Phylogenetic Tracking of LA-MRSA ST398 Intra-Farm Transmission among Animals, Humans and the Environment on German Dairy Farms
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and MRSA Isolation
2.2. DNA Extraction and WGS
2.3. Bioinformatics and Phylogenetic Analyses
3. Results
3.1. Genotypic Characterization of LA-MRSA Isolates
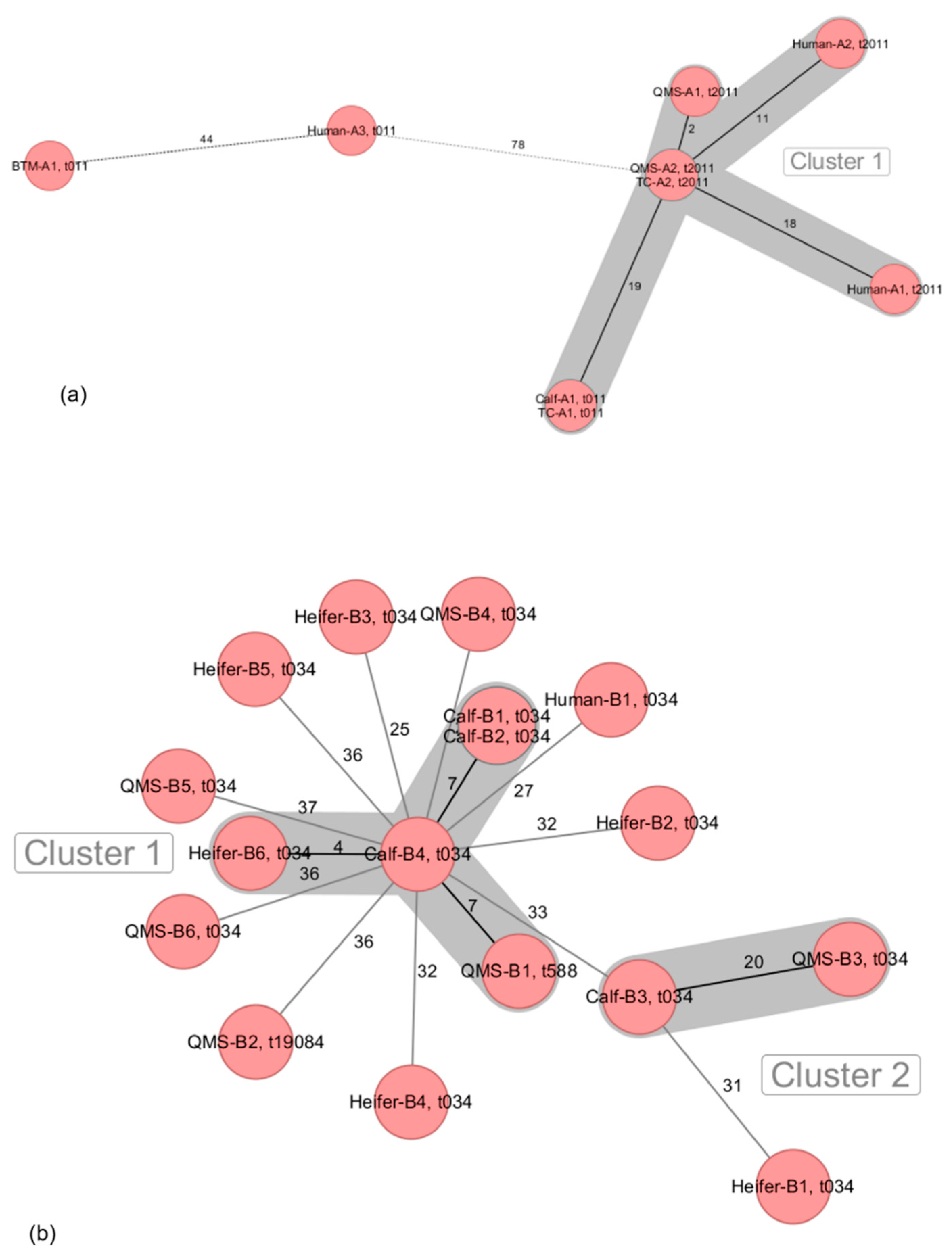
3.2. Intra-Farm Phylogenetic Relationship of LA-MRSA ST398 Isolates
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tenhagen, B.A.; Alt, K.; Pfefferkorn, B.; Wiehle, L.; Kasbohrer, A.; Fetsch, A. Short communication: Methicillin-resistant Staphylococcus aureus in conventional and organic dairy herds in Germany. J. Dairy Sci. 2018, 101, 3380–3386. [Google Scholar] [CrossRef]
- Feßler, A.T.; Olde Riekerink, R.G.; Rothkamp, A.; Kadlec, K.; Sampimon, O.C.; Lam, T.J.; Schwarz, S. Characterization of methicillin-resistant Staphylococcus aureus CC398 obtained from humans and animals on dairy farms. Vet. Microbiol. 2012, 160, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, K.; Entorf, M.; Peters, T. Occurrence and Characteristics of Livestock-Associated Methicillin-Resistant Staphylococcus aureus in Quarter Milk Samples From Dairy Cows in Germany. Front. Microbiol. 2019, 10, 1295. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.A.; Zadoks, R.N. Methicillin resistant S. aureus in human and bovine mastitis. J. Mammary Gland Biol. Neoplasia 2011, 16, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.K.; Nam, H.M.; Jang, G.C.; Lee, H.S.; Jung, S.C.; Kim, T.S. Transmission and persistence of methicillin-resistant Staphylococcus aureus in milk, environment, and workers in dairy cattle farms. Foodborne Pathog. Dis. 2013, 10, 731–736. [Google Scholar] [CrossRef]
- Goerge, T.; Lorenz, M.B.; van Alen, S.; Hubner, N.O.; Becker, K.; Kock, R. MRSA colonization and infection among persons with occupational livestock exposure in Europe: Prevalence, preventive options and evidence. Vet. Microbiol. 2017, 200, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Wieler, L.H.; Witte, W. Livestock-Associated MRSA: The Impact on Humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef]
- Cuny, C.; Abdelbary, M.; Layer, F.; Werner, G.; Witte, W. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 2015, 177, 219–223. [Google Scholar] [CrossRef]
- Lienen, T.; Schnitt, A.; Hammerl, J.A.; Maurischat, S.; Tenhagen, B.A. Genomic Distinctions of LA-MRSA ST398 on Dairy Farms From Different German Federal States With a Low Risk of Severe Human Infections. Front. Microbiol. 2020, 11, 575321. [Google Scholar] [CrossRef]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef]
- Schnitt, A.; Lienen, T.; Wichmann-Schauer, H.; Cuny, C.; Tenhagen, B.A. The occurrence and distribution of livestock-associated methicillin-resistant Staphylococcus aureus ST398 on German dairy farms. J. Dairy Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Barkema, H.W.; Schukken, Y.H.; Zadoks, R.N. Invited Review: The role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J. Dairy Sci. 2006, 89, 1877–1895. [Google Scholar] [CrossRef]
- Tenhagen, B.A.; Vossenkuhl, B.; Käsbohrer, A.; Alt, K.; Kraushaar, B.; Guerra, B.; Schroeter, A.; Fetsch, A. Methicillin-resistant Staphylococcus aureus in cattle food chains—Prevalence, diversity, and antimicrobial resistance in Germany. J. Anim Sci. 2014, 92, 2741–2751. [Google Scholar] [CrossRef]
- Vandendriessche, S.; Vanderhaeghen, W.; Soares, F.V.; Hallin, M.; Catry, B.; Hermans, K.; Butaye, P.; Haesebrouck, F.; Struelens, M.J.; Denis, O. Prevalence, risk factors and genetic diversity of methicillin-resistant Staphylococcus aureus carried by humans and animals across livestock production sectors. J. Antimicrob. Chemother. 2013, 68, 1510–1516. [Google Scholar] [CrossRef]
- Krukowski, H.; Bakuła, Z.; Iskra, M.; Olender, A.; Bis-Wencel, H.; Jagielski, T. The first outbreak of methicillin-resistant Staphylococcus aureus in dairy cattle in Poland with evidence of on-farm and intrahousehold transmission. J. Dairy Sci. 2020, 103, 10577–10584. [Google Scholar] [CrossRef] [PubMed]
- Carfora, V.; Giacinti, G.; Sagrafoli, D.; Marri, N.; Giangolini, G.; Alba, P.; Feltrin, F.; Sorbara, L.; Amoruso, R.; Caprioli, A.; et al. Methicillin-resistant and methicillin-susceptible Staphylococcus aureus in dairy sheep and in-contact humans: An intra-farm study. J. Dairy Sci. 2016, 99, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Pletinckx, L.J.; Verhegghe, M.; Crombe, F.; Dewulf, J.; De Bleecker, Y.; Rasschaert, G.; Butaye, P.; Goddeeris, B.M.; De Man, I. Evidence of possible methicillin-resistant Staphylococcus aureus ST398 spread between pigs and other animals and people residing on the same farm. Prev. Vet. Med. 2013, 109, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Zoppi, S.; Dondo, A.; Di Blasio, A.; Vitale, N.; Carfora, V.; Goria, M.; Chiavacci, L.; Giorgi, I.; D’Errico, V.; Irico, L.; et al. Livestock-Associated Methicillin-Resistant Staphylococcus aureus and Related Risk Factors in Holdings of Veal Calves in Northwest Italy. Microb. Drug Resist. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.E.; Ronco, T.; Stegger, M.; Sieber, R.N.; Fertner, M.E.; Martin, H.L.; Farre, M.; Toft, N.; Larsen, A.R.; Pedersen, K. LA-MRSA CC398 in Dairy Cattle and Veal Calf Farms Indicates Spillover From Pig Production. Front. Microbiol. 2019, 10, 2733. [Google Scholar] [CrossRef]
- Ronco, T.; Klaas, I.C.; Stegger, M.; Svennesen, L.; Astrup, L.B.; Farre, M.; Pedersen, K. Genomic investigation of Staphylococcus aureus isolates from bulk tank milk and dairy cows with clinical mastitis. Vet. Microbiol. 2018, 215, 35–42. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Leopold, S.R.; Goering, R.V.; Witten, A.; Harmsen, D.; Mellmann, A. Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef]
- Locatelli, C.; Cremonesi, P.; Caprioli, A.; Carfora, V.; Ianzano, A.; Barberio, A.; Morandi, S.; Casula, A.; Castiglioni, B.; Bronzo, V.; et al. Occurrence of methicillin-resistant Staphylococcus aureus in dairy cattle herds, related swine farms, and humans in contact with herds. J. Dairy Sci. 2017, 100, 608–619. [Google Scholar] [CrossRef]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Koutsoumanis, K.; Lindqvist, R.; Nørrung, B.; et al. Risk for the development of Antimicrobial Resistance (AMR) due to feeding of calves with milk containing residues of antibiotics. EFSA J. 2017, 15, e04665. [Google Scholar] [CrossRef]
- Spohr, M.; Rau, J.; Friedrich, A.; Klittich, G.; Fetsch, A.; Guerra, B.; Hammerl, J.A.; Tenhagen, B.A. Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health 2011, 58, 252–261. [Google Scholar] [CrossRef]
- Sergelidis, D.; Angelidis, A.S. Methicillin-resistant Staphylococcus aureus: A controversial food-borne pathogen. Lett. Appl. Microbiol. 2017, 64, 409–418. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Szafrańska, A.K.; Junker, V.; Steglich, M.; Nübel, U. Rapid cell division of Staphylococcus aureus during colonization of the human nose. BMC Genom. 2019, 20, 229. [Google Scholar] [CrossRef]


| Farm | Dairy Herd Size | Number of LA-MRSA 1 Isolates | Source of LA-MRSA | Spa Types |
|---|---|---|---|---|
| A | 350 | 9 | BTM 2, QMS 3, TC 4, Calf, Human | t011, t2011 |
| B | 700 | 17 | QMS, Calf, Heifer, Human | t034, t588, t19084 |
| C | 970 | 23 | BTM, QMS, Calf, Human | t011, t034, t571 |
| D | 280 | 19 | BTM, QMS, TC, Calf, Heifer, Human | t011, t034, t1928 |
| E | 94 | 25 | BTM, QMS, TC, Calf, AF 5, Heifer, EN 6 | t034 |
| F | 27 | 22 | BTM, QMS, TC, Calf, Pig, EN | t011, t1451 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lienen, T.; Schnitt, A.; Cuny, C.; Maurischat, S.; Tenhagen, B.-A. Phylogenetic Tracking of LA-MRSA ST398 Intra-Farm Transmission among Animals, Humans and the Environment on German Dairy Farms. Microorganisms 2021, 9, 1119. https://doi.org/10.3390/microorganisms9061119
Lienen T, Schnitt A, Cuny C, Maurischat S, Tenhagen B-A. Phylogenetic Tracking of LA-MRSA ST398 Intra-Farm Transmission among Animals, Humans and the Environment on German Dairy Farms. Microorganisms. 2021; 9(6):1119. https://doi.org/10.3390/microorganisms9061119
Chicago/Turabian StyleLienen, Tobias, Arne Schnitt, Christiane Cuny, Sven Maurischat, and Bernd-Alois Tenhagen. 2021. "Phylogenetic Tracking of LA-MRSA ST398 Intra-Farm Transmission among Animals, Humans and the Environment on German Dairy Farms" Microorganisms 9, no. 6: 1119. https://doi.org/10.3390/microorganisms9061119
APA StyleLienen, T., Schnitt, A., Cuny, C., Maurischat, S., & Tenhagen, B.-A. (2021). Phylogenetic Tracking of LA-MRSA ST398 Intra-Farm Transmission among Animals, Humans and the Environment on German Dairy Farms. Microorganisms, 9(6), 1119. https://doi.org/10.3390/microorganisms9061119






