Tick Ecdysteroid Hormone, Global Microbiota/Rickettsia Signaling in the Ovary versus Carcass during Vitellogenesis in Part-Fed (Virgin) American Dog Ticks, Dermacentor variabilis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ticks and In Vivo Injection of Ecdysteroids
2.2. Tissue Dissection and DNA Extraction
2.3. Microbiota Composition
2.4. Bioinformatics Data Processing
2.5. Quantitative PCR (qPCR) for Total Bacteria
2.6. Quantification of Rickettsia by qPCR
2.7. Sequencing and Phylogenetic Analyses of Rickettsia
2.8. Data Analysis
3. Results
3.1. Overall Distribution of Bacteria within Different Organs
3.2. Bacterial Quantification
3.3. Rickettsia Quantification
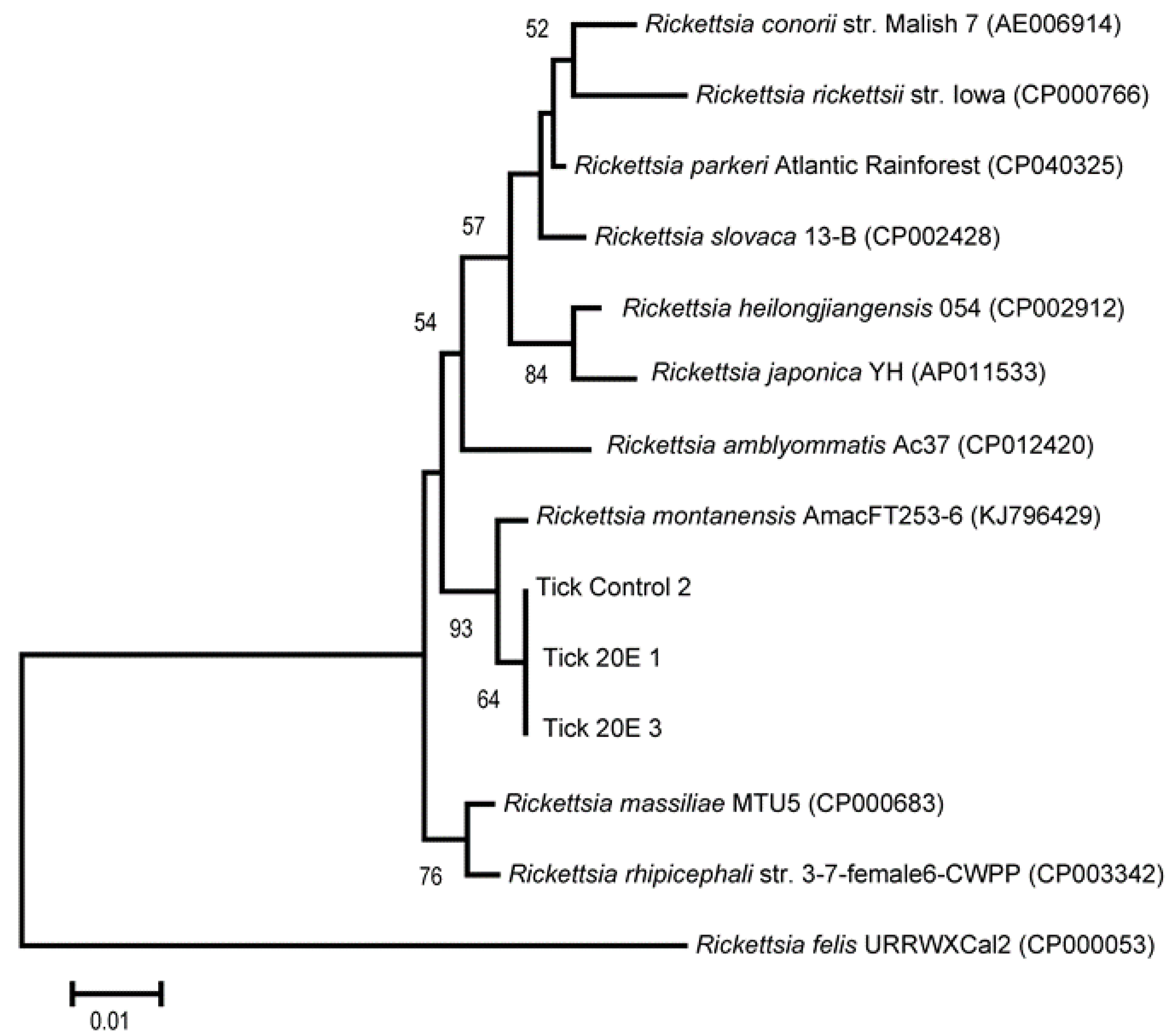
3.4. Sequencing of 23S-5S Gene and Phylogenic Analyses of Rickettsia
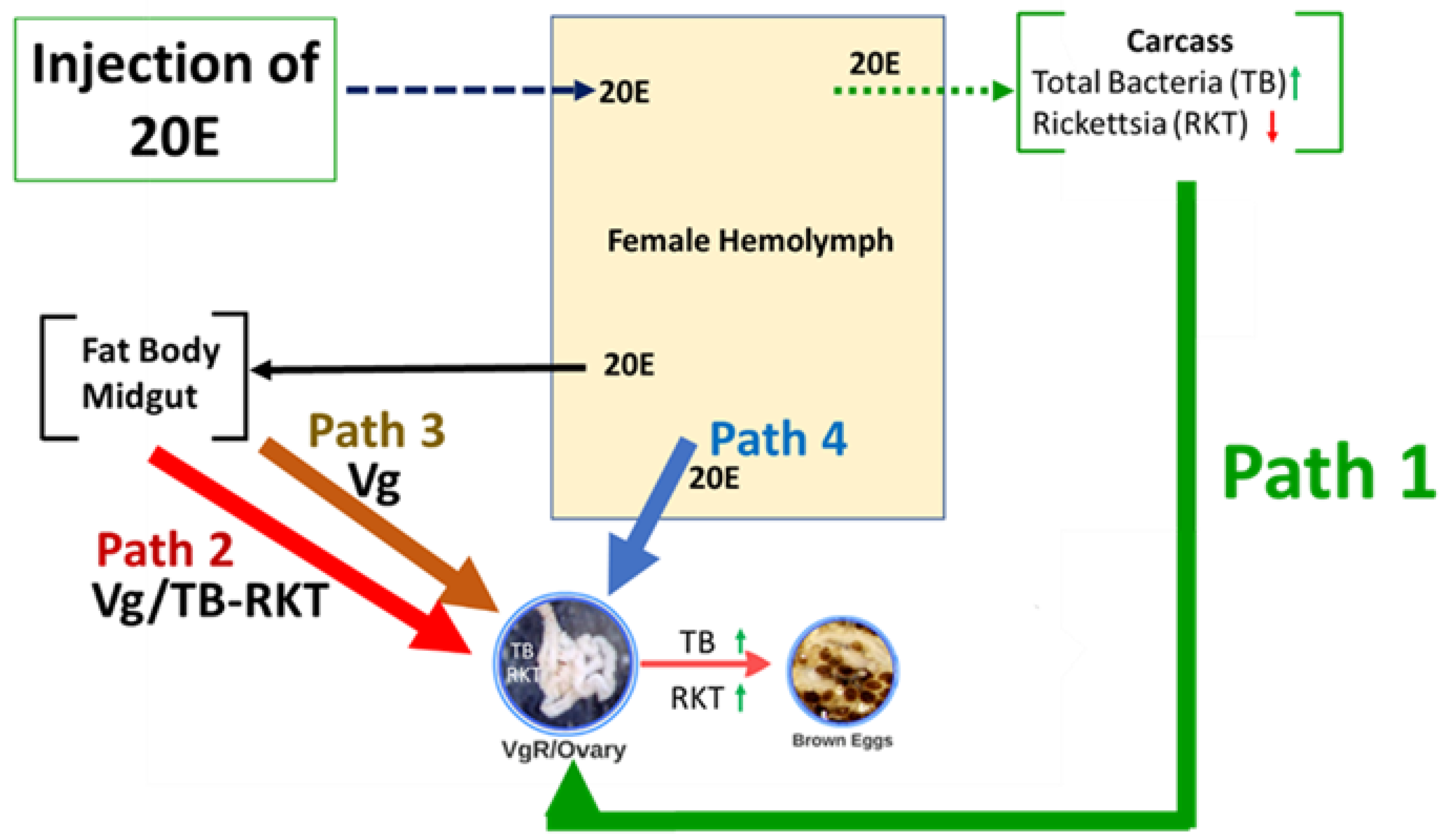
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- CDC. Tickborne Diseases of the United States: A Reference Manual for Health Care Providers, 5th ed.; CDC: New York, NY, USA, 2018. [Google Scholar]
- Werner, S.L.; Banda, B.K.; Burnsides, C.L.; Stuber, A.J. Zoonosis: Update on existing and emerging vector-borne illnesses in the USA. Curr. Emerg. Hosp. Med. Rep. 2019, 7, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Macaluso, K.; Paddock, C. Tick-borne spotted fever group ricketsioses and Rickettsia species. In Biology of Ticks; Sonenshine, D.E., Roe, R.M., Eds.; Oxford University Press: New York, NY, USA, 2014. [Google Scholar]
- Madison-Antenucci, S.; Kramer, L.D.; Gebhardt, L.L.; Kauffman, E. Emerging tick-borne diseases. Clin. Microbiol. Rev. 2020, 33, 33. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Paddock, C.D.; Raoult, D. Tick-borne Rickettsioses around the world: Emerging diseases challenging old concepts. Clin. Microbiol. Rev. 2005, 18, 719–756. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Paddock, C.D.; Socolovschi, C.; Labruna, M.B.; Mediannikov, O.; Kernif, T.; Abdad, M.Y.; Stenos, J.; Bitam, I.; Fournier, P.-E. Update on tick-borne rickettsioses around the world: A geographic approach. Clin. Microbiol. Rev. 2013, 26, 657–702. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski, M.; Rymaszewska, A. Expansion of tick-borne rickettsioses in the world. Microorganisms 2020, 8, 1906. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.; Artsob, H. Spotted fever group Rickettsiae: A brief review and a Canadian perspective. Zoonoses Public Health 2012, 59, 65–79. [Google Scholar] [CrossRef]
- Randolph, S.E. The relative contributions of transovarial and transstadial transmission to the maintenance of tick-borne diseases. In Lyme Borreliosis; Springer: Berlin/Heidelberg, Germany, 1994; pp. 131–134. [Google Scholar]
- Sonenshine, D.E.; Hynes, W.L.; Ceraul, S.M.; Mitchell, R.; Benzine, T. Host blood proteins and peptides in the midgut of the tick Dermacentor variabilis contributeto bacterial control. Exp. Appl. Acarol. 2005, 36, 207–223. [Google Scholar] [CrossRef]
- He, K.; Lin, K.; Ding, S.; Wang, G.; Li, F. The vitellogenin receptor has an essential role in vertical transmission of rice stripe virus during oogenesis in the small brown plant hopper. Pest Manag. Sci. 2019, 75, 1370–1382. [Google Scholar] [CrossRef]
- Germond, J.; Brownluedi, M.; Walker, P.; Debony, E.; Wahli, W. Sequence homologies within the 5’end region of the 4 estrogen-controlled vitellogenin genes in Xenopus and chicken. Experientia 1984, 40, 624. [Google Scholar]
- Spieth, J.; Nettleton, M.; Zucker-Aprison, E.; Lea, K.; Blumenthal, T. Vitellogenin motifs conserved in nematodes and vertebrates. J. Mol. Evol. 1991, 32, 429–438. [Google Scholar] [CrossRef]
- Wahli, W. Evolution and expression of vitellogenin genes. Trends Genet. 1988, 4, 227–232. [Google Scholar] [CrossRef]
- Hamblin, M.T.; Marx, J.L.; Wolfner, M.F.; Hagedorn, H.H. The vitellogenin gene family of Aedes aegypti. Mem. Inst. Oswaldo Cruz 1987, 82, 109–114. [Google Scholar] [CrossRef]
- Khalil, S.M.; Donohue, K.V.; Thompson, D.M.; Jeffers, L.A.; Ananthapadmanaban, U.; Sonenshine, D.E.; Mitchell, R.D.; Roe, R.M. Full-length sequence, regulation and developmental studies of a second vitellogenin gene from the American dog tick, Dermacentor variabilis. J. Insect Physiol. 2011, 57, 400–408. [Google Scholar] [CrossRef]
- Mitchell, R.D.; Ross, E.; Osgood, C.; Sonenshine, D.E.; Donohue, K.V.; Khalil, S.M.; Thompson, D.M.; Roe, R.M. Molecular characterization, tissue-specific expression and RNAi knockdown of the first vitellogenin receptor from a tick. Insect Biochem. Mol. Biol. 2007, 37, 375–388. [Google Scholar] [CrossRef]
- Roe, R.M.; Donohue, K.V.; Khalil, S.M.; Bissinger, B.W.; Zhu, J.; Sonenshine, D.E. Hormonal Regulation of Metamorphosis and Reproduction in Ticks; Sonenshine, D.E., Michael, R.R., Eds.; Oxford University Press: New York, NY, USA, 2014; p. 416. [Google Scholar]
- Xavier, M.A.; Tirloni, L.; Pinto, A.F.; Diedrich, J.K.; Yates, J.R.; Mulenga, A.; Logullo, C.; da Silva Vaz, I.; Seixas, A.; Termignoni, C. A proteomic insight into vitellogenesis during tick ovary maturation. Sci. Rep. 2018, 8, 4698. [Google Scholar] [CrossRef]
- Gulia-Nuss, M.; Nuss, A.B.; Meyer, J.M.; Sonenshine, D.E.; Roe, R.M.; Waterhouse, R.M.; Sattelle, D.B.; De La Fuente, J.; Ribeiro, J.M.; Megy, K. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 2016, 7, 10507. [Google Scholar] [CrossRef]
- Roe, R.M.; Donohue, K.V.; Khalil, S.M.; Sonenshine, D.E. Hormonal regulation of metamorphosis and reproduction in ticks. Front. Biosci. 2008, 13, 7250–7268. [Google Scholar] [CrossRef][Green Version]
- Boldbaatar, D.; Battsetseg, B.; Matsuo, T.; Hatta, T.; Umemiya-Shirafuji, R.; Xuan, X.; Fujisaki, K. Tick vitellogenin receptor reveals critical role in oocyte development and transovarial transmission of Babesia parasite. Biochem. Cell Biol. 2008, 86, 331–344. [Google Scholar] [CrossRef]
- Smith, A.D.; Kaufman, W.R. Molecular characterization of the vitellogenin receptor from the tick, Amblyomma hebraeum (Acari: Ixodidae). Insect Biochem. Mol. Biol. 2013, 43, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Seixas, A.; Alzugaray, M.F.; Tirloni, L.; Parizi, L.F.; Pinto, A.F.M.; Githaka, N.W.o.; Konnai, S.; Ohashi, K.; Yates, J.R., III; Termignoni, C. Expression profile of Rhipicephalus microplus vitellogenin receptor during oogenesis. Ticks Tick Borne Dis. 2018, 9, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Gouin, E.; Gantelet, H.; Egile, C.; Lasa, I.; Ohayon, H.; Villiers, V.; Gounon, P.; Sansonetti, P.; Cossart, P. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 1999, 112, 1697–1708. [Google Scholar] [CrossRef]
- Sonenshine, D.E.; Macaluso, K.R. Microbial invasion vs. tick immune regulation. Front. Cell. Infect. Microbiol. 2017, 7, 390. [Google Scholar] [CrossRef]
- Solano-Gallego, L.; Sainz, Á.; Roura, X.; Estrada-Peña, A.; Miró, G. A review of canine babesiosis: The European perspective. Parasites Vectors 2016, 9, 336. [Google Scholar] [CrossRef]
- Hussein, H.E.; Johnson, W.C.; Taus, N.S.; Suarez, C.E.; Scoles, G.A.; Ueti, M.W. Silencing expression of the Rhipicephalus microplus vitellogenin receptor gene blocks Babesia bovis transmission and interferes with oocyte maturation. Parasit Vectors 2019, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Khalil, S.M.; Jeffers, L.A.; Ananthapadmanaban, U.; Sonenshine, D.E.; Mitchell, R.D.; Osgood, C.J.; Apperson, C.S.; Roe, R.M. In vivo role of 20-hydroxyecdysone in the regulation of the vitellogenin mRNA and egg development in the American dog tick, Dermacentor variabilis (Say). J. Insect Physiol. 2005, 51, 1105–1116. [Google Scholar] [CrossRef]
- Rosell, R.; Coons, L.B. The role of the fat body, midgut and ovary in vitellogenin production and vitellogenesis in the female tick, Dermacentor variabilis. Int. J. Parasitol. 1992, 22, 341–349. [Google Scholar] [CrossRef]
- Mitchell, R.D., III; Sonenshine, D.E.; Pérez de León, A.A. Vitellogenin receptor as a target for tick control: A mini-review. Front. Physiol. 2019, 10, 618. [Google Scholar] [CrossRef] [PubMed]
- Sonenshine, D.E. Biology of Ticks; Oxford University Press: New York, NY, USA, 1991. [Google Scholar]
- Ponnusamy, L.; Gonzalez, A.; Van Treuren, W.; Weiss, S.; Parobek, C.M.; Juliano, J.J.; Knight, R.; Roe, R.M.; Apperson, C.S.; Meshnick, S.R. Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum. Appl. Environ. Microbiol. 2014, 80, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, V.; Gaïa, N.; Girard, M.; Schrenzel, J. Decontamination of 16S rRNA gene amplicon sequence datasets based on bacterial load assessment by qPCR. BMC Microbiol. 2016, 16, 1–8. [Google Scholar] [CrossRef]
- Browning, R.; Adamson, S.; Karim, S. Choice of a stable set of reference genes for qRT-PCR analysis in Amblyomma maculatum (Acari: Ixodidae). J. Med. Entomol. 2014, 49, 1339–1346. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Ponnusamy, L.; Sutton, H.T.; Meshnick, S.R.; Nicholson, W.L.; Apperson, C.S. Development and validation of an improved PCR method using the 23S-5S intergenic spacer for detection of rickettsiae in Dermacentor variabilis ticks and tissue samples from humans and laboratory animals. J. Clin. Microbiol. 2016, 54, 972–979. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pelzer, E.S.; Allan, J.A.; Theodoropoulos, C.; Ross, T.; Beagley, K.W.; Knox, C.L. Hormone-dependent bacterial growth, persistence and biofilm formation–a pilot study investigating human follicular fluid collected during IVF cycles. PLoS ONE 2012, 7, e49965. [Google Scholar] [CrossRef]
- Negri, I. Wolbachia as an “infectious” extrinsic factor manipulating host signaling pathways. Front. Endocrinol. 2012, 2, 115. [Google Scholar] [CrossRef]
- Bressac, C.; Rousset, F. The reproductive incompatibility system in Drosophila simulans: DAPI-staining analysis of the Wolbachia symbionts in sperm cysts. J. Invertebr. Pathol. 1993, 61, 226–230. [Google Scholar] [CrossRef]
- Hadfield, S.J.; Axton, J.M. Reproduction: Germ cells colonized by endosymbiotic bacteria. Nature 1999, 402, 482. [Google Scholar] [CrossRef]
- Clark, M.E.; Veneti, Z.; Bourtzis, K.; Karr, T.L. Wolbachia distribution and cytoplasmic incompatibility during sperm development: The cyst as the basic cellular unit of CI expression. Mech. Dev. 2003, 120, 185–198. [Google Scholar] [CrossRef]
- Herren, J.K.; Paredes, J.C.; Schüpfer, F.; Lemaitre, B. Vertical transmission of a Drosophila endosymbiont via cooption of the yolk transport and internalization machinery. MBio 2013, 4, e00532-12. [Google Scholar] [CrossRef]
- Salmela, H.; Amdam, G.V.; Freitak, D. Transfer of immunity from mother to offspring is mediated via egg-yolk protein vitellogenin. PLoS Pathog. 2015, 11, e1005015. [Google Scholar] [CrossRef]
- Guo, Y.; Hoffmann, A.; Xu, X.-Q.; Mo, P.-W.; Huang, H.-J.; Gong, J.-T.; Ju, J.-F.; Hong, X.-Y. Vertical transmission of Wolbachia is associated with host vitellogenin in Laodelphax striatellus. Front. Microbiol. 2018, 9, 2016. [Google Scholar] [CrossRef]
- Huo, Y.; Liu, W.; Zhang, F.; Chen, X.; Li, L.; Liu, Q.; Zhou, Y.; Wei, T.; Fang, R.; Wang, X. Transovarial transmission of a plant virus is mediated by vitellogenin of its insect vector. PLoS Pathog. 2014, 10, e1003949. [Google Scholar] [CrossRef]
- Huo, Y.; Yu, Y.; Liu, Q.; Liu, D.; Zhang, M.; Liang, J.; Chen, X.; Zhang, L.; Fang, R. Rice stripe virus hitchhikes the vector insect vitellogenin ligand-receptor pathway for ovary entry. Philos. Trans. R. Soc. Lond. B Biol. Sci. B 2019, 374, 20180312. [Google Scholar] [CrossRef]
- Tufail, M.; Takeda, M. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 2008, 54, 1447–1458. [Google Scholar] [CrossRef]
- Tufail, M.; Takeda, M. Insect vitellogenin/lipophorin receptors: Molecular structures, role in oogenesis, and regulatory mechanisms. J. Insect Physiol. 2009, 55, 88–104. [Google Scholar] [CrossRef] [PubMed]
- Van Treuren, W.; Ponnusamy, L.; Brinkerhoff, R.J.; Gonzalez, A.; Parobek, C.M.; Juliano, J.J.; Andreadis, T.G.; Falco, R.C.; Ziegler, L.B.; Hathaway, N. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl. Environ. Microbiol. 2015, 81, 6200–6209. [Google Scholar] [CrossRef]
- Thapa, S.; Zhang, Y.; Allen, M.S. Bacterial microbiomes of Ixodes scapularis ticks collected from Massachusetts and Texas, USA. BMC Microbiol. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Abraham, N.M.; Liu, L.; Jutras, B.L.; Yadav, A.K.; Narasimhan, S.; Gopalakrishnan, V.; Ansari, J.M.; Jefferson, K.K.; Cava, F.; Jacobs-Wagner, C. Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc. Natl. Acad. Sci. USA 2017, 114, E781–E790. [Google Scholar] [CrossRef] [PubMed]
- Travanty, N.V.; Ponnusamy, L.; Kakumanu, M.L.; Nicholson, W.L.; Apperson, C.S. Diversity and structure of the bacterial microbiome of the American dog tick, Dermacentor variabilis, is dominated by the endosymbiont Francisella. Symbiosis 2019, 79, 239–250. [Google Scholar] [CrossRef]
- Duron, O.; Morel, O.; Noël, V.; Buysse, M.; Binetruy, F.; Lancelot, R.; Loire, E.; Ménard, C.; Bouchez, O.; Vavre, F. Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr. Biol. 2018, 28, 1896–1902.e5. [Google Scholar] [CrossRef] [PubMed]
- Mira, A.; Moran, N.A. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb. Ecol. 2002, 44, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Gerhart, J.G.; Moses, A.S.; Raghavan, R. A Francisella-like endosymbiont in the Gulf Coast tick evolved from a mammalian pathogen. Sci. Rep. 2016, 6, 1–6. [Google Scholar] [CrossRef]
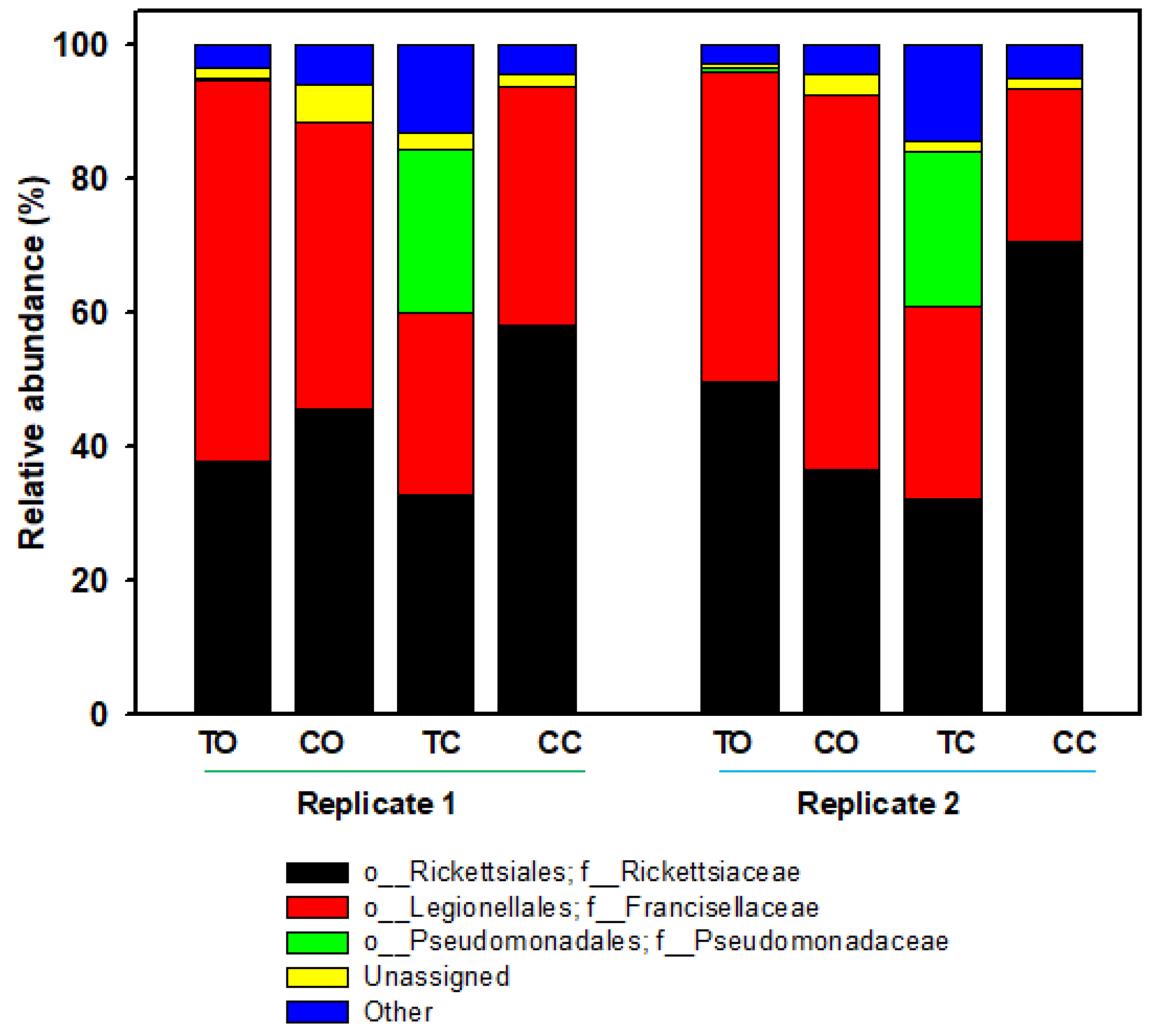
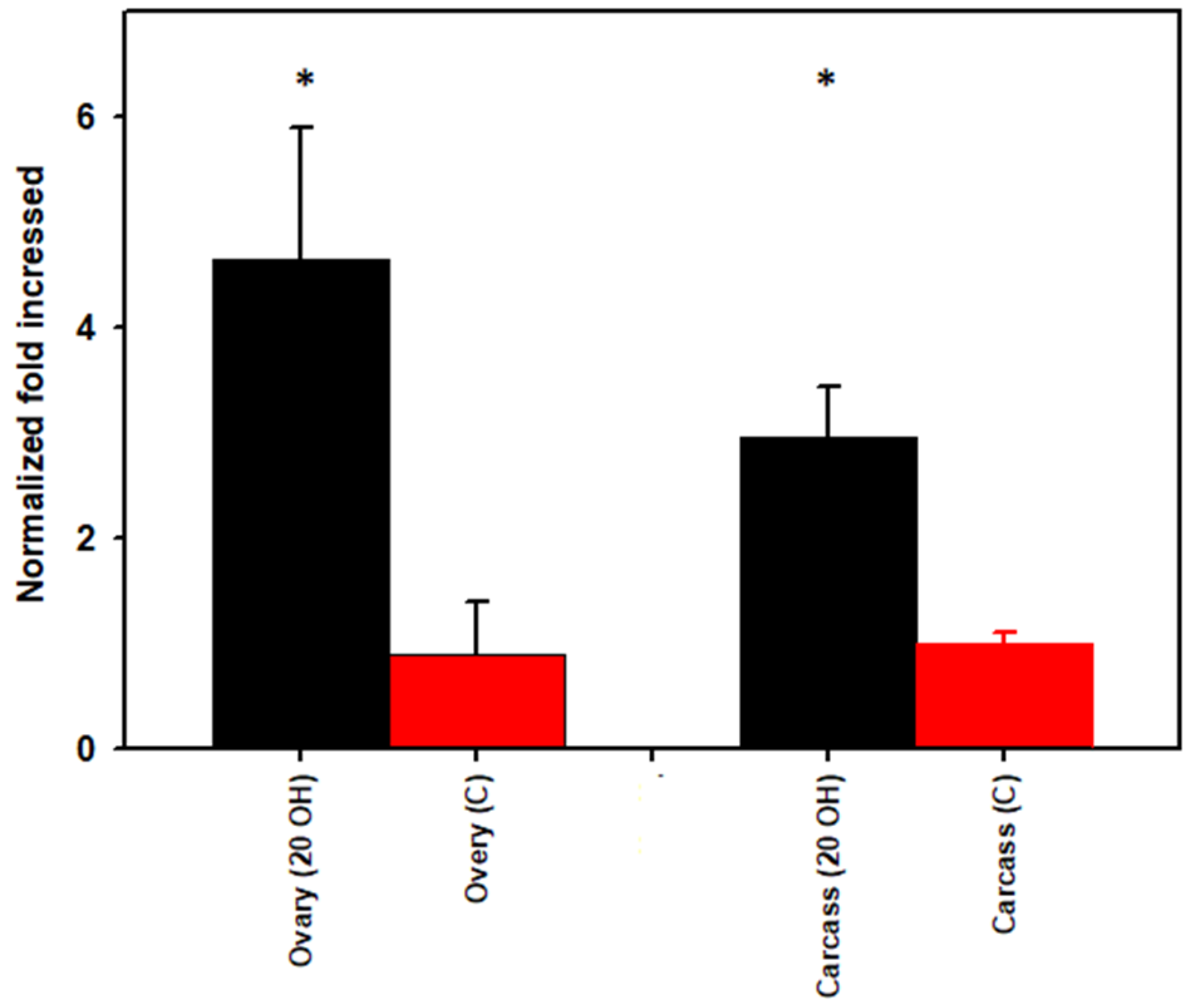
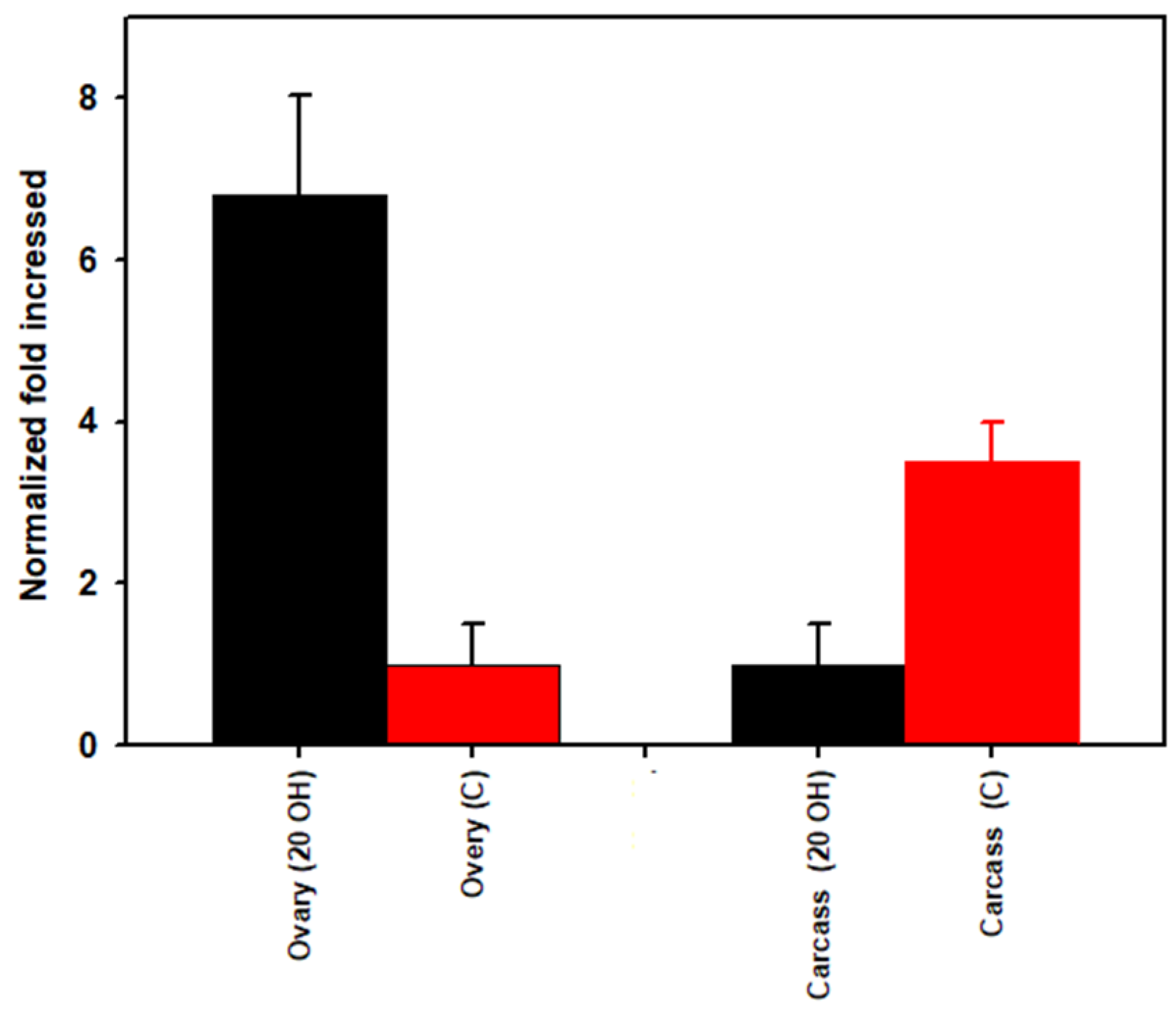
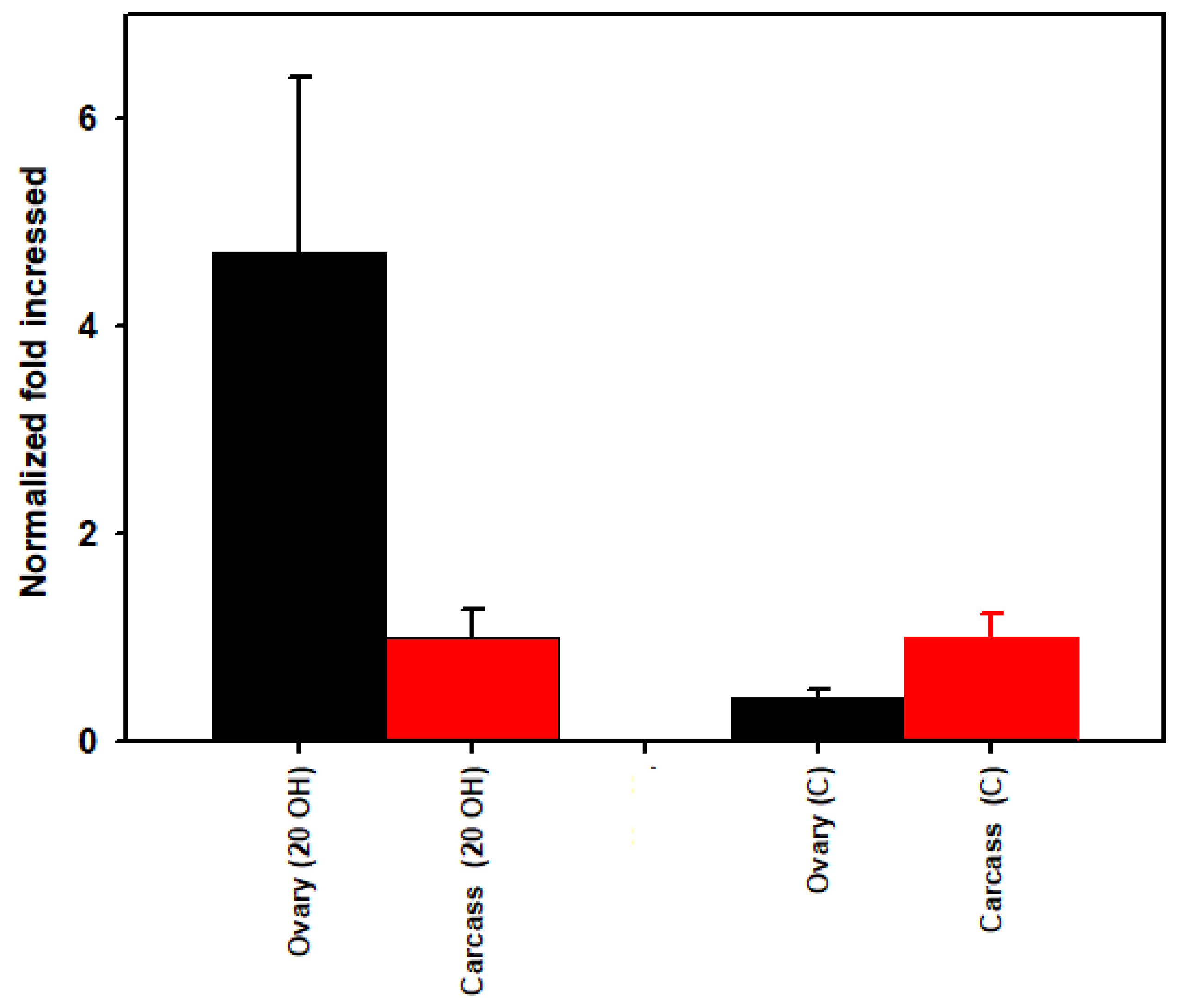
| 20E Ovaries | Control Ovaries | 20E Carcass | Control Carcass | Taxonomy | ||||
|---|---|---|---|---|---|---|---|---|
| Reads | % | Reads | % | Reads | % | Reads | % | |
| 22,507.5 | 43.82 | 12,803.5 | 41.24 | 10,135 | 32.51 | 38,957 | 64.38 | Rickettsia |
| 26,622.5 | 51.53 | 15,504.5 | 49.25 | 8686 | 27.95 | 17,403.5 | 29.30 | Francisella |
| 208.5 | 0.41 | 1.5 | 0.004 | 7425.5 | 23.76 | 5 | 0.008 | Pseudomonas |
| 665 | 1.28 | 1355 | 4.41 | 661.5 | 2.08 | 939.5 | 1.58 | Unassigned # |
| 1525 | 2.95 | 1577.5 | 5.09 | 4246.5 | 13.71 | 2861 | 4.74 | Others * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponnusamy, L.; Sutton, H.; Mitchell, R.D., III; Sonenshine, D.E.; Apperson, C.S.; Roe, R.M. Tick Ecdysteroid Hormone, Global Microbiota/Rickettsia Signaling in the Ovary versus Carcass during Vitellogenesis in Part-Fed (Virgin) American Dog Ticks, Dermacentor variabilis. Microorganisms 2021, 9, 1242. https://doi.org/10.3390/microorganisms9061242
Ponnusamy L, Sutton H, Mitchell RD III, Sonenshine DE, Apperson CS, Roe RM. Tick Ecdysteroid Hormone, Global Microbiota/Rickettsia Signaling in the Ovary versus Carcass during Vitellogenesis in Part-Fed (Virgin) American Dog Ticks, Dermacentor variabilis. Microorganisms. 2021; 9(6):1242. https://doi.org/10.3390/microorganisms9061242
Chicago/Turabian StylePonnusamy, Loganathan, Haley Sutton, Robert D. Mitchell, III, Daniel E. Sonenshine, Charles S. Apperson, and Richard Michael Roe. 2021. "Tick Ecdysteroid Hormone, Global Microbiota/Rickettsia Signaling in the Ovary versus Carcass during Vitellogenesis in Part-Fed (Virgin) American Dog Ticks, Dermacentor variabilis" Microorganisms 9, no. 6: 1242. https://doi.org/10.3390/microorganisms9061242
APA StylePonnusamy, L., Sutton, H., Mitchell, R. D., III, Sonenshine, D. E., Apperson, C. S., & Roe, R. M. (2021). Tick Ecdysteroid Hormone, Global Microbiota/Rickettsia Signaling in the Ovary versus Carcass during Vitellogenesis in Part-Fed (Virgin) American Dog Ticks, Dermacentor variabilis. Microorganisms, 9(6), 1242. https://doi.org/10.3390/microorganisms9061242







