Microbiome of Odontogenic Abscesses
Abstract
1. Introduction
- Does the oral microbiome of these patients show differences compared to the oral microbiome of healthy patients described in the literature?
- Which bacteria can be detected in pus using 16S rRNA gene next-generation sequencing analysis?
2. Materials and Methods
2.1. Nucleic Acid Extraction of Samples, Library Construction, 16S rRNA Amplicon Sequencing
2.2. Bioinformatics Analysis
2.3. Statistical Analysis
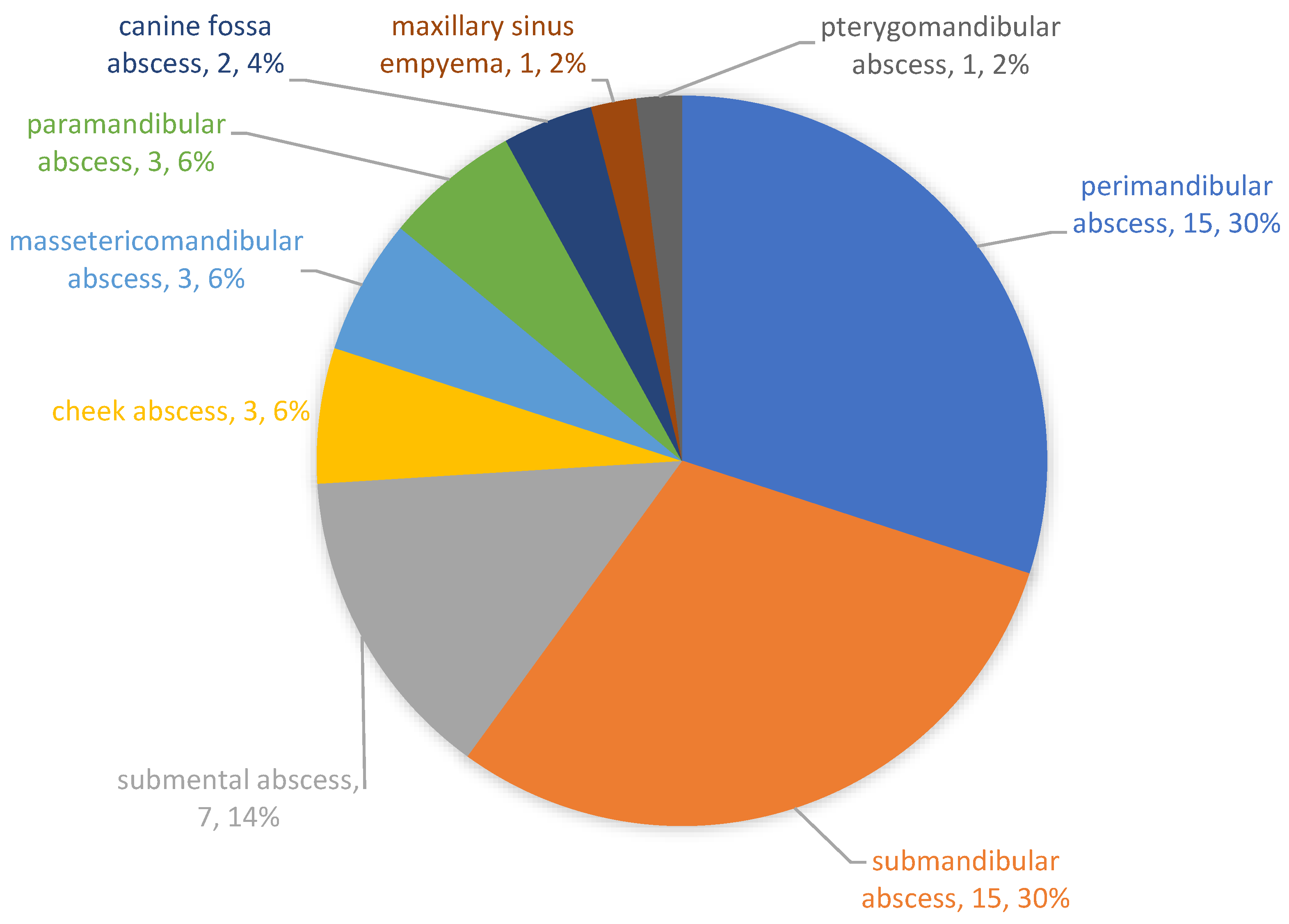
3. Results
4. Discussion
4.1. Culture-Based versus Molecular Detection Methods
4.2. Microbiome of the Saliva
4.3. Microbiome of the Pus
4.4. Pathogenicity of the Pus Microbiome
4.5. Phylum-Level Diversity and the Importance of Streptococcus
5. Conclusions
- The oral microbiome of patients with odontogenic abscesses was comparable to that of healthy subjects described in the literature, although very individual. However, the individually variable microbiome could possibly contain more bacteria with increased pathogenic potential.
- Odontogenic infections are mainly polymicrobial (96%) and rarely mono-infections (4%). Similar to saliva, pus showed its own microbiome, with a mean number of 31.44 (±12.09) genera.
- Odontogenic abscesses are mainly caused by anaerobic bacterial strains. Aerobic and facultative anaerobic bacteria seem to play a minor role compared to previously published results described by other authors.
- The most abundant genera in the pus were Prevotella, Porphyromonas and Fusobacterium, followed by Veillonella, Parvimonas, Streptococcus, Mogibacterium and Filifactor.
- The pus microbiomes likely have a much higher pathogenic potential than the oral microbiomes derived from saliva.
- Microbiome analysis detects significantly more bacteria than conventional culture-based methods and shows results even in the case of culture-negative samples. Molecular methods are expected to become the gold standard in medical microbiology diagnostics, particularly for polymicrobial infections with a predominance of anaerobic bacteria.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Nawas, B.; Karbach, J. S3-Leitlinie (Langversion): Odontogene Infektionen. In Leitlinien Zahnmedizin; Arbeitsgemeinschaft der Wissenschaftlichen Medizinischen Fachgesellschaften: Berlin, Germany, 2016; Available online: www.awmf.org (accessed on 10 March 2020).
- Al-Nawas, B.; Maeurer, M. Severe versus Local Odontogenic Bacterial Infections: Comparison of Microbial Isolates. Eur. Surg. Res. 2007, 40, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.F.; Rôças, I.N. Microbiology and Treatment of Acute Apical Abscesses. Clin. Microbiol. Rev. 2013, 26, 255–273. [Google Scholar] [CrossRef]
- Warnke, P.H.; Becker, S.T.; Springer, I.N.; Haerle, F.; Ullmann, U.; Russo, P.A.; Wiltfang, J.; Fickenscher, H.; Schubert, S. Penicillin compared with other advanced broad spectrum antibiotics regarding antibacterial activity against oral pathogens isolated from odontogenic abscesses. J. Cranio-Maxillofac. Surg. 2008, 36, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Böttger, S.; Lautenbacher, K.; Domann, E.; Howaldt, H.-P.; Attia, S.; Streckbein, P.; Wilbrand, J.-F. Indication for an additional postoperative antibiotic treatment after surgical incision of serious odontogenic abscesses. J. Cranio-Maxillofac. Surg. 2020, 48, 229–234. [Google Scholar] [CrossRef]
- Flynn, T.R. The swollen face. Severe odontogenic infections. Emerg. Med. Clin. N. Am. 2000, 18, 481–519. [Google Scholar] [CrossRef]
- Eckert, A.W.; Maurer, P.; Wilhelms, D.; Schubert, J. Soft tissue infections in oral, maxillofacial, and plastic surgery. Bacterial spectra and antibiotics. Mund. Kiefer Gesichtschir. 2005, 9, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Bali, R.K.; Sharma, P.; Gaba, S.; Kaur, A.; Ghanghas, P. A review of complications of odontogenic infections. Natl. J. Maxillofac. Surg. 2015, 6, 136–143. [Google Scholar] [CrossRef]
- Palma, D.M.; Giuliano, S.; Cracchiolo, A.N.; Falcone, M.; Ceccarelli, G.; Tetamo, R.; Venditti, M. Clinical features and outcome of patients with descending necrotizing mediastinitis: Prospective analysis of 34 cases. Infection 2015, 44, 77–84. [Google Scholar] [CrossRef]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Wiese, K.G.; Merten, H.A.; Wiltfang, J.; Luhr, H.G. Clinical studies on the pathophysiology of odontogenic abscesses. Mund Kiefer Gesichtschir. 1999, 3, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.F., Jr.; Rôças, I.N. As-yet-uncultivated oral bacteria: Breadth and association with oral and extra-oral diseases. J. Oral Microbiol. 2013, 5, 21077. [Google Scholar] [CrossRef] [PubMed]
- Böttger, S.; Zechel-Gran, S.; Streckbein, P.; Knitschke, M.; Hain, T.; Weigel, M.; Wilbrand, J.-F.; Domann, E.; Howaldt, H.-P.; Attia, S. A New Type of Chronic Wound Infection after Wisdom Tooth Extraction: A Diagnostic Approach with 16S-rRNA Gene Analysis, Next-Generation Sequencing, and Bioinformatics. Pathogens 2020, 9, 798. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.F.; Rôças, I.N. The Oral Microbiota in Health and Disease: An Overview of Molecular Findings. Methods Mol. Biol. 2017, 1537, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Voelkerding, K.V.; Dames, S.A.; Durtschi, J.D. Next-Generation Sequencing: From Basic Research to Diagnostics. Clin. Chem. 2009, 55, 641–658. [Google Scholar] [CrossRef]
- Echen, H.; Ejiang, W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front. Microbiol. 2014, 5, 508. [Google Scholar] [CrossRef]
- Keller, P.M.; Hombach, M.; Bloemberg, G.V. 16S-rRNA-Gen-basierte Identifikation bakterieller Infektionen. Biospektrum 2010, 16, 755–758. [Google Scholar]
- Griessl, T.; Zechel-Gran, S.; Olejniczak, S.; Weigel, M.; Hain, T.; Domann, E. High-resolution taxonomic examination of the oral microbiome after oil pulling with standardized sunflower seed oil and healthy participants: A pilot study. Clin. Oral Investig. 2021, 25, 2689–2703. [Google Scholar] [CrossRef]
- Regier, Y.; Komma, K.; Weigel, M.; Kraiczy, P.; Laisi, A.; Pulliainen, A.T.; Hain, T.; Kempf, V.A.J. Combination of microbiome analysis and serodiagnostics to assess the risk of pathogen transmission by ticks to humans and animals in central Germany. Parasites Vectors 2019, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef]
- Sizova, M.V.; Hohmann, T.; Hazen, A.; Paster, B.J.; Halem, S.R.; Murphy, C.M.; Panikov, N.S.; Epstein, S.S. New Approaches for Isolation of Previously Uncultivated Oral Bacteria. Appl. Environ. Microbiol. 2011, 78, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Gibbons, R.J.; Dale, A.C.; Bortnick, L.; Rosenthal, E.; Macdonald, J.B. The microbiota of the gingival crevice area of man. I. Total microscopic and viable counts and counts of specific organisms. Arch. Oral Biol. 1963, 8, 275–280. [Google Scholar] [CrossRef]
- Heim, N.; Faron, A.; Wiedemeyer, V.; Reich, R.; Martini, M. Microbiology and antibiotic sensitivity of head and neck space infections of odontogenic origin. Differences in inpatient and outpatient management. J. Cranio-Maxillofac. Surg. 2017, 45, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Flynn, T.R.; Shanti, R.M.; Hayes, C. Severe Odontogenic Infections, Part 2: Prospective Outcomes Study. J. Oral Maxillofac. Surg. 2006, 64, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Chandra, H.J.; Rao, B.H.S.; Manzoor, A.P.M.; Arun, A.B. Characterization and Antibiotic Sensitivity Profile of Bacteria in Orofacial Abscesses of Odontogenic Origin. J. Maxillofac. Oral Surg. 2016, 16, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Opitz, D.; Camerer, C.; Camerer, D.-M.; Raguse, J.-D.; Menneking, H.; Hoffmeister, B.; Adolphs, N. Incidence and management of severe odontogenic infections—A retrospective analysis from 2004 to 2011. J. Cranio-Maxillofac. Surg. 2015, 43, 285–289. [Google Scholar] [CrossRef]
- Lee, Y.Q.; Kanagalingam, J. Bacteriology of deep neck abscesses: A retrospective review of 96 consecutive cases. Singap. Med. J. 2011, 52, 351–355. [Google Scholar] [CrossRef]
- Celakovsky, P.; Kalfert, D.; Smatanova, K.; Tuček, L.; Čermáková, E.; Mejzlik, J.; Kotulek, M.; Vrbacky, A.; Matoušek, P.; Stanikova, L.; et al. Bacteriology of deep neck infections: Analysis of 634 patients. Aust. Dent. J. 2015, 60, 212–215. [Google Scholar] [CrossRef]
- Poeschl, P.W.; Spusta, L.; Russmueller, G.; Seemann, R.; Hirschl, A.; Poeschl, E.; Klug, C.; Ewers, R. Antibiotic susceptibility and resistance of the odontogenic microbiological spectrum and its clinical impact on severe deep space head and neck infections. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2010, 110, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.F.; Rôças, I.N. Community as the unit of pathogenicity: An emerging concept as to the microbial pathogenesis of apical periodontitis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2009, 107, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Munson, M.A.; Pitt-Ford, T.; Chong, B.; Weightman, A.; Wade, W.G. Molecular and Cultural Analysis of the Microflora Associated with Endodontic Infections. J. Dent. Res. 2002, 81, 761–766. [Google Scholar] [CrossRef]
- Jenkinson, H.F.; Lamont, R.J. Oral microbial communities in sickness and in health. Trends Microbiol. 2005, 13, 589–595. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef]
- Chen, T.; Yu, W.-H.; Izard, J.; Baranova, O.V.; Lakshmanan, A.; Dewhirst, F.E. The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010, baq013. [Google Scholar] [CrossRef]
- Zaura, E.; Keijser, B.J.F.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Bik, E.M.; Long, C.D.; Armitage, G.C.; Loomer, P.; Emerson, J.; Mongodin, E.F.; Nelson, K.E.; Gill, S.R.; Fraser-Liggett, C.M.; Relman, D.A. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010, 4, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, Y.; Zhang, M.; Wang, G.; Qi, Z.; Bridgewater, L.; Zhao, L.; Tang, Z.; Pang, X. A Filifactor alocis-centered co-occurrence group associates with periodontitis across different oral habitats. Sci. Rep. 2015, 5, 9053. [Google Scholar] [CrossRef]
- Nibali, L.; Sousa, V.; Davrandi, M.; Spratt, D.; Alyahya, Q.; Dopico, J.; Donos, N. Differences in the periodontal microbiome of successfully treated and persistent aggressive periodontitis. J. Clin. Periodontol. 2020, 47, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.F.; Rôças, I.N.; Souto, R.; de Uzeda, M.; Colombo, A.P. Microbiological evaluation of acute periradicular abscesses by DNA-DNA hybridization. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2001, 92, 451–457. [Google Scholar] [CrossRef]
- Van Winkelhoff, A.J.; Carlee, A.W.; de Graaff, J. Bacteroides endodontalis and other black-pigmented Bacteroides species in odontogenic abscesses. Infect. Immun. 1985, 49, 494–497. [Google Scholar] [CrossRef]
- Mashima, I.; Kamaguchi, A.; Nakazawa, F. The Distribution and Frequency of Oral Veillonella spp. in the Tongue Biofilm of Healthy Young Adults. Curr. Microbiol. 2011, 63, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, M.; Wang, Y.; Zhou, X.; Peng, X.; Ren, B.; Li, M.; Cheng, L. Effect of Veillonella parvula on the physiological activity of Streptococcus mutans. Arch. Oral Biol. 2020, 109, 104578. [Google Scholar] [CrossRef] [PubMed]
- Rôças, I.; Siqueira, J. Culture-Independent Detection of Eikenella corrodens and Veillonella parvula in Primary Endodontic Infections. J. Endod. 2006, 32, 509–512. [Google Scholar] [CrossRef]
- Ter Steeg, P.F.; Van Der Hoeven, J.S. Growth stimulation of Treponema denticola by periodontal microorganisms. Antonie Leeuwenhoek 1990, 57, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Stehlikova, Z.; Tlaskal, V.; Galanova, N.; Roubalova, R.; Kreisinger, J.; Dvorak, J.; Prochazkova, P.; Kostovcikova, K.; Bartova, J.; Libanska, M.; et al. Oral Microbiota Composition and Antimicrobial Antibody Response in Patients with Recurrent Aphthous Stomatitis. Microorganisms 2019, 7, 636. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pu, Y.; Lu, H.; Zhao, N.; Wang, Y.; Guo, Y.; Guo, C. Porphyromonas, Treponema, and Mogibacterium promote IL8/IFNγ/TNFα-based pro-inflammation in patients with medication-related osteonecrosis of the jaw. J. Oral Microbiol. 2020, 13, 1851112. [Google Scholar] [CrossRef]
- Gomes, B.P.; Berber, V.B.; Kokaras, A.S.; Chen, T.; Paster, B.J. Microbiomes of Endodontic-Periodontal Lesions before and after Chemomechanical Preparation. J. Endod. 2015, 41, 1975–1984. [Google Scholar] [CrossRef]
- Gomes, B.P.; Louzada, L.M.; Almeida-Gomes, R.F.; Pinheiro, E.T.; Sousa, E.L.; Jacinto, R.C.; Arruda-Vasconcelos, R. Investigation of Filifactor alocis in primary and in secondary endodontic infections: A molecular study. Arch. Oral Biol. 2020, 118, 104826. [Google Scholar] [CrossRef]
- Aruni, A.W.; Mishra, A.; Dou, Y.; Chioma, O.; Hamilton, B.N.; Fletcher, H.M. Filifactor alocis—A new emerging periodontal pathogen. Microbes Infect. 2015, 17, 517–530. [Google Scholar] [CrossRef]
- Aruni, W.; Chioma, O.; Fletcher, H. Filifactor alocis: The Newly Discovered Kid on the Block with Special Talents. J. Dent. Res. 2014, 93, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.C.; Ebersole, J.L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 2005, 38, 72–122. [Google Scholar] [CrossRef]
- Jun, H.-K.; Jung, Y.-J.; Choi, B.-K. Treponema denticola, Porphyromonas gingivalis, and Tannerella forsythia induce cell death and release of endogenous danger signals. Arch. Oral Biol. 2017, 73, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Mysak, J.; Podzimek, S.; Sommerova, P.; Lyuya-Mi, Y.; Bartova, J.; Janatova, T.; Prochazkova, J.; Duskova, J. Porphyromonas gingivalis: Major Periodontopathic Pathogen Overview. J. Immunol. Res. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Williams, B.L.; McCann, G.F.; Schoenknecht, F.D. Bacteriology of dental abscesses of endodontic origin. J. Clin. Microbiol. 1983, 18, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Khemaleelakul, S.; Baumgartner, J.; Pruksakorn, S. Identification of bacteria in acute endodontic infections and their antimicrobial susceptibility. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2002, 94, 746–755. [Google Scholar] [CrossRef]
- Plum, A.W.; Mortelliti, A.J.; Walsh, R.E. Microbial Flora and Antibiotic Resistance in Odontogenic Abscesses in Upstate New York. Ear Nose Throat J. 2018, 97, E27–E31. [Google Scholar] [CrossRef]
- Shakya, N.; Sharma, D.; Newaskar, V.; Agrawal, D.; Shrivastava, S.; Yadav, R. Epidemiology, Microbiology and Antibiotic Sensitivity of Odontogenic Space Infections in Central India. J. Maxillofac. Oral Surg. 2018, 17, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Heimdahl, A.; Von Konow, L.; Satoh, T.; Nord, C.E. Clinical appearance of orofacial infections of odontogenic origin in relation to microbiological findings. J. Clin. Microbiol. 1985, 22, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Bahl, R.; Sandhu, S.; Sahai, N.; Gupta, M.; Singh, K. Odontogenic infections: Microbiology and management. Contemp. Clin. Dent. 2014, 5, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Kuriyama, T.; Williams, D.W.; Yanagisawa, M.; Iwahara, K.; Shimizu, C.; Nakagawa, K.; Yamamoto, E.; Karasawa, T. Antimicrobial susceptibility of 800 anaerobic isolates from patients with dentoalveolar infection to 13 oral antibiotics. Oral Microbiol. Immunol. 2007, 22, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-Y.; Huang, Y.-T.; Liao, C.-H.; Yen, L.-C.; Lin, H.-Y.; Hsueh, P.-R. Increasing Trends in Antimicrobial Resistance among Clinically Important Anaerobes and Bacteroides fragilis Isolates Causing Nosocomial Infections: Emerging Resistance to Carbapenems. Antimicrob. Agents Chemother. 2008, 52, 3161–3168. [Google Scholar] [CrossRef] [PubMed]
- Sherrard, L.J.; Graham, K.A.; McGrath, S.J.; McIlreavey, L.; Hatch, J.; Muhlebach, M.S.; Wolfgang, M.C.; Gilpin, D.F.; Elborn, J.S.; Schneiders, T.; et al. Antibiotic resistance in Prevotella species isolated from patients with cystic fibrosis. J. Antimicrob. Chemother. 2013, 68, 2369–2374. [Google Scholar] [CrossRef]











| Genus (Saliva Samples) | Mean | S.dev | Median | MM | Genus (Pus Samples) | Mean | S.dev. | Median | MM |
|---|---|---|---|---|---|---|---|---|---|
| Prevotella | 17.65 | 13.95 | 16.39 | 26.67 | Prevotella | 27.12 | 17.00 | 27.91 | 48.76 |
| Streptococcus | 17.26 | 13.85 | 14.95 | 24.34 | Fusobacterium | 16.03 | 18.92 | 9.13 | 15.95 |
| Veillonella | 9.33 | 8.63 | 6.67 | 10.85 | Porphyromonas | 12.94 | 13.95 | 11.26 | 19.67 |
| Rothia | 8.43 | 11.95 | 4.40 | 7.16 | Streptococcus | 6.57 | 20.07 | 1.15 | 2.00 |
| Neisseria | 7.18 | 10.99 | 2.87 | 4.68 | Peptostreptococcus | 4.91 | 6.97 | 0.17 | 0.30 |
| Haemophilus | 5.85 | 8.47 | 2.25 | 3.66 | Parvimonas | 4.79 | 6.58 | 2.16 | 3.78 |
| Actinomyces | 4.39 | 3.41 | 3.99 | 6.50 | Veillonella | 3.31 | 3.28 | 2.83 | 4.95 |
| Alloprevotella | 4.14 | 8.01 | 0.86 | 1.39 | Alloprevotella | 2.36 | 5.14 | 0.42 | 0.74 |
| Lactobacillus | 4.10 | 6.02 | 1.94 | 3.16 | Mogibacterium | 1.45 | 1.78 | 1.02 | 1.79 |
| Porphyromonas | 2.63 | 4.72 | 0.90 | 1.47 | Filifactor | 1.36 | 2.17 | 0.47 | 0.82 |
| Fusobacterium | 2.35 | 2.91 | 1.47 | 2.39 | Atopobium | 1.32 | 2.09 | 0.14 | 0.24 |
| Atopobium | 2.23 | 2.58 | 1.32 | 2.15 | Erysipelothrix | 1.28 | 3.26 | 0.00 | 0.00 |
| Gemella | 1.28 | 2.05 | 0.45 | 0.73 | Slackia | 1.24 | 1.86 | 0.24 | 0.42 |
| Leptotrichia | 1.14 | 1.34 | 0.42 | 0.69 | Treponema | 1.18 | 3.10 | 0.05 | 0.08 |
| Bifidobacterium | 1.09 | 1.97 | 0.23 | 0.37 | Oribacterium | 0.99 | 2.70 | 0.02 | 0.03 |
| Peptostreptococcus | 0.93 | 1.64 | 0.14 | 0.24 | Lachnospira | 0.96 | 2.34 | 0.00 | 0.00 |
| Capnocytophaga | 0.92 | 1.46 | 0.24 | 0.39 | Erysipelotrichaceae bacterium | 0.85 | 2.03 | 0.00 | 0.00 |
| Treponema | 0.83 | 1.96 | 0.22 | 0.35 | Selenomonas | 0.82 | 4.37 | 0.01 | 0.02 |
| Selenomonas | 0.71 | 1.03 | 0.29 | 0.48 | Coriobacterium | 0.79 | 2.36 | 0.06 | 0.10 |
| Mogibacterium | 0.55 | 0.96 | 0.19 | 0.31 | Family_XIII_ge | 0.74 | 1.33 | 0.00 | 0.00 |
| Parvimonas | 0.50 | 0.93 | 0.12 | 0.20 | Mycoplasma | 0.71 | 2.21 | 0.00 | 0.01 |
| Oribacterium | 0.49 | 0.65 | 0.18 | 0.29 | Lachnospiraceae bacterium | 0.66 | 1.61 | 0.00 | 0.00 |
| Campylobacter | 0.46 | 0.49 | 0.34 | 0.55 | Fretibacterium | 0.51 | 2.16 | 0.01 | 0.01 |
| Erysipelothrix | 0.42 | 0.80 | 0.00 | 0.00 | Gemella | 0.48 | 1.42 | 0.01 | 0.01 |
| Lachnospira | 0.41 | 0.90 | 0.00 | 0.00 | Bacteroides | 0.47 | 2.80 | 0.00 | 0.00 |
| Lachnoanaerobaculum | 0.37 | 0.66 | 0.08 | 0.12 | Pyramidobacter | 0.46 | 2.24 | 0.00 | 0.00 |
| Lachnospiraceae bacterium | 0.36 | 0.89 | 0.01 | 0.01 | Dialister | 0.41 | 0.89 | 0.04 | 0.08 |
| Pasteurella | 0.30 | 0.88 | 0.00 | 0.00 | Tannerella | 0.33 | 0.84 | 0.00 | 0.01 |
| Mycoplasma | 0.29 | 0.67 | 0.04 | 0.06 | Campylobacter | 0.31 | 0.70 | 0.02 | 0.04 |
| Slackia | 0.25 | 0.45 | 0.08 | 0.14 | Neisseria | 0.31 | 0.75 | 0.01 | 0.01 |
| Erysipelotrichaceae bacterium | 0.24 | 0.53 | 0.00 | 0.00 | Ruminococcus | 0.30 | 1.37 | 0.00 | 0.00 |
| Filifactor | 0.24 | 0.46 | 0.02 | 0.04 | Eubacterium | 0.29 | 0.51 | 0.01 | 0.01 |
| Fretibacterium | 0.24 | 0.44 | 0.06 | 0.10 | Haemophilus | 0.28 | 0.80 | 0.01 | 0.01 |
| Tannerella | 0.23 | 0.36 | 0.08 | 0.13 | Actinomyces | 0.25 | 1.21 | 0.01 | 0.02 |
| Family_XIII_ge | 0.23 | 0.49 | 0.00 | 0.00 | Catonella | 0.21 | 0.58 | 0.03 | 0.06 |
| Coriobacterium | 0.21 | 0.35 | 0.09 | 0.14 | Olsenella | 0.20 | 0.64 | 0.00 | 0.00 |
| Olsenella | 0.17 | 0.42 | 0.03 | 0.06 | Rikenella | 0.19 | 0.41 | 0.00 | 0.01 |
| Dialister | 0.14 | 0.28 | 0.04 | 0.07 | Staphylococcus | 0.18 | 0.79 | 0.01 | 0.02 |
| Ruminococcus | 0.13 | 0.27 | 0.02 | 0.03 | Clostridium | 0.17 | 0.73 | 0.00 | 0.00 |
| Desulfovibrio | 0.09 | 0.27 | 0.00 | 0.00 | Acidaminococcus | 0.16 | 0.79 | 0.00 | 0.00 |
| Escherichia-Shigella (E.coli) | 0.09 | 0.48 | 0.00 | 0.00 | Clostridiales bacterium | 0.16 | 0.52 | 0.00 | 0.00 |
| SR1_(Absconditabacteria)_ge | 0.09 | 0.37 | 0.00 | 0.00 | Bacteroidales_S24-7 | 0.16 | 1.00 | 0.00 | 0.00 |
| Corynebacterium | 0.08 | 0.15 | 0.03 | 0.05 | Peptococcus | 0.13 | 0.38 | 0.00 | 0.00 |
| Catonella | 0.08 | 0.14 | 0.02 | 0.04 | Rothia | 0.12 | 0.36 | 0.01 | 0.02 |
| Bacteroides | 0.07 | 0.38 | 0.00 | 0.00 | Stenotrophomonas | 0.12 | 0.82 | 0.00 | 0.00 |
| Mollicutes | 0.07 | 0.21 | 0.00 | 0.00 | Lactobacillus | 0.12 | 0.31 | 0.02 | 0.03 |
| Sphaerochaeta | 0.07 | 0.36 | 0.00 | 0.00 | Desulfovibrio | 0.11 | 0.55 | 0.00 | 0.00 |
| Rikenella | 0.07 | 0.15 | 0.01 | 0.01 | Delftia | 0.09 | 0.41 | 0.00 | 0.01 |
| Peptococcus | 0.05 | 0.10 | 0.00 | 0.00 | Bacteroidales bacterium | 0.08 | 0.44 | 0.00 | 0.00 |
| Flavobacterium | 0.04 | 0.08 | 0.00 | 0.00 | Pasteurella | 0.07 | 0.36 | 0.00 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Böttger, S.; Zechel-Gran, S.; Schmermund, D.; Streckbein, P.; Wilbrand, J.-F.; Knitschke, M.; Pons-Kühnemann, J.; Hain, T.; Weigel, M.; Howaldt, H.-P.; et al. Microbiome of Odontogenic Abscesses. Microorganisms 2021, 9, 1307. https://doi.org/10.3390/microorganisms9061307
Böttger S, Zechel-Gran S, Schmermund D, Streckbein P, Wilbrand J-F, Knitschke M, Pons-Kühnemann J, Hain T, Weigel M, Howaldt H-P, et al. Microbiome of Odontogenic Abscesses. Microorganisms. 2021; 9(6):1307. https://doi.org/10.3390/microorganisms9061307
Chicago/Turabian StyleBöttger, Sebastian, Silke Zechel-Gran, Daniel Schmermund, Philipp Streckbein, Jan-Falco Wilbrand, Michael Knitschke, Jörn Pons-Kühnemann, Torsten Hain, Markus Weigel, Hans-Peter Howaldt, and et al. 2021. "Microbiome of Odontogenic Abscesses" Microorganisms 9, no. 6: 1307. https://doi.org/10.3390/microorganisms9061307
APA StyleBöttger, S., Zechel-Gran, S., Schmermund, D., Streckbein, P., Wilbrand, J.-F., Knitschke, M., Pons-Kühnemann, J., Hain, T., Weigel, M., Howaldt, H.-P., Domann, E., & Attia, S. (2021). Microbiome of Odontogenic Abscesses. Microorganisms, 9(6), 1307. https://doi.org/10.3390/microorganisms9061307








