Different Reactions in Each Enterotype Depending on the Intake of Probiotic Yogurt Powder
Abstract
1. Introduction
2. Materials and Methods
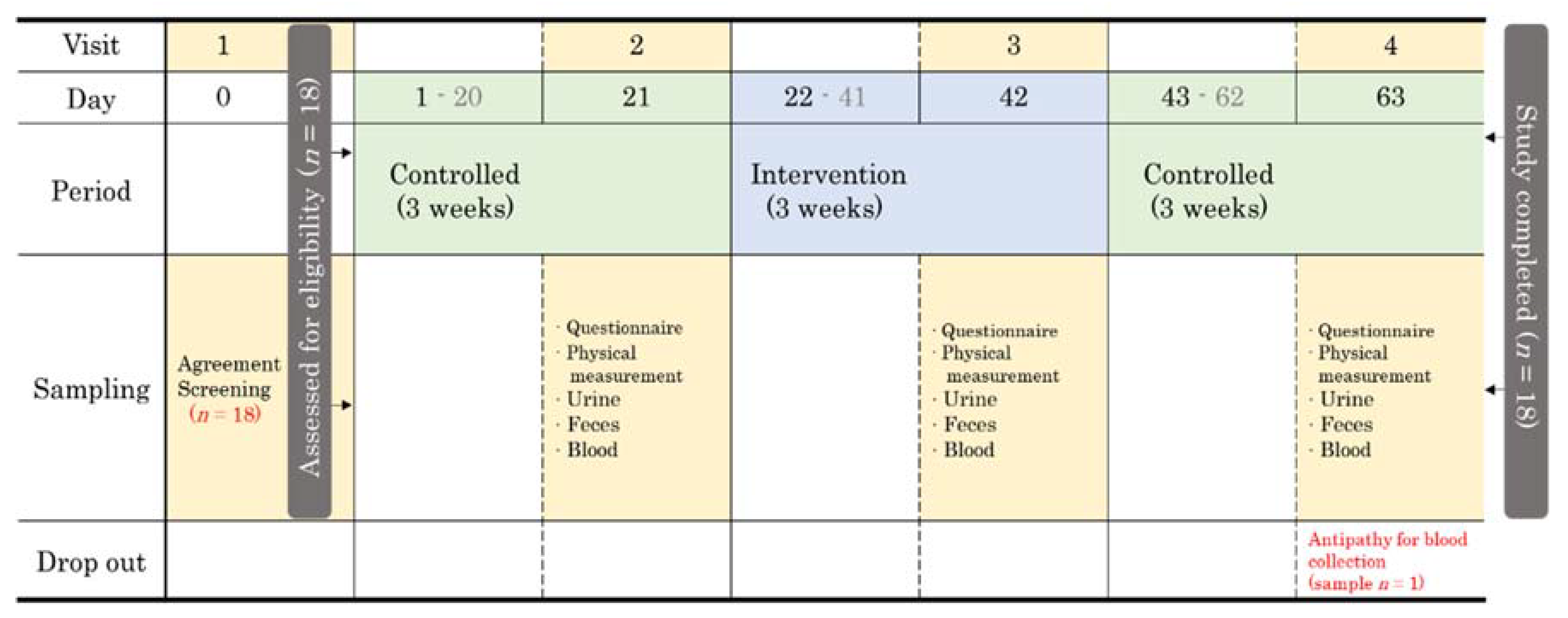
2.1. Study Schedule and Design
2.2. Participant Recruitment and Screening
2.3. YP Supplements and Intake Method
2.4. Sample Collection
2.5. Gut Microbiota Analysis
2.5.1. Fecal DNA Extraction
2.5.2. Polymerase Chain Reaction (PCR) Amplification of the 16S rRNA Genes
2.5.3. 16S rRNA Gene Sequencing Data Processing and Identification of Microbial Taxa
2.6. Statistical Analyses
3. Results
3.1. Basic Demographic Characteristics of Participants
3.2. Blood Parameters and Human Body Measurements
3.3. Changes in Stool Conditions
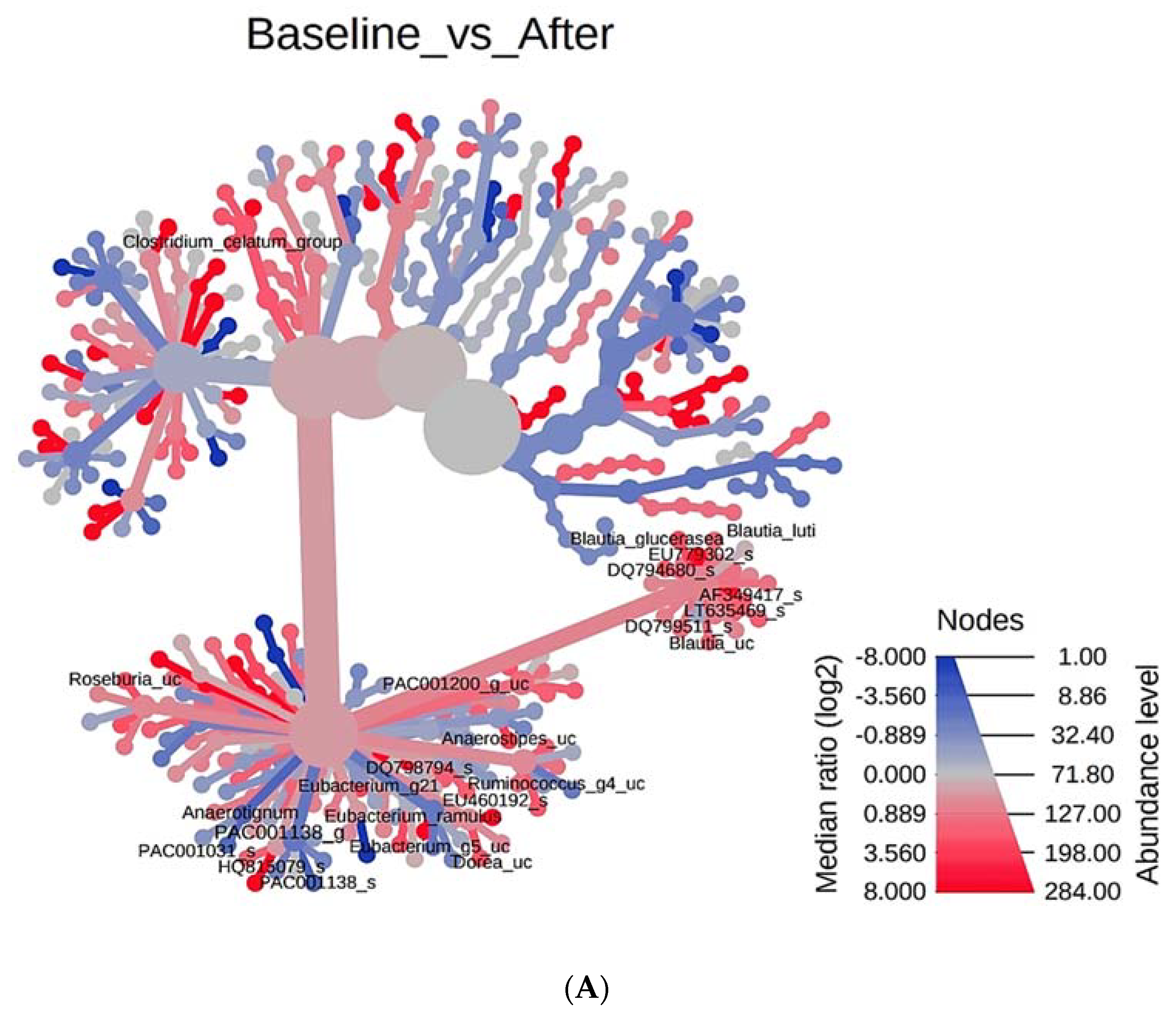
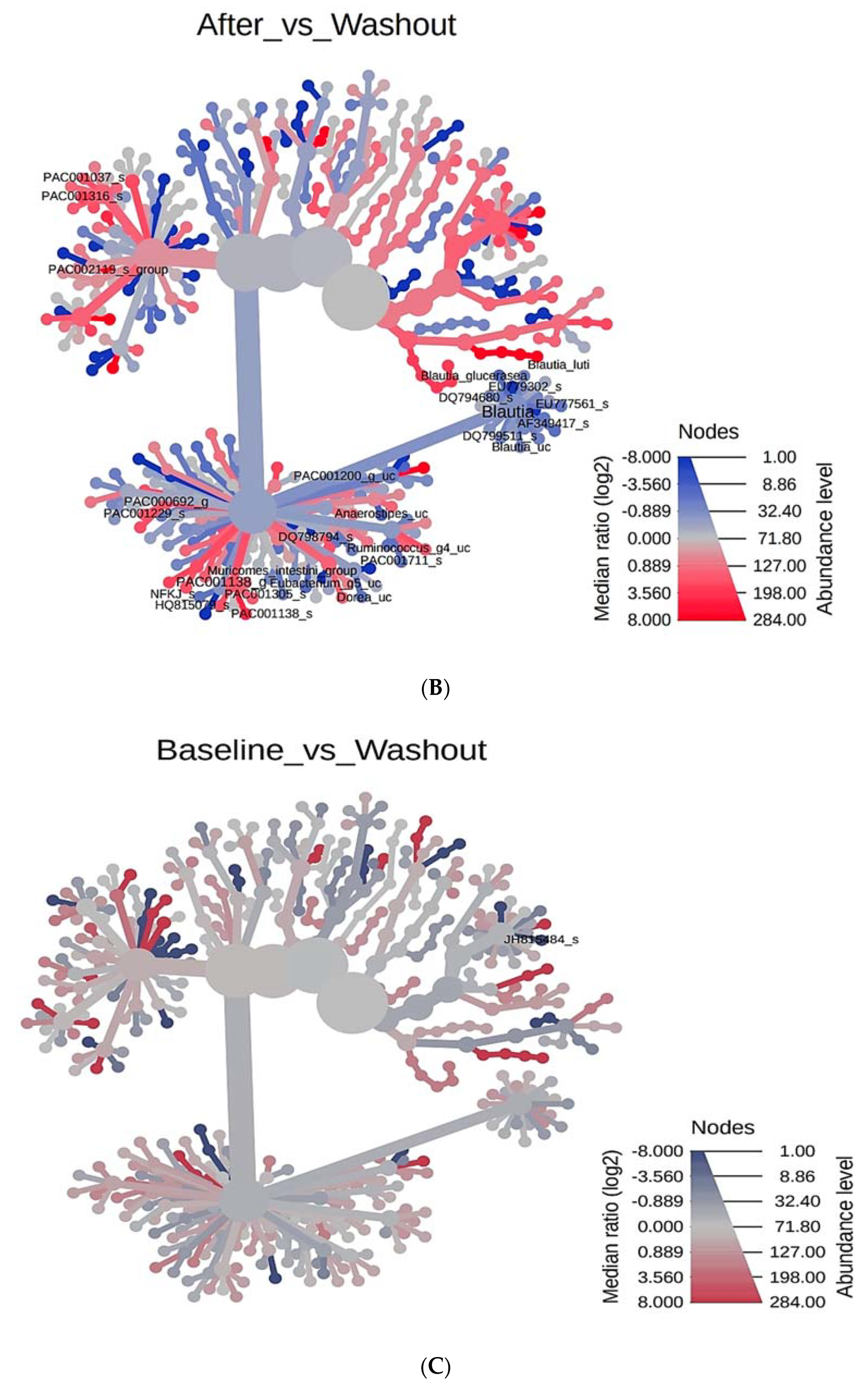
3.4. Phylogenetic Tree Analysis According to PYP Intake
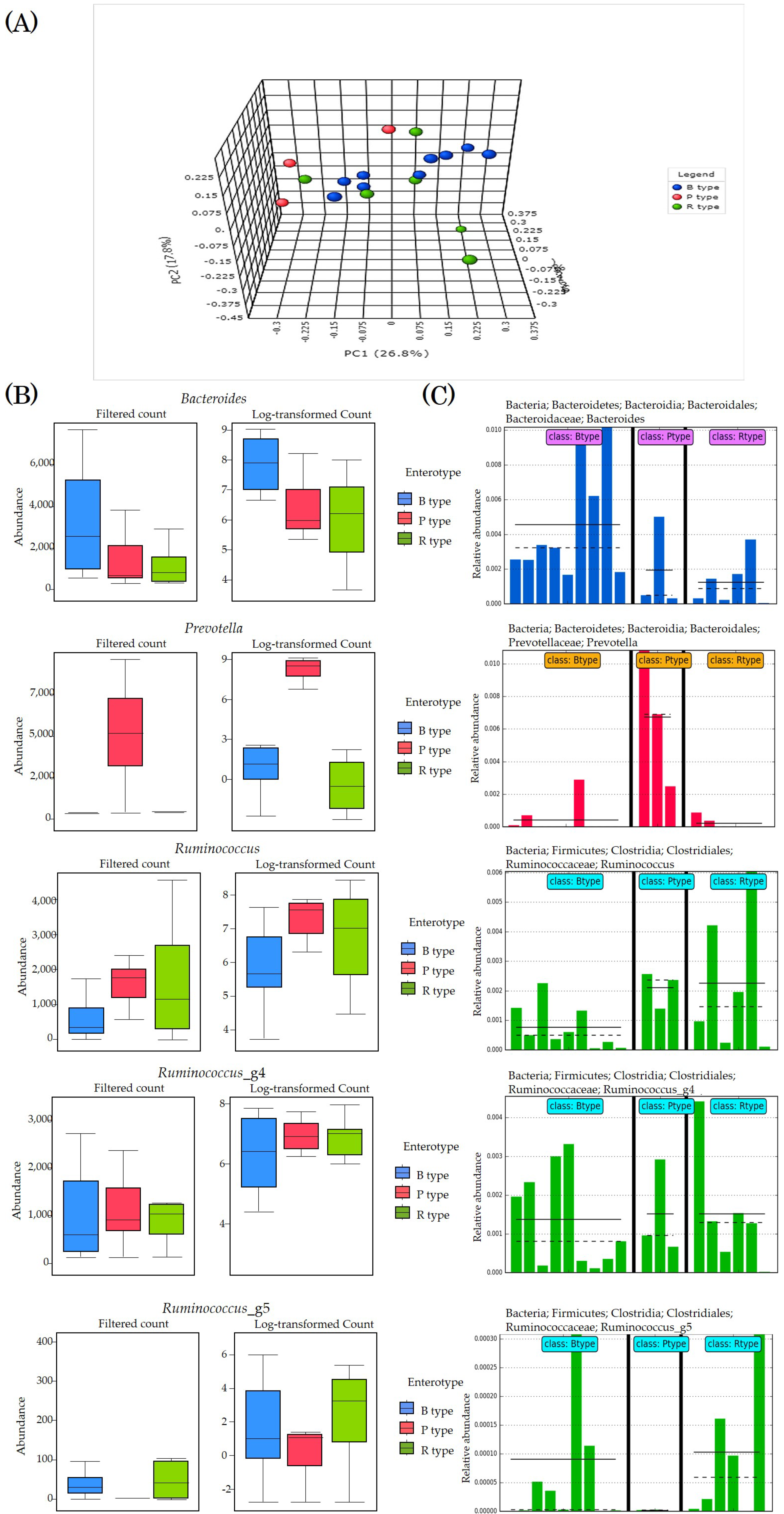
3.5. Enterotyping
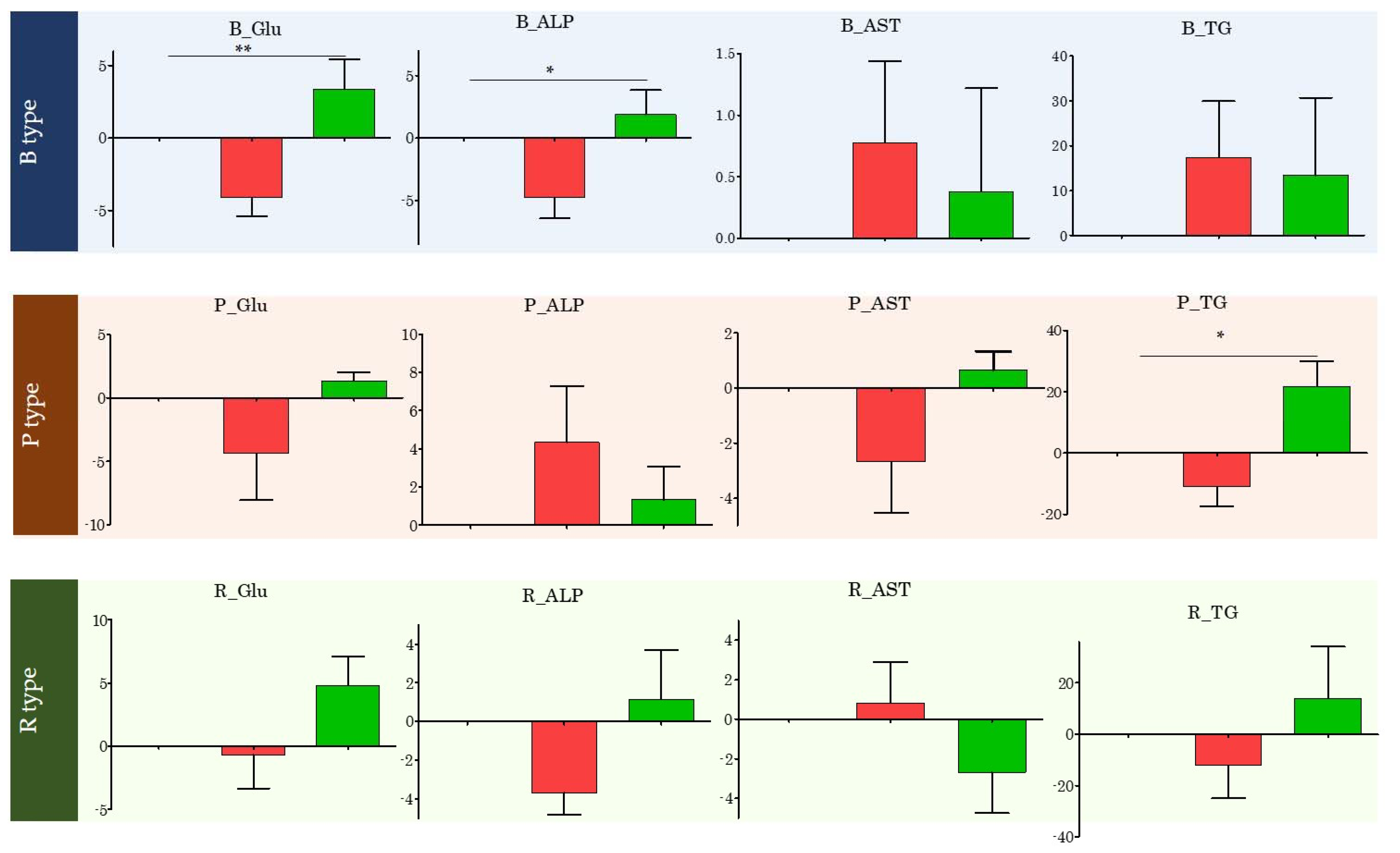
3.6. Changes in Specific Blood Index Values Based on Enterotype
3.7. Enterotype-Based Changes in Intestinal Microflora According to PYP Intake
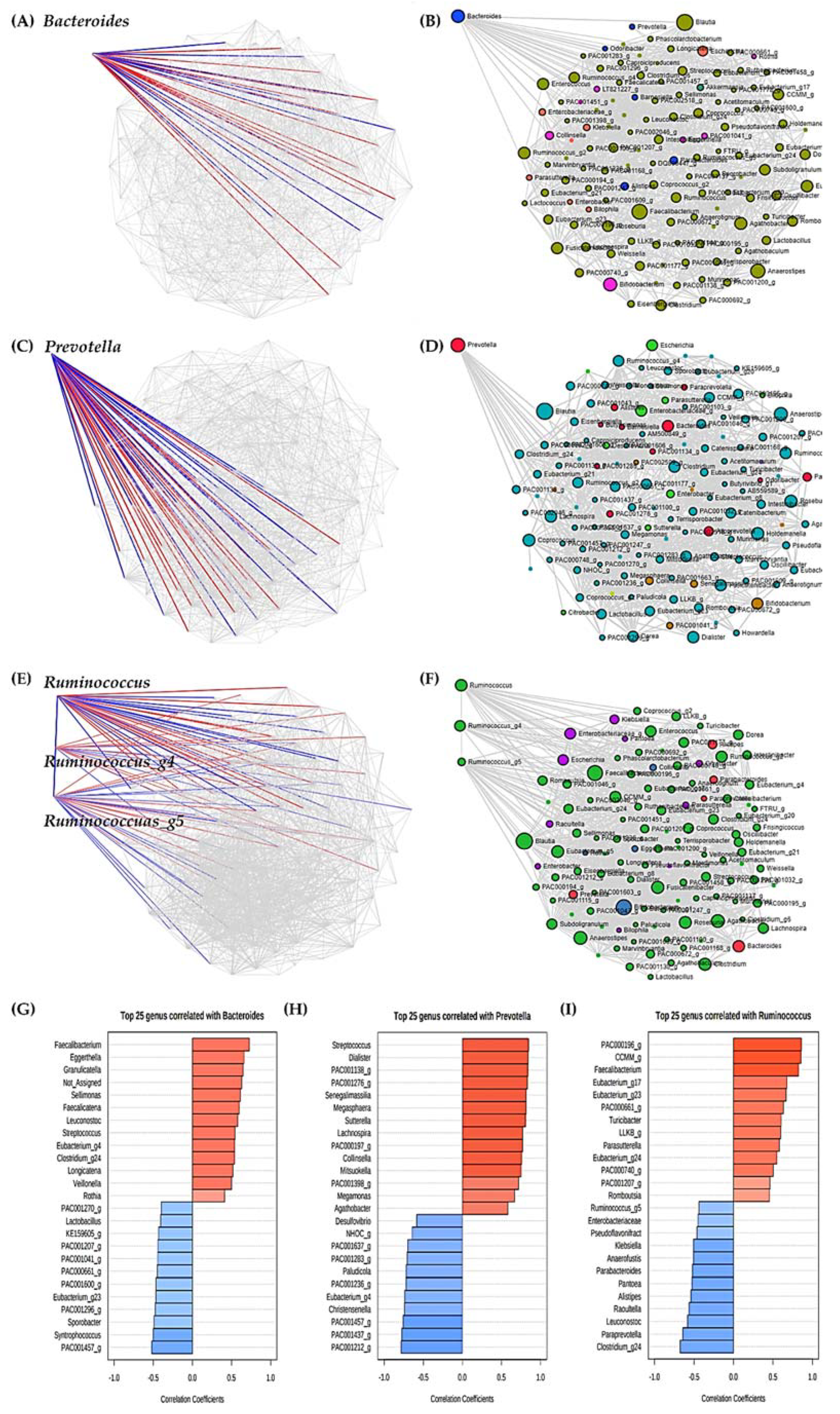
3.8. Analysis of Significant Strains Showing Correlation by Enterotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- SongHee, L.; HeeSang, Y.; Hee-Gyoo, K.; SangSun, K.; SungHee, H. Association between Altered Blood Parameters and Gut Microbiota after Synbiotic Intake in Healthy, Elderly Korean Women. Nutrients 2020, 12, 3112. [Google Scholar]
- Korem, T.; Zeevi, D.; Suez, J.; Weinberger, A.; Avnit-Sagi, T.; Pompan-Lotan, M.; Matot, E.; Jona, G.; Harmelin, A.; Cohen, N.; et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 2015, 349, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Zeevi, D.; Korem, T.; Godneva, A.; Bar, N.; Kurilshikov, A.; Lotan-Pompan, M.; Weinberger, A.; Fu, J.; Wijmenga, C.; Zhernakova, A.; et al. Structural variation in the gut microbiome associates with host health. Nature 2019, 568, 43–48. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Unno, T.; Kim, B.Y.; Park, M.S. Sex Differences in Gut Microbiota. World J. Men’s Health 2020, 38, 48–60. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Forslund, S.K.; Gudmundsdottir, V.; Petersen, A.O.; Hildebrand, F.; Hyotylainen, T.; Nielsen, T.; Hansen, T.; Bork, P.; Ehrlich, S.D.; et al. A computational framework to integrate high-throughput ’-omics’ datasets for the identification of potential mechanistic links. Nat. Protoc. 2018, 13, 2781–2800. [Google Scholar] [CrossRef]
- Kim, J.; Lim, H.; Jo, H.H. Do Aging and Low Fertility Reduce Carbon Emissions in Korea? Evidence from IPAT Augmented EKC Analysis. Int. J. Environ. Res. Public Health 2020, 17, 2972. [Google Scholar] [CrossRef]
- Kontis, V.; Bennett, J.E.; Mathers, C.D.; Li, G.; Foreman, K.; Ezzati, M. Future life expectancy in 35 industrialised countries: Projections with a Bayesian model ensemble. Lancet 2017, 389, 1323–1335. [Google Scholar] [CrossRef]
- Salazar, N.; Valdes-Varela, L.; Gonzalez, S.; Gueimonde, M.; de Los Reyes-Gavilan, C.G. Nutrition and the gut microbiome in the elderly. Gut Microbes 2017, 8, 82–97. [Google Scholar] [CrossRef]
- Zhang, X.; Zhong, H.; Li, Y.; Shi, Z.; Ren, H.; Zhang, Z.; Zhou, X.; Tang, S.; Han, X.; Lin, Y.; et al. Sex-and age-related trajectories of the adult human gut microbiota shared across populations of different ethnicities. Nat. Aging 2021, 1, 87–100. [Google Scholar] [CrossRef]
- Brotman, R.M.; Shardell, M.D.; Gajer, P.; Fadrosh, D.; Chang, K.; Silver, M.; Viscidi, R.P.; Burke, A.E.; Ravel, J.; Gravitt, P.E. Association between the vaginal microbiota, menopause status and signs of vulvovaginal atrophy. Menopause 2014, 21, 450. [Google Scholar] [CrossRef] [PubMed]
- Telle-Hansen, V.H.; Holven, K.B.; Ulven, S.M. Impact of a Healthy Dietary Pattern on Gut Microbiota and Systemic Inflammation in Humans. Nutrients 2018, 10, 1783. [Google Scholar] [CrossRef]
- Patel, R.; Shahane, A. The epidemiology of Sjogren’s syndrome. Clin. Epidemiol. 2014, 6, 247–255. [Google Scholar] [PubMed]
- Vieira, A.T.; Castelo, P.M.; Ribeiro, D.A.; Ferreira, C.M. Influence of Oral and Gut Microbiota in the Health of Menopausal Women. Front. Microbiol. 2017, 8, 1884. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, N.; Duncan, S.H.; Young, P.; Belenguer, A.; McWilliam Leitch, C.; Scott, K.P.; Flint, H.J.; Louis, P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014, 8, 1323–1335. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Cardona, F.; Tinahones, F.J.; Queipo-Ortuno, M.I. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front. Microbiol. 2014, 5, 190. [Google Scholar] [CrossRef]
- Ganesan, K.; Chung, S.K.; Vanamala, J.; Xu, B. Causal Relationship between Diet-Induced Gut Microbiota Changes and Diabetes: A Novel Strategy to Transplant Faecalibacterium prausnitzii in Preventing Diabetes. Int. J. Mol. Sci. 2018, 19, 3720. [Google Scholar] [CrossRef]
- Christ, A.; Lauterbach, M.; Latz, E. Western Diet and the Immune System: An Inflammatory Connection. Immunity 2019, 51, 794–811. [Google Scholar] [CrossRef]
- Statovci, D.; Aguilera, M.; MacSharry, J.; Melgar, S. The impact of western diet and nutrients on the microbiota and immune response at mucosal interfaces. Front. Immunol. 2017, 8, 838. [Google Scholar] [CrossRef]
- Ortega, M.A.; Fraile-Martinez, O.; Naya, I.; Garcia-Honduvilla, N.; Alvarez-Mon, M.; Bujan, J.; Asunsolo, A.; de la Torre, B. Type 2 Diabetes Mellitus Associated with Obesity (Diabesity). The Central Role of Gut Microbiota and Its Translational Applications. Nutrients 2020, 12, 2749. [Google Scholar] [CrossRef]
- Garcia-Montero, C.; Fraile-Martinez, O.; Gomez-Lahoz, A.M.; Pekarek, L.; Castellanos, A.J.; Noguerales-Fraguas, F.; Coca, S.; Guijarro, L.G.; Garcia-Honduvilla, N.; Asunsolo, A.; et al. Nutritional Components in Western Diet Versus Mediterranean Diet at the Gut Microbiota-Immune System Interplay. Implications for Health and Disease. Nutrients 2021, 13, 699. [Google Scholar] [CrossRef]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, M.S.; Lee, M.S.; Park, Y.S.; Lee, H.J.; Kang, S.-A.; Lee, H.S.; Lee, K.-E.; Yang, H.J.; Kim, M.J.; et al. Korean diet: Characteristics and historical background. J. Ethn. Foods 2016, 3, 26–31. [Google Scholar] [CrossRef]
- Kwon, Y.J.; Lee, H.; Yoon, Y.; Kim, H.M.; Chu, S.H.; Lee, J.W. Development and Validation of a Questionnaire to Measure Adherence to the Mediterranean Diet in Korean Adults. Nutrients 2020, 12, 1102. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Jung, S.; Kim, S.A.; Kang, M.S.; Kim, M.S.; Joung, H.; Hwang, G.S.; Shin, D.M. Differential Effects of Typical Korean Versus American-Style Diets on Gut Microbial Composition and Metabolic Profile in Healthy Overweight Koreans: A Randomized Crossover Trial. Nutrients 2019, 11, 2450. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Kim, J.M.; Park, M.S.; Bae, J.W.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 2011, 77, 2264–2274. [Google Scholar] [CrossRef] [PubMed]
- Merra, G.; Noce, A.; Marrone, G.; Cintoni, M.; Tarsitano, M.G.; Capacci, A.; De Lorenzo, A. Influence of Mediterranean Diet on Human Gut Microbiota. Nutrients 2020, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Del Chierico, F.; Vernocchi, P.; Dallapiccola, B.; Putignani, L. Mediterranean diet and health: Food effects on gut microbiota and disease control. Int. J. Mol. Sci. 2014, 15, 11678–11699. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, Y.; Cho, Y.; Jun, B.; Oh, K.W. Trends in the prevalence of major cardiovascular disease risk factors among Korean adults: Results from the Korea National Health and Nutrition Examination Survey, 1998–2012. Int. J. Cardiol. 2014, 174, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.-S.; Ju, S.-Y. Trends in nutrient intakes and consumption while eating-out among Korean adults based on Korea National Health and Nutrition Examination Survey (1998–2012) data. Nutr. Res. Pract. 2014, 8, 670. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Shin, S.; Ha, K.; Hwang, Y.; Park, Y.H.; Kang, M.S.; Joung, H. Effect of a balanced Korean diet on metabolic risk factors among overweight/obese Korean adults: A randomized controlled trial. Eur. J. Nutr. 2020, 59, 3023–3035. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.; Bryan, J.; Hodgson, J.; Murphy, K. Definition of the Mediterranean Diet; a Literature Review. Nutrients 2015, 7, 9139–9153. [Google Scholar] [CrossRef]
- Noh, H.; Jang, H.H.; Kim, G.; Zouiouich, S.; Cho, S.Y.; Kim, H.J.; Kim, J.; Choe, J.S.; Gunter, M.J.; Ferrari, P.; et al. Taxonomic Composition and Diversity of the Gut Microbiota in Relation to Habitual Dietary Intake in Korean Adults. Nutrients 2021, 13, 366. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.Y.; Rho, M.; Song, Y.M.; Lee, K.; Sung, J.; Ko, G. Stability of gut enterotypes in Korean monozygotic twins and their association with biomarkers and diet. Sci. Rep. 2014, 4, 7348. [Google Scholar] [CrossRef]
- Panek, M.; Cipcic Paljetak, H.; Baresic, A.; Peric, M.; Matijasic, M.; Lojkic, I.; Vranesic Bender, D.; Krznaric, Z.; Verbanac, D. Methodology challenges in studying human gut microbiota—Effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci. Rep. 2018, 8, 5143. [Google Scholar] [CrossRef]
- Claesson, M.J.; O’Toole, P.W. Evaluating the latest high-throughput molecular techniques for the exploration of microbial gut communities. Gut Microbes 2010, 1, 277–278. [Google Scholar] [CrossRef]
- Salipante, S.J.; Kawashima, T.; Rosenthal, C.; Hoogestraat, D.R.; Cummings, L.A.; Sengupta, D.J.; Harkins, T.T.; Cookson, B.T.; Hoffman, N.G. Performance comparison of Illumina and ion torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl. Environ. Microbiol. 2014, 80, 7583–7591. [Google Scholar] [CrossRef]
- Sinclair, L.; Osman, O.A.; Bertilsson, S.; Eiler, A. Microbial community composition and diversity via 16S rRNA gene amplicons: Evaluating the illumina platform. PLoS ONE 2015, 10, e0116955. [Google Scholar] [CrossRef]
- D’Amore, R.; Ijaz, U.Z.; Schirmer, M.; Kenny, J.G.; Gregory, R.; Darby, A.C.; Shakya, M.; Podar, M.; Quince, C.; Hall, N. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genom. 2016, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Hang, C.J.; Lin, T.L.; Tsai, Y.L.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar]
- Roberfroid, M.; Gibson, G.R.; Hoyles, L.; McCartney, A.L.; Rastall, R.; Rowland, I.; Wolvers, D.; Watzl, B.; Szajewska, H.; Stahl, B.; et al. Prebiotic effects: Metabolic and health benefits. Br. J. Nutr. 2010, 104 (Suppl. 2), S1–S63. [Google Scholar] [CrossRef] [PubMed]
- Bertazzoni, E.; Donelli, G.; Midtvedt, T.; Nicoli, J.; Sanz, Y. Probiotics and clinical effects: Is the number what counts? J. Chemother. 2013, 25, 193–212. [Google Scholar] [CrossRef] [PubMed]
- Taleb, S.; Boulaba, K.; Yousfi, A.; Taleb, N.; Difallah, B.; Negrichi, S. Associations between body mass index, waist circumference, waist circumference to-height ratio, and hypertension in an Algerian adult population. Environ. Sci. Pollut. Res. Int. 2020, 1–9. [Google Scholar] [CrossRef]
- Wang, K.; Yu, X.; Li, Y.; Guo, Y.; Ge, L.; Pu, F.; Ma, X.; Cui, W.; Marrota, F.; He, F.; et al. Bifidobacterium bifidum TMC3115 Can Characteristically Influence Glucose and Lipid Profile and Intestinal Microbiota in the Middle-Aged and Elderly. Probiotics Antimicrob. Proteins 2019, 11, 1182–1194. [Google Scholar] [CrossRef] [PubMed]
- Vuljanic, D.; Dojder, A.; Spoljaric, V.; Saracevic, A.; Dukic, L.; Lenicek-Krleza, J.; Vlasic-Tanaskovic, J.; Maradin, I.; Grzunov, A.; Vogrinc, Z.; et al. Analytical verification of 12 most commonly used urine dipsticks in Croatia: Comparability, repeatability and accuracy. Biochem. Med. 2019, 29, 010708. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.Y.; Song, E.J.; Kim, S.H.; Lee, J.; Nam, Y.D. Comparison of DNA extraction methods for human gut microbial community profiling. Syst. Appl. Microbiol. 2018, 41, 151–157. [Google Scholar] [CrossRef]
- Dassi, E.; Ballarini, A.; Covello, G.; Htm, C.M.B.; Quattrone, A.; Jousson, O.; De Sanctis, V.; Bertorelli, R.; Denti, M.A.; Segata, N. Enhanced microbial diversity in the saliva microbiome induced by short-term probiotic intake revealed by 16S rRNA sequencing on the IonTorrent PGM platform. J. Biotechnol. 2014, 190, 30–39. [Google Scholar] [CrossRef]
- Ogata, K.; Takeshita, T.; Shibata, Y.; Matsumi, R.; Kageyama, S.; Asakawa, M.; Yamashita, Y. Effect of coffee on the compositional shift of oral indigenous microbiota cultured in vitro. J. Oral Sci. 2019, 61, 418–424. [Google Scholar] [CrossRef]
- Adamiak, J.; Otlewska, A.; Tafer, H.; Lopandic, K.; Gutarowska, B.; Sterflinger, K.; Piñar, G. First evaluation of the microbiome of built cultural heritage by using the Ion Torrent next generation sequencing platform. Int. Biodeterior. Biodegrad. 2018, 131, 11–18. [Google Scholar] [CrossRef]
- Malapelle, U.; Vigliar, E.; Sgariglia, R.; Bellevicine, C.; Colarossi, L.; Vitale, D.; Pallante, P.; Troncone, G. Ion Torrent next-generation sequencing for routine identification of clinically relevant mutations in colorectal cancer patients. J. Clin. Pathol. 2015, 68, 64–68. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Dave, V.; Street, K.; Francis, S.; Bradman, A.; Riley, L.; Eskenazi, B.; Holland, N. Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatr. Res. 2016, 79, 846–854. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Robles-Vera, I.; Callejo, M.; Ramos, R.; Duarte, J.; Perez-Vizcaino, F. Impact of Vitamin D Deficit on the Rat Gut Microbiome. Nutrients 2019, 11, 2564. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F. Probiotics in inflammatory bowel disease—Therapeutic rationale and role. Adv. Drug Deliv. Rev. 2004, 56, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Leber, B.; Tripolt, N.J.; Blattl, D.; Eder, M.; Wascher, T.C.; Pieber, T.R.; Stauber, R.; Sourij, H.; Oettl, K.; Stadlbauer, V. The influence of probiotic supplementation on gut permeability in patients with metabolic syndrome: An open label, randomized pilot study. Eur. J. Clin. Nutr. 2012, 66, 1110–1115. [Google Scholar] [CrossRef] [PubMed]
- Summers, C.; Rankin, S.M.; Condliffe, A.M.; Singh, N.; Peters, A.M.; Chilvers, E.R. Neutrophil kinetics in health and disease. Trends Immunol. 2010, 31, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Prete, A.; Cariello, R.; Tuccillo, C.; Cotticelli, G.; Del Vecchio Blanco, C.; Loguercio, C. Gut microbiota and probiotics in chronic liver diseases. Dig. Liver Dis. 2011, 43, 431–438. [Google Scholar]
- Gratz, S.W.; Mykkanen, H.; El-Nezami, H.S. Probiotics and gut health: A special focus on liver diseases. World J. Gastroenterol. 2010, 16, 403–410. [Google Scholar] [CrossRef]
- Mazani, M.; Nemati, A.; Amani, M.; Haedari, K.; Mogadam, R.A.; Baghi, A.N. The effect of probiotic yoghurt consumption on oxidative stress and inflammatory factors in young females after exhaustive exercise. J. Pak. Med. Assoc. 2018, 68, 1748–1754. [Google Scholar]
- Pane, M.; Amoruso, A.; Deidda, F.; Graziano, T.; Allesina, S.; Mogna, L. Gut Microbiota, Probiotics, and Sport: From Clinical Evidence to Agonistic Performance. J. Clin. Gastroenterol. 2018, 52 (Suppl. 1), S46–S49. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Koropatkin, N.M.; Cameron, E.A.; Martens, E.C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 2012, 10, 323–335. [Google Scholar] [CrossRef]
- Brodmann, T.; Endo, A.; Gueimonde, M.; Vinderola, G.; Kneifel, W.; de Vos, W.M.; Salminen, S.; Gomez-Gallego, C. Safety of Novel Microbes for Human Consumption: Practical Examples of Assessment in the European Union. Front. Microbiol. 2017, 8, 1725. [Google Scholar] [CrossRef]
- Wu, S.; Li, T.; Niu, H.; Zhu, Y.; Liu, Y.; Duan, Y.; Sun, Q.; Yang, X. Effects of glucose oxidase on growth performance, gut function, and cecal microbiota of broiler chickens. Poult. Sci. 2019, 98, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.S.; Streidl, T.; Hitch, T.C.A.; Wortmann, E.; Deptula, P.; Kofoed, M.V.W.; Riedel, T.; Neumann-Schaal, M.; Hansen, M.; Nielsen, D.S.; et al. Sporofaciens musculi gen. nov. sp. nov. a novel bacterium isolated from the caecum of an obese mouse. Int. J. Syst. Evol. Microbiol. 2019, 71, 004673. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, A.; Benno, Y.; Nakase, T. Phylogenetic evidence for the transfer of Eubacterium lentum to the genus Eggerthella as Eggerthella lenta gen. nov. comb. nov. Int. J. Syst. Bacteriol. 1999, 49 Pt 4, 1725–1732. [Google Scholar] [CrossRef]
- Traisaeng, S.; Batsukh, A.; Chuang, T.H.; Herr, D.R.; Huang, Y.F.; Chimeddorj, B.; Huang, C.M. Leuconostoc mesenteroides fermentation produces butyric acid and mediates Ffar2 to regulate blood glucose and insulin in type 1 diabetic mice. Sci. Rep. 2020, 10, 7928. [Google Scholar] [CrossRef]
- Ferreira-Halder, C.V.; Faria, A.V.S.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef]
- Little, R.; Wine, E.; Kamath, B.M.; Griffiths, A.M.; Ricciuto, A. Gut microbiome in primary sclerosing cholangitis: A review. World J. Gastroenterol. 2020, 26, 2768–2780. [Google Scholar] [CrossRef]
- Abe, K.; Takahashi, A.; Fujita, M.; Imaizumi, H.; Hayashi, M.; Okai, K.; Ohira, H. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS ONE 2018, 13, e0198757. [Google Scholar] [CrossRef]
- Allegretti, J.R.; Kassam, Z. Fecal Microbiota Transplantation in Patients with Primary Sclerosing Cholangitis: The Next Steps in This Promising Story. Am. J. Gastroenterol. 2019, 114, 1354–1355. [Google Scholar] [CrossRef]
- Fehlner-Peach, H.; Magnabosco, C.; Raghavan, V.; Scher, J.U.; Tett, A.; Cox, L.M.; Gottsegen, C.; Watters, A.; Wiltshire-Gordon, J.D.; Segata, N.; et al. Distinct Polysaccharide Utilization Profiles of Human Intestinal Prevotella copri Isolates. Cell Host Microbe 2019, 26, 680–690.e5. [Google Scholar] [CrossRef]
- De Filippo, C.; Di Paola, M.; Ramazzotti, M.; Albanese, D.; Pieraccini, G.; Banci, E.; Miglietta, F.; Cavalieri, D.; Lionetti, P. Diet, Environments, and Gut Microbiota. A Preliminary Investigation in Children Living in Rural and Urban Burkina Faso and Italy. Front. Microbiol. 2017, 8, 1979. [Google Scholar] [CrossRef] [PubMed]
- Mobeen, F.; Sharma, V.; Tulika, P. Enterotype Variations of the Healthy Human Gut Microbiome in Different Geographical Regions. Bioinformation 2018, 14, 560–573. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Enterotype bacteroides is associated with a high risk in patients with diabetes: A pilot study. J. Diabetes Res. 2020, 2020, 1–11. [Google Scholar] [CrossRef]
- Guo, W.L.; Chen, M.; Pan, W.L.; Zhang, Q.; Xu, J.X.; Lin, Y.; Li, L.; Liu, B.; Bai, W.; Zhang, Y.; et al. Hypoglycemic and hypolipidemic mechanism of organic chromium derived from chelation of Grifola frondosa polysaccharide-chromium (III) and its modulation of intestinal microflora in high fat-diet and STZ-induced diabetic mice. Int. J. Biol. Macromol. 2020, 145, 1208–1218. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Hernandez, J.; Moreno, Y.; Chuan, C.; Hernandez, M. In vivo study of the survival of Lactobacillus delbruecki subsp. bulgaricus CECT 4005T and Streptococcus thermophilus CECT 801 by DVC-FISH after consumption of fermented milk. J. Food Sci. 2012, 77, M593–M597. [Google Scholar] [CrossRef] [PubMed]
- Junjua, M.; Kechaou, N.; Chain, F.; Awussi, A.A.; Roussel, Y.; Perrin, C.; Roux, E.; Langella, P.; Bermúdez-Humarán, L.G.; le Roux, Y.; et al. A large scale in vitro screening of Streptococcus thermophilus strains revealed strains with a high anti-inflammatory potential. LWT 2016, 70, 78–87. [Google Scholar] [CrossRef]
- del Campo, R.; Bravo, D.; Canton, R.; Ruiz-Garbajosa, P.; Garcia-Albiach, R.; Montesi-Libois, A.; Yuste, F.J.; Abraira, V.; Baquero, F. Scarce evidence of yogurt lactic acid bacteria in human feces after daily yogurt consumption by healthy volunteers. Appl. Environ. Microbiol. 2005, 71, 547–549. [Google Scholar] [CrossRef] [PubMed]
- Elli, M.; Callegari, M.L.; Ferrari, S.; Bessi, E.; Cattivelli, D.; Soldi, S.; Morelli, L.; Goupil Feuillerat, N.; Antoine, J.M. Survival of yogurt bacteria in the human gut. Appl. Environ. Microbiol. 2006, 72, 5113–5117. [Google Scholar] [CrossRef] [PubMed]
- Kilic, A.O.; Pavlova, S.I.; Ma, W.G.; Tao, L. Analysis of Lactobacillus phages and bacteriocins in American dairy products and characterization of a phage isolated from yogurt. Appl. Environ. Microbiol. 1996, 62, 2111–2116. [Google Scholar] [CrossRef]
- Hamaoka, N.; Yasokawa, D.; Okumura, Y.; Nakagawa, R.; Tanaka, T. Manufacturing process of a novel Mozzarella cheese using originally isolated lactic acid bacteria, Streptococcus salivarius ssp. thermophilus, from Hokkaido. J. Jpn. Soc. Food Sci. Technol. 2017, 64, 132–138. [Google Scholar] [CrossRef]
- Hussain, S.K.; Dong, T.S.; Agopian, V.; Pisegna, J.R.; Durazo, F.A.; Enayati, P.; Sundaram, V.; Benhammou, J.N.; Noureddin, M.; Choi, G.; et al. Dietary Protein, Fiber and Coffee Are Associated with Small Intestine Microbiome Composition and Diversity in Patients with Liver Cirrhosis. Nutrients 2020, 12, 1395. [Google Scholar] [CrossRef]
- El Khoury, D.; Cuda, C.; Luhovyy, B.L.; Anderson, G.H. Beta glucan: Health benefits in obesity and metabolic syndrome. J. Nutr. Metab. 2012, 2012, 851362. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef]
- Png, C.W.; Linden, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef]
- Maffei, V.J.; Kim, S.; Blanchard, E.t.; Luo, M.; Jazwinski, S.M.; Taylor, C.M.; Welsh, D.A. Biological Aging and the Human Gut Microbiota. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 1474–1482. [Google Scholar] [CrossRef]
- La Reau, A.J.; Meier-Kolthoff, J.P.; Suen, G. Sequence-based analysis of the genus Ruminococcus resolves its phylogeny and reveals strong host association. Microb. Genom. 2016, 2, e000099. [Google Scholar] [CrossRef]
- Qiu, X.; Zhao, X.; Cui, X.; Mao, X.; Tang, N.; Jiao, C.; Wang, D.; Zhang, Y.; Ye, Z.; Zhang, H. Characterization of fungal and bacterial dysbiosis in young adult Chinese patients with Crohn’s disease. Ther. Adv. Gastroenterol. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Wu, H.; Wu, S.D.; Lu, N.; Wang, Y.T.; Liu, H.N.; Dong, L.; Liu, T.T.; Shen, X.Z. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation. J. Gastroenterol. Hepatol. 2018, 33, 1844–1852. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Gorvitovskaia, A.; Holmes, S.P.; Huse, S.M. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 2016, 4, 15. [Google Scholar] [CrossRef]
- Zupancic, M.L.; Cantarel, B.L.; Liu, Z.; Drabek, E.F.; Ryan, K.A.; Cirimotich, S.; Jones, C.; Knight, R.; Walters, W.A.; Knights, D.; et al. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS ONE 2012, 7, e43052. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Ye, Y.; Zhou, Y.; Fodor, A.A. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE 2012, 7, e34242. [Google Scholar]
- Liang, C.; Tseng, H.C.; Chen, H.M.; Wang, W.C.; Chiu, C.M.; Chang, J.Y.; Lu, K.Y.; Weng, S.L.; Chang, T.H.; Chang, C.H.; et al. Diversity and enterotype in gut bacterial community of adults in Taiwan. BMC Genom. 2017, 18 (Suppl. 1), 932. [Google Scholar] [CrossRef]
- Cheng, M.; Kang, N. Stereotypes about enterotype: The old and new ideas. Genom. Proteom. Bioinform. 2019, 17, 4–12. [Google Scholar] [CrossRef]
- Wu, X.; Unno, T.; Kang, S.; Park, S. A Korean-Style Balanced Diet Has a Potential Connection with Ruminococcaceae Enterotype and Reduction of Metabolic Syndrome Incidence in Korean Adults. Nutrients 2021, 13, 495. [Google Scholar] [CrossRef] [PubMed]
- Wutthi-In, M.; Cheevadhanarak, S.; Yasom, S.; Kerdphoo, S.; Thiennimitr, P.; Phrommintikul, A.; Chattipakorn, N.; Kittichotirat, W.; Chattipakorn, S. Gut Microbiota Profiles of Treated Metabolic Syndrome Patients and Their Relationship with Metabolic Health. Sci. Rep. 2020, 10, 10085. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.Y.; Cha, J.M.; Oh, J.K.; Tan, P.L.; Kim, S.H.; Kwak, M.S.; Jeon, J.W.; Shin, H.P. Probiotics Ameliorate Stool Consistency in Patients with Chronic Constipation: A Randomized, Double-Blind, Placebo-Controlled Study. Dig. Dis. Sci. 2018, 63, 2754–2764. [Google Scholar] [CrossRef] [PubMed]



| Variables | (n=) | ||
|---|---|---|---|
| Age (Years) | 50–59 | 2 | |
| 60–69 | 11 | ||
| 70–79 | 4 | ||
| 80–89 | 1 | ||
| Menopausal transition | Menstruating | 0 | |
| Post-Menopausal | 18 | ||
| Smoking | No (Non-smoking) | 18 | |
| Yes (Smoking) | Current smoker | 0 | |
| Ex-smoker | 0 | ||
| Alcohol | No | 14 | |
| Yes | Daily | 4 | |
| Weekly | 0 | ||
| Occasionally | 0 | ||
| Exercise | No | 1 | |
| Yes | Daily | 17 | |
| Weekly | 0 | ||
| Occasionally | 0 | ||
| Usual diet | Western diet | 0 | |
| Mediterranean diet | 7 | ||
| Mixed Korean diet | 11 | ||
| Indices | 1st (Before) | 2nd (After) | 3rd (Washout) | 1st vs. 2nd | 2nd vs. 3rd | 1st vs. 3rd | |||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | p-Value | |||
| Glucose | 84.83 | 8.85 | 81.83 | 10.34 | 85.18 | 9.91 | 0.027 | 0.012 | 0.765 |
| BUN (mg/dL) | 17.47 | 5.46 | 18.09 | 4.15 | 16.02 | 3.73 | 0.358 | 0.010 | 0.152 |
| Creatinine (mg/dL) | 0.61 | 0.09 | 0.56 | 0.08 | 0.57 | 0.09 | 0.003 | 0.805 | 0.001 |
| Uric acid (mg/dL) | 4.74 | 0.96 | 4.74 | 0.72 | 4.50 | 0.72 | 1.000 | 0.058 | 0.084 |
| TP (g/dL) | 7.46 | 0.28 | 7.28 | 0.38 | 7.59 | 0.40 | 0.014 | 0.000 | 0.106 |
| ALB (g/dL) | 4.16 | 0.22 | 4.25 | 0.22 | 4.26 | 0.29 | 0.045 | 0.921 | 0.039 |
| ALP (U/L) | 74.67 | 17.04 | 71.78 | 16.35 | 74.26 | 16.17 | 0.034 | 0.242 | 0.556 |
| AST (U/L) | 25.00 | 7.43 | 25.22 | 8.48 | 24.71 | 6.31 | 0.791 | 0.470 | 0.641 |
| ALT (U/L) | 17.11 | 4.93 | 18.28 | 5.03 | 17.94 | 5.14 | 0.106 | 0.729 | 0.388 |
| T-BILC (mg/dL) | 0.63 | 0.21 | 0.62 | 0.11 | 0.61 | 0.14 | 0.683 | 0.833 | 0.721 |
| Cholesterol (mg/dL) | 214.89 | 38.61 | 218.17 | 44.83 | 225.59 | 42.98 | 0.620 | 0.262 | 0.145 |
| Triglyceride (mg/dL) | 118.06 | 33.68 | 120.83 | 40.30 | 135.06 | 62.40 | 0.738 | 0.168 | 0.080 |
| GGT (U/L) | 16.72 | 3.14 | 17.89 | 3.26 | 18.35 | 5.01 | 0.016 | 0.537 | 0.081 |
| HDL cholesterol (mg/dL) | 54.21 | 9.33 | 55.77 | 11.19 | 55.50 | 11.39 | 0.278 | 0.808 | 0.485 |
| LDH (U/L) | 197.00 | 37.18 | 194.39 | 38.40 | 185.53 | 43.94 | 0.384 | 0.054 | 0.057 |
| CRP (mg/dL) | 0.14 | 0.19 | 0.15 | 0.26 | 0.28 | 0.72 | 0.579 | 0.289 | 0.310 |
| LDL cholesterol (mg/dL) | 109.75 | 21.20 | 119.59 | 26.96 | 118.51 | 24.47 | 0.019 | 0.669 | 0.061 |
| A/G ratio | 1.27 | 0.16 | 1.44 | 0.25 | 1.31 | 0.18 | 0.001 | 0.013 | 0.083 |
| B/C ratio | 29.22 | 9.70 | 32.61 | 8.76 | 28.71 | 8.43 | 0.013 | 0.013 | 0.870 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; You, H.; Lee, M.; Kim, D.; Jung, S.; Park, Y.; Hyun, S. Different Reactions in Each Enterotype Depending on the Intake of Probiotic Yogurt Powder. Microorganisms 2021, 9, 1277. https://doi.org/10.3390/microorganisms9061277
Lee S, You H, Lee M, Kim D, Jung S, Park Y, Hyun S. Different Reactions in Each Enterotype Depending on the Intake of Probiotic Yogurt Powder. Microorganisms. 2021; 9(6):1277. https://doi.org/10.3390/microorganisms9061277
Chicago/Turabian StyleLee, Songhee, Heesang You, Minho Lee, Doojin Kim, Sunghee Jung, Youngsook Park, and Sunghee Hyun. 2021. "Different Reactions in Each Enterotype Depending on the Intake of Probiotic Yogurt Powder" Microorganisms 9, no. 6: 1277. https://doi.org/10.3390/microorganisms9061277
APA StyleLee, S., You, H., Lee, M., Kim, D., Jung, S., Park, Y., & Hyun, S. (2021). Different Reactions in Each Enterotype Depending on the Intake of Probiotic Yogurt Powder. Microorganisms, 9(6), 1277. https://doi.org/10.3390/microorganisms9061277








