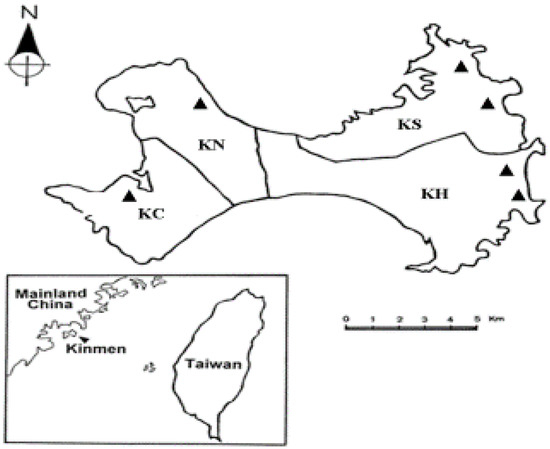
Figure 1.
Map of Kinmen Island showing the various collection sites (indicated as ▲) for ticks removed from trapped rodents.
All these ticks were subsequently cleaned and stored in separate glass vials containing 75% ethanol. All tick specimens of I. granulatus were identified to the species level on the basis of their morphological characteristics, as described previously [28]. Briefly, the external features of the I. granulatus ticks were recorded by using a stereo-microscope (SMZ 1500, Nikon, Tokyo, Japan) equipped with a fiber lamp and photographed for species identification.
2.2. DNA Extraction from Tick Specimens
Total genomic DNA was extracted from individual tick specimens used in this study. Briefly, tick specimens were cleaned by sonication for 3–5 min in 75% ethanol solution and then washed twice in sterile distilled water. Afterwards, the individual tick specimen was homogenized in a microcentrifuge tube filled with 180 μL lysing buffer solution (DNeasy Blood &Tissue Kit, catalogue no. 69506, Qiagen, Taipei, Taiwan) and then homogenized with a TissueLyser II apparatus (catalogue no. 85300, Qiagen, Taipei, Taiwan Branch), instructed by the manufacturer. The homogenate was centrifuged at room temperature and the supernatant fluid was further processed by a DNeasy Tissue Kit, as instructed by the manufacturer. After filtration, the filtrated fluid was collected and the DNA concentration was determined spectrophotometrically with a DNA calculator (Epoch, Biotek, Taipei, Taiwan Branch) and the extracted DNA was stored at −80 °C for further investigations.
2.3. DNA Amplification by Nested Polymerase Chain Reaction
DNA samples extracted from each tick specimens were used as a template for PCR amplification. Two primer sets based on the citrate synthase gene (gltA) were used for amplification. Initially, the primer set of RpCS.877p (5′-GGGGGCCTGCTCACGGCGG-3′) and RpCs.1258n (5′-ATTGCAAAAAGTACAGTGAACA-3′) was used to amplify the primary product of gltA. Nested PCR was then performed using the species-specific primer sets: RpCS.896p (5′-GGCTAATGAAGCAGTGATAA-3′) and RpCS.1233n (5′-GCGACGGTATACCCATAGC-3′) for amplifying a product approximately 338 bp [11]. All PCR reagents and Taq polymerase were obtained and used as recommended by the supplier (Takara Shuzo Co., Ltd., Kyoto, Japan). Briefly, each 25 μL reaction mixture contained 3 μL DNA template, 1.5 μL forward and reverse primers, 2.5 μL 10X PCR buffer (Mg2+), 2 μL dNTP mixture (10 mM each), 1 unit of Taq DNA polymerase and was filled-up with adequate volume of ddH2O. In contrast, adequate amounts of sterile distilled water were added for serving as a negative control. PCR amplification was performed with a thermocycler (Veriti, Applied Bioosystems, Taipei, Taiwan) and was denatured at 95 °C for 5 min and then amplified for 35 cycles with the conditions of denaturation at 95 °C for 30 s, annealing at 54 °C for 30 s, extension at 72 °C for 1 min, and followed by a final extension step at 72 °C for 3 min. For the nested-PCR, the following conditions were used: denaturation at 95 °C for 5 min and then amplified for 40 cycles with the conditions of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, extension at 72 °C for 1 min, and followed by a final extension step at 72 °C for 3 min.
For outer membrane protein B gene (ompB), same quantities as in gltA for the reaction mixture were used. The primer sets of rompB-OF (5′-GTAACCGGAAGTAATCGTTCGTAA-3′) and rompB-OR (5′-GCTTTATAACCAGCTAAACCACC-3′) was used to amplify the primary product of ompB. Nested PCR was then performed using the species-specific primer sets: rompB SPG-IF (5′-GTTTAATACGTGCTGCTAACCAA-3′) and rompB SPG/TG-IR (5′-GGTTTGGCCCATATACCATAAG-3′) for amplifying a product approximately 420-bp [11]. The PCR conditions were used same as gltA amplification except the annealing temperature of 50 °C and 52 °C for the initial and nested PCR cycles, respectively.
All amplified PCR products were electrophoresed on 1.5% agarose gels in Tris-Borate-EDTA (TBE) buffer and visualized under ultraviolet (UV) light after staining with ethidium bromide. A 100-bp DNA ladder (GeneRuler, Thermo Scientific, Taipei, Taiwan) was used as the standard marker for comparison. A negative control of distilled water was included in parallel with each amplification.
2.4. Sequence Alignments and Phylogenetic Analysis
Approximately 10-μL of each selected samples with clear bands on the agarose gel was submitted for DNA sequencing (Mission Biotech Co., Ltd., Taipei, Taiwan). After purification (QIAquick PCR Purification Kit, catalog No. 28104), sequencing reaction was performed with 25 cycles under the same conditions and same primer set of nested amplification by dye-deoxy terminator reaction method using the Big Dye Terminator Cycle Sequencing Kit in an ABI Prism 377-96 DNA Sequencer (Applied Biosystems, Foster City, CA, USA). The resulting sequences were initially edited by BioEdit software (V5.3) and aligned with the CLUSTAL W software [34]. Thereafter, the aligned sequences of Rickettsia gltA gene from four Taiwan strains were analyzed by comparing with other 19 strains of Rickettsia sequences from the different biological and geographical origin that are available from GenBank. Further analysis based on ompB genes of two Taiwan strains belonging to the SFG Rickettsia was performed by comparing with other 14 strains of Rickettsia sequences documented in GenBank. Phylogenetic analysis was performed by neighbor-joining (NJ) compared with maximum likelihood (ML) methods to estimate the phylogeny of the entire alignment using MEGA X software package [35]. The genetic distance values of inter- and intra-species variations were also analyzed by the Kimura two-parameter model [36]. All phylogenetic trees were constructed and performed with 1000 bootstrap replications to evaluate the reliability of the construction, as described previously [37].
2.5. Nucleotide Sequence Accession Numbers
The nucleotide sequences of PCR-amplified gltA genes of four Rickettsia strains from I. granulatus ticks of Taiwan determined in this study have been registered and assigned the following GenBank accession numbers: Ig-9103-KH-56 (MT847612), Ig-9103-KH-47 (MT847613), Ig-9106-KH-100 (MT847614) and Ig-9012-KS-495-N (MT847619). The GenBank accession numbers for the PCR-amplified ompB genes of two Rickettsia strains from I. granulatus ticks of Taiwan were also assigned as: Ig-9103-KH-47 (MZ198103) and Ig-9106-KH-100 (MZ198104). For phylogenetic analysis, the nucleotide sequences of gltA and ompB genes from other 19 and 14 Rickettsia strains were included for comparison, respectively. Their GenBank accession numbers are shown in Table 1.

Table 1.
Phylogenetic analysis based on gltA and ompB genes of Rickettsia used in this study.
3. Results
3.1. Detection of Rickettsia in I. granulatus Ticks from Taiwan
The existence of Rickettsia in I. granulatus ticks was detected by nested PCR assay targeting the gltA gene. In general, a total of 12 out of 247 (4.86%) I. granulatus ticks were detected with Rickettsia infection from Kinmen Island (Table 2). Based on the life stage of ticks, the Rickettsia infection was detected in nymphs, males and females of I. granulatus ticks with an infection rate of 3.81%, 0% and 6.84%, respectively (Table 2). The geographical prevalence of Rickettsia infection was detected only in Kinhu (7.21%) and Kinsha (3.92%) townships (Table 2).

Table 2.
Detection of Rickettsia infection in various life-cycle stages of Ixodes granulatus ticks collected from various sites of Kinmen Island, Taiwan by nested-PCR assay targeting the citrate synthase (gltA) gene of Rickettsia.
3.2. Sequence Alignment and Genetic Analysis of Rickettsia Detected in Ticks
To clarify the genetic identity of Rickettsia in I. granulatus ticks of Taiwan, the sequences of gltA gene fragments from four Taiwan strains of Rickettsia performed by this study were aligned and compared with the downloaded sequences of 19 other Rickettsia strains from different biological and geographical origin documented in GenBank. Results indicate that all these Rickettsia strains detected in I. granulatus ticks of Taiwan were genetically affiliated to the genospecies of R. felis and R. parkeri with the highly sequence similarity of 96.8–100% and 98.7–99.2% respectively (Table 3).

Table 3.
Intra- and inter-species analysis of genetic distance values a based on the gltA gene sequences between Rickettsia strains of Taiwan and other strains of Rickettsia documented in GenBank.
In addition, intra- and inter-species analysis based on the genetic distance (GD) values of gltA gene indicated a lower levels (GD < 0.032 and <0.013) of genetic divergence within the Rickettsia strains of Taiwan, as compared with the type strain of R. felis and R. parkeri, respectively (Table 3). Further analysis based on the GD values of ompB gene indicated a 100% similarity of genetic divergence (GD < 0.000) within the Rickettsia strains of Taiwan, as compared with the IG-1 strain (GD < 0.016) from Lanyu of Taiwan and the type strain of R. parkeri (GD < 0.024), respectively (Table 4).

Table 4.
Intra- and inter-species analysis of genetic distance values a based on the ompB gene sequences between Rickettsia strains of Taiwan and other strains of Rickettsia documented in GenBank.
3.3. Phylogenetic Analysis of Rickettsia Detected in Ticks
Phylogenetic relationships based on the sequence alignment of gltA and ompB genes were performed to demonstrate the genetic relationships among 23 and 16 strains of Rickettsia investigated in this study, respectively. Phylogenetic trees constructed by neighbour-joining (NJ) and maximum likelihood (ML) methods were used to analyze the phylogenetic relationships of Rickettsia strains. Bootstrap analysis was used to analyze the repeatability of the clustering of specimens represented in phylogenetic trees.
4. Discussion
This study reports the first molecular detection and genetic identification of Rickettsia infection in I. granulatus ticks of Taiwan. In this study, the Rickettsia species detected in I. granulatus ticks from Taiwan are genetically affiliated to the genospecies of R. parkeri and R. felis, which is different from the previous studies which described a R. honei-like organism in I. granulatus ticks identified from Thailand, Taiwan and Japan [20,30,31]. Although the R. felis was first reported in I. granulatus ticks from Taiwan (Figure 2, Figure 3 and Figure 4), it had been detected in the scrub typhus mite (Leptotrombidium deliense) in Taiwan [38]. In addition, the R. parkeri is considered as an emerging tick-borne pathogen of Spotted Fever Group Rickettsia and it is mainly transmitted by Amblyomma ticks in South America (Uruguay, Argentina and Brazil) [39]. Thus, our study demonstrates the first molecular evidence confirming the presence of Rickettsia species detected in I. granulatus ticks of Taiwan and provides the first convincing sequences (GenBank accession numbers: MT847612-4 and MT847619) of Rickettsia species discovered in I. granulatus ticks of Taiwan.
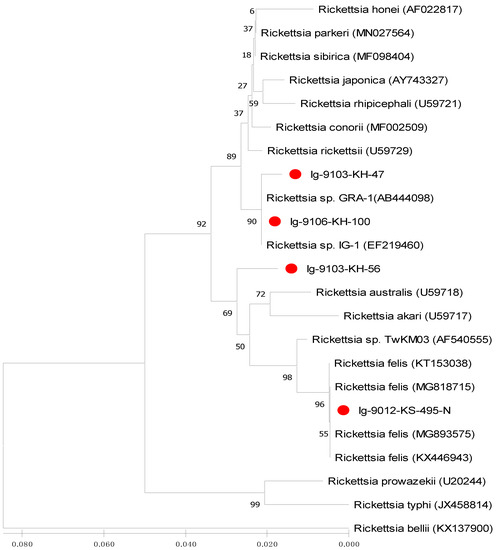
Figure 2.
Phylogenetic relationships based on the citrate synthase gene (gltA) sequences of Rickettsia between 4 specimens (indicated as ●) collected from Ixodes granulatus ticks of Kinmen Island and 19 other Rickettsia specimens identified from various biological and geographical origins. The tree was constructed and analyzed by neighbour-joining (NJ) method using 1000 bootstraps replicates. Numbers at the nodes indicate the percentages of reliability of each branch of the tree. Branch length is drawn proportional to the estimated sequence divergence.
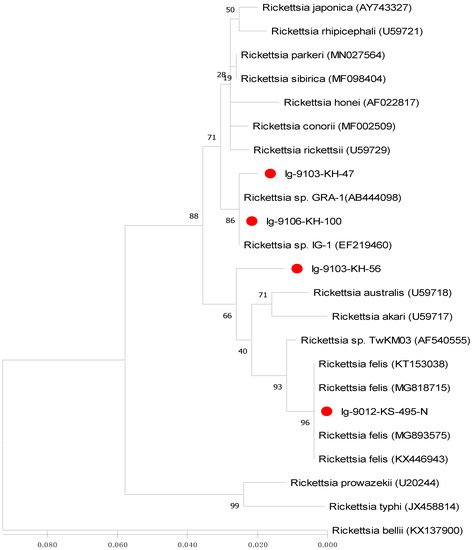
Figure 3.
Phylogenetic relationships based on the citrate synthase gene (gltA) sequences of Rickettsia between 4 specimens (indicated as ●) collected from Ixodes granulatus ticks of Kinmen Island and 19 other Rickettsia specimens identified from various biological and geographical origins. The tree was constructed and analyzed by maximum likelihood (ML) method using 1000 bootstraps replicates. Numbers at the nodes indicate the percentages of reliability of each branch of the tree. Branch length is drawn proportional to the estimated sequence divergence.
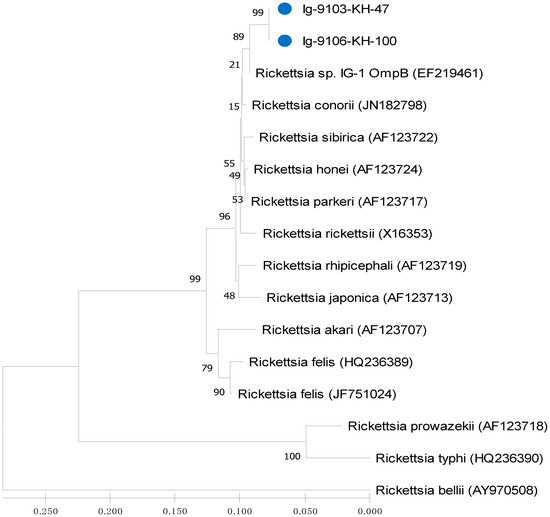
Figure 4.
Phylogenetic relationships based on the outer membrane protein B (ompB) sequences of Rickettsia between 2 specimens (indicated as ●) collected from Ixodes granulatus ticks of Taiwan and 14 other Rickettsia specimens identified from various biological and geographical origins. The tree was constructed and analyzed by neighbour-joining (NJ) method using 1000 bootstraps replicates. Numbers at the nodes indicate the percentages of reliability of each branch of the tree. Branch length is drawn proportional to the estimated sequence divergence.
The confirmed mechanism for the transmission of R. felis (flea/mite-borne) into ticks remains elusive. Due to the ability of parasitizing on the same rodent hosts, it is possible that the horizontal transmission of Rickettsia occurred between tick-flea or tick-mite interactions. Actually, ticks may accidentally feed on rodents that were previously fed by infected fleas/mites, and these ticks may acquire the Rickettsia infection through feeding blood from the same parasitized hosts [40]. Another non-systemic mode of transmission by a co-feeding mechanism may also contribute to the ticks feeding closely with another infected flea/mite on the same host that may facilitate the transmission of pathogens from an infected vector to a new vector [41]. Due to close contact with humans, these observations may highlight the epidemiological significance of rodents serving as a sentinel and reservoir host for the Rickettsia transmission in nature.
Phylogenetic relationships among Rickettsia in I. granulatus ticks can be determined by analyzing the sequence homogeneity of the gltA gene of Rickettsia. Indeed, sequence analysis based on the gltA gene of Rickettsia strains among various species from different origins had been shown to be useful for evaluating the genetic relatedness of Rickettsia detected from various biological and geographical sources [16,22,23,32,33]. Although the Rickettsia has been divided into various genospecies, most Rickettsia species are primarily found in arthropod hosts [33]. In this study, the phylogenetic analysis based on the sequences of gltA gene from I. granulatus ticks of Taiwan demonstrated a highly genetic homogeneity affiliated to the genospecies of R. parkeri and R. felis (Figure 2 and Figure 4, Table 3). The R. parkeri strains are mainly associated with the Rickettsia strain from humans (GenBank accession no. MN027564) and the R. felis strains are mainly affiliated to the Rickettsia strains from cat flea and lice (GenBank accession no. MG893575 and MG818715)). The phylogenetic trees constructed by either NJ or ML analysis strongly support the discrimination recognizing the separation of different genospecies between the Rickettsia strains in I. granulatus ticks collected from Taiwan and other genospecies of Rickettsia from different geographic and biological origins. Accordingly, these results demonstrate that genetic identities of Rickettsia strains detected in I. granulatus ticks collected from Taiwan were verified as a monophyletic group affiliated to the spotted fever (R. parkeri) and transitional (R. felis) groups of Rickettsia.
Whether global climate change may also increase the expansion of Ixodes ticks, which may enhance the transmission of tick-borne pathogens in Taiwan, remains unknown. Indeed, a previous 10-year study on rickettsias conducted in Germany demonstrated that the Rickettsia infection rate significantly increased over the years from 33.3% in 2005 to 50.8% in 2015 [10]. In addition, the previous study also discovered that the I. ricinus tick is reported to have spread into the previously unidentified northern areas of Sweden, Finland and Norway [42,43]. Because of the close association of I. granulatus ticks with residential area of humans, there is a serious concern regarding whether the Rickettsia species within this tick species can be transmitted into humans.
In this study, the Rickettsia infection in I. granulatus ticks was detected only in the Kinhu (KH) and Kinsa (KS) townships. The collection sites of KH and KS are mainly vegetable gardens as compared to the grassy areas in Kining and Kincheng townships. This difference of habitat may contribute to the capture rate of rodents in relation to the collection number of I. granulatus ticks from rodent hosts (Table 2). In addition, there is no detection of Rickettsia infection in male ticks is attributed to the small sample size of collected males. Indeed, the male tick is rarely collected from captured rodents and this phenomenon requires further investigation.
Although our results have demonstrated the PCR detection of Rickettsia infection in I. granulatus ticks, the vector competence should be further confirmed by cultivating of Rickettsia organisms from this tick species. Nevertheless, further investigations on the identification of vector ticks and genetic diversity of tick-borne Rickettsia may help to illustrate the spread of vector ticks and the risk of transmission of tick-borne rickettsial infections in Taiwan.
5. Conclusions
This study provides the first genetic identification of the Rickettsia infection detected in I. granulatus ticks collected from Kinmen Island of Taiwan. Based on the phylogenetic analyses, the I. granulatus ticks were found infected with the genospecies of R. parkeri-like and R. felis. Further investigations on the vector competence of these ticks may help to understand the potential risk and threat to human populations in Taiwan.
Author Contributions
Conceptualization, C.-M.S. and L.-L.C.; investigation, L.-L.C. and P.-W.Y.; formal analysis, C.-M.S. and L.-L.C.; writing of manuscript, C.-M.S. and L.-L.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by grant from the Ministry of Science and Technology (MOST 109-2314-B-037-077), Taipei, Taiwan, Republic of China.
Institutional Review Board Statement
The collection of ticks from rodents was approved by the Institutional Animal Care and Use Committee (IACUC) of Kaohsiung Medical University (IACUC Approval No: 106142).
Acknowledgments
We would like to appreciate the administrative help for the collection of ticks from the Public Health Bureau, Kinmen county of Taiwan.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fournier, P.E.; Dumler, J.S.; Greub, G.; Zhang, J.; Wu, Y.; Raoult, D. Gene sequence-based criteria for identification of new Rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J. Clin. Microbiol. 2003, 41, 5456–5465. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Paddock, C.D.; Raoult, D. Tick-borne rickettsioses around the world: Emerging diseases challenging old concepts. Clin. Microbiol. Rev. 2005, 18, 719–756. [Google Scholar] [CrossRef] [PubMed]
- Gillesoie, J.J.; Williams, K.; Shukla, M.; Snyder, E.E.; Nordberg, E.K.; Ceraul, S.M.; Dharmanolla, C.; Rainey, D.; Soneja, J.; Shallom, J.M.; et al. Rickettsia phylogenimics: Unwinding the intricacies of obligate intracellular life. PLoS ONE 2008, 3, 2018. [Google Scholar]
- Azad, A.F.; Beard, C.B. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 1998, 4, 179–186. [Google Scholar] [CrossRef]
- Bremer, W.G.; Schaefer, J.J.; Wagner, E.R.; Ewing, S.A.; Rikihisa, Y.; Needham, G.R.; Jittapalapong, S.; Moore, D.L.; Stich, R.W. Transstadial and intrastadial experimental transmission of Ehrlichia canis by male Rhipicephalus sanguineus. Vet. Parasitol. 2005, 131, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez, S.E.; Troyo, A. A review of the genus Rickettsia in Central America. Res. Rep. Trop. Med. 2018, 9, 103–112. [Google Scholar] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Satjanadumrong, J.; Robinson, M.T.; Hughes, T.; Blacksel, S.D. Distribution and ecological drivers of spotted fever group Rickettsia in Asia. EcoHealth 2019, 16, 611–626. [Google Scholar] [CrossRef]
- Loginov, D.S.; Loginova, Y.F.; Dycka, F.; Böttinger, K.; Vechtova, P.; Sterba, J. Tissue-specific signatures in tick cell line MS profiles. Parasit. Vectors 2019, 12, 212. [Google Scholar] [CrossRef]
- Blazejak, K.; Janecek, E.; Strube, C. A 10-year surveillance of Rickettsiales (Rickettsia spp. and Anaplasma phagocytophilum) in the city of Hanover, Germany, reveals Rickettsia spp. as emerging pathogens in ticks. Parasit. Vectors 2017, 10, 588. [Google Scholar] [CrossRef]
- Choi, Y.J.; Jang, W.J.; Kim, J.H.; Ryu, J.S.; Lee, S.H.; Park, K.H.; Paik, H.S.; Koh, Y.S.; Choi, M.S.; Kim, I.S. Spotted fever group and typhus group rickettsioses in humans, South Korea. Emerg. Infect. Dis. 2005, 11, 237–244. [Google Scholar] [CrossRef]
- Teglas, M.; Matern, E.; Lein, S.; Foley, P.; Mahan, S.M.; Foley, J. Ticks and tick-borne disease in Guatemalan cattle and horses. Vet. Parasitol. 2005, 131, 119–127. [Google Scholar] [CrossRef]
- Unsworth, N.B.; Stenos, J.; Graves, S.R.; Faa, A.G.; Cox, G.E.; Dyer, J.R.; Boutlis, C.S.; Lane, A.M.; Shaw, M.D.; Robson, J.; et al. Flinders Island Spotted Fever rickettsiosis caused by marmionii strain of Rickettsia honei, Eastern Australia. Emerg. Infect. Dis. 2007, 13, 566–573. [Google Scholar] [CrossRef]
- Wood, H.; Drebot, M.A.; Dewailly, E.; Dillon, L.; Dimitrova, K.; Forde, M.; Grolla, A.; Lee, E.; Loftis, A.; Makowski, K.; et al. Short Report: Seroprevalence of seven zoonotic pathogens in pregnant women from the Caribbean. Am. J. Trop. Med. Hyg. 2014, 91, 642–644. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.G.; Junior, J.M.; Foster, R.J.; Harmsen, B.J.; Sanchez, E.; Martins, T.F.; Quigley, H.; Marcili, A.; Labruna, M.B. Ticks and rickettsiae from wildlife in Belize, Central America. Parasit. Vectors 2016, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Blanton, L.S.; Walker, D.H. Rickettsiae as emerging infectious agents. Clin. Lab. Med. 2017, 37, 383–400. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Jin, Y.; Fan, M.; Xu, G.; Liu, Q.; Raoult, D. Genotypic and antigenic identification of two new strains of spotted fever group rickettsiae isolated from China. J. Clin. Microbiol. 1993, 31, 83–88. [Google Scholar] [CrossRef]
- Uchida, T.; Uchiyama, T.; Kumano, K.; Walker, D.H. Rickettsia japonica sp. nov., the etiological agent of spotted fever group rickettsiosis in Japan. Int. J. Syst. Bacteriol. 1992, 42, 303–305. [Google Scholar] [CrossRef]
- Stenos, J.; Roux, V.; Walker, D.H.; Raoult, D. Rickettsia honei sp. nov., the aetiological agent of Flinders Island spotted fever in Australia. Int. J. Syst. Bacteriol. 1998, 48, 1309–1404. [Google Scholar] [CrossRef]
- Kollars, T.M., Jr.; Tippayachai, B.; Bodhidatta, D. Short report: Thai tick typhus, Rickettsia honei, and a unique rickettsia detected in Ixodes granulatus (Ixodidae: Acari) from Thailand. Am. J. Trop. Med. Hyg. 2001, 65, 535–537. [Google Scholar] [CrossRef]
- Fujita, H.; Fournier, P.E.; Takada, N.; Saito, T.; Raoult, D. Rickettsia asiatica sp. nov. isolated in Japan. Int. J. Syst. Evol. Microbiol. 2006, 56, 2365–2368. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fournier, P.E.; Takada, N.; Fujita, H.; Raoult, D. Rickettsia tamurae sp. nov. isolated from Amblyomma testudinarium ticks. Int. J. Syst. Evol. Microbiol. 2006, 56, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Sun, Y.; Ju, W.; Wang, X.; Cao, W.; Wu, M. Vector competence of the tick Ixodes sinensis (Acari: Ixodidae) for Rickettsia monacensis. Parasit. Vectors 2014, 7, 512. [Google Scholar] [CrossRef] [PubMed]
- Springer, A.; Montenegro, V.M.; Schicht, S.; Wolfel, S.; Schaper, S.R.; Chitimia-Dobler, L.; Siebert, S.; Strube, C. Detection of Rickettsia monacensis and Rickettsia amblyommatis in ticks collected from dogs in Costa Rica and Nicaragua. Ticks Tick Borne Dis. 2018, 9, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- da Paixao Seva, A.; Martins, T.F.; Munoz-Leal, S.; Rodrigues, A.C.; Pinter, A.; Luz, H.R.; Angerami, R.N.; Labruna, M.B. A human case of spotted fever caused by Rickettsia parkeri strain Atlantic rainforest and its association to the tick Amblyomma ovale. Parasit. Vectors 2019, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Balashov, Y.S. Bloodsucking ticks (Ixodoidea)-vectors of diseases of man and animals. Misc. Publ. Entomol. Soc. Am. 1972, 8, 268–305. [Google Scholar]
- Wilson, N. New distributional records of ticks from Southeast Asia and the Pacific (Metastigmata: Argasidae, Ixodidae). Orient. Insects 1970, 4, 37–46. [Google Scholar] [CrossRef]
- Chao, L.L.; Wu, W.J.; Shih, C.M. First detection and molecular identification of Borrelia burgdorferi- like spirochetes in Ixodes granulatus ticks collected on Kinmen Island of Taiwan. Am. J. Trop. Med. Hyg. 2009, 80, 389–394. [Google Scholar] [CrossRef]
- Chao, L.L.; Wu, W.J.; Shih, C.M. Molecular detection of Borrelia valaisiana-related spirochetes from Ixodes granulatus ticks in Taiwan. Exp. Appl. Acarol. 2010, 52, 393–407. [Google Scholar] [CrossRef]
- Fujita, H.; Kadosaka, T.; Nitta, Y.; Ando, S.; Takano, A.; Watanabe, H.; Kawabata, H. Rickettsia sp. in Ixodes granulatus ticks, Japan. Emerg. Infect. Dis. 2008, 14, 1963–1965. [Google Scholar] [CrossRef]
- Tsai, K.H.; Wang, H.C.; Chen, C.H.; Huang, J.H.; Lu, H.Y.; Su, C.L.; Shu, P.Y. Isolation and identification of a novel spotted fever group rickettsia, strain IG-1, from Ixodes granulatus ticks collected on Orchid Island (Lanyu), Taiwan. Am. J. Trop. Med. Hyg. 2008, 79, 256–261. [Google Scholar] [CrossRef]
- Robinson, M.T.; Satjannadumrong, J.; Hugbes, T.; Stenos, J.; Blacksell, S.D. Diagnosis of spotted fever group Rickettsia infections: The Asian perspective. Epidemiol. Infect. 2019, 147, e286. [Google Scholar] [CrossRef]
- Roux, V.; Rydkina, E.; Eremeeva, M.; Raoult, D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int. J. Syst. Bacteriol. 1997, 47, 252–261. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Bio. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 52, 1119–1134. [Google Scholar]
- Tsui, P.Y.; Tsai, K.H.; Weng, M.H.; Hung, Y.W.; Liu, Y.T.; Hu, K.Y.; Lien, J.C.; Lin, P.Y.; Shaio, M.F.; Wang, H.C.; et al. Molecular detection and characterization of spotted fever group Rickettsiae in Taiwan. Am. J. Trop. Med. Hyg. 2007, 77, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.B.; Bechelli, J.; Smalley, C.; Karim, S.; Walker, D.H. Vector tick transmission model of spotted fever rickettsiosis. Am. J. Pathol. 2019, 189, 115–123. [Google Scholar] [CrossRef]
- Blair, P.J.; Jiang, J.; Schoeler, G.B.; Moron, C.; Anaya, E.; Cespedes, M.; Cruz, C.; Felices, V.; Guevara, C.; Mendoza, L.; et al. Characterization of spotted fever group rickettsiae in flea and tick specimens from northern Peru. J. Clin. Microbiol. 2004, 42, 4961–4967. [Google Scholar] [CrossRef]
- Randolph, S.E.; Gern, L.; Nuttall, P.A. Co-feeding ticks epidemiological significance for tick-borne pathogen transmission. Parasitol. Today 1996, 12, 472–479. [Google Scholar] [CrossRef]
- EI-Sayed, A.; Kamel, M. Climate changes and their role in emergence and re-emergence of diseases. Environ. Sci. Pollut. Res. 2020, 27, 22336–22352. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Pena, A. Ticks as vectors: Taxonomy, biology and ecology. Rev. Sci. Tech. Off. Int. Epiz. 2015, 34, 53–65. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).