Identification of Microorganisms from Several Surfaces by MALDI-TOF MS: P. aeruginosa Is Leading in Biofilm Formation
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Collection
2.2. Isolation and Identification of Bacterial Strains
2.3. Detection of Biofilm Formation
2.3.1. Substrate Materials and Surface Treatments
2.3.2. Congo Red Agar Method (CRA)
2.3.3. Microtiter Plate Method (MtP)
2.3.4. Tube Method (TM)
2.3.5. Colony-Forming Unit Method (CFU)
2.3.6. Scanning Electron Microscopy (SEM)
2.4. Statistical Analysis
2.5. Phylogenic Analysis of 16S rRNA of the Bacterial Spectra
3. Results
3.1. Isolation, Cultivation, and Identification of Bacteria
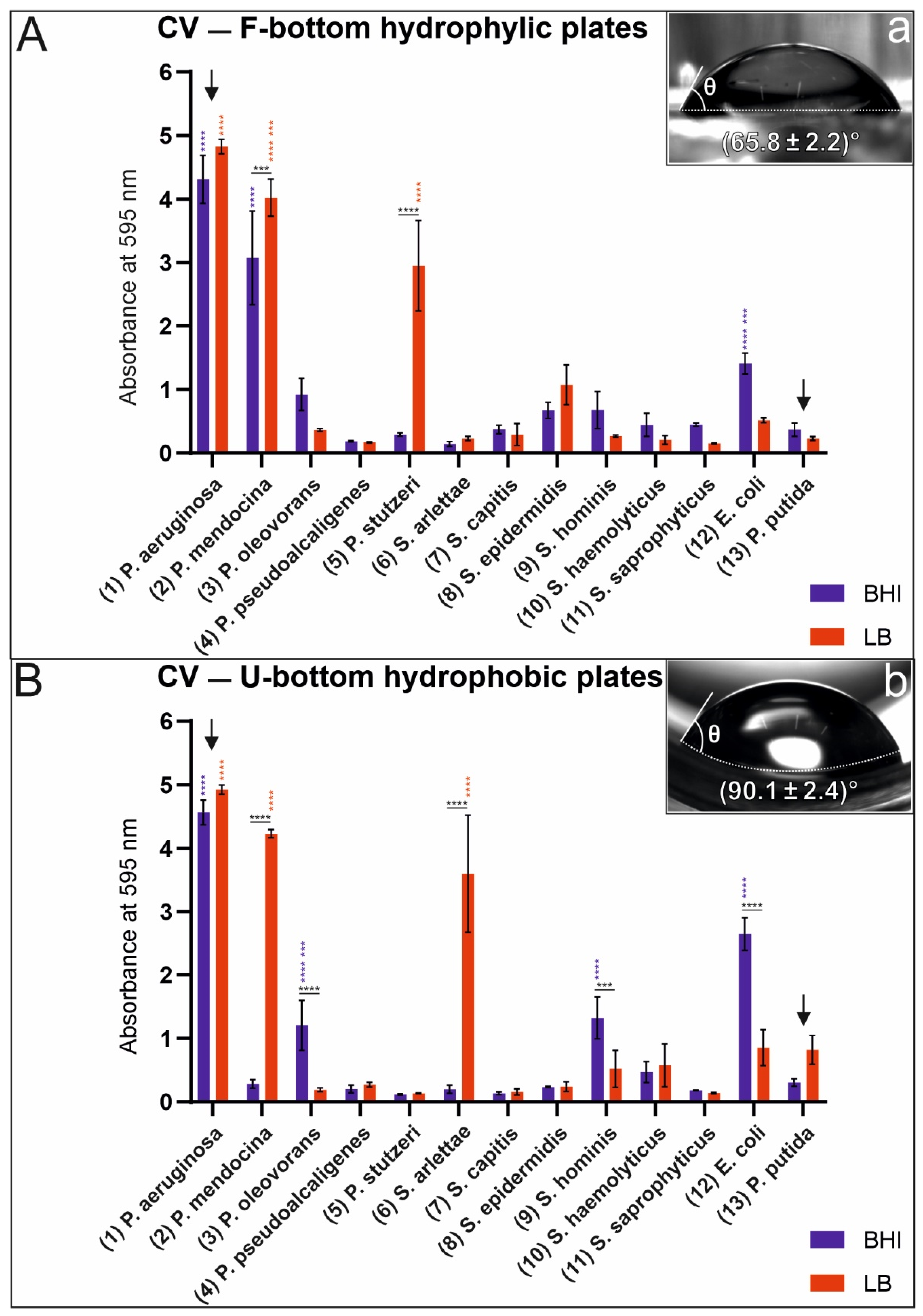
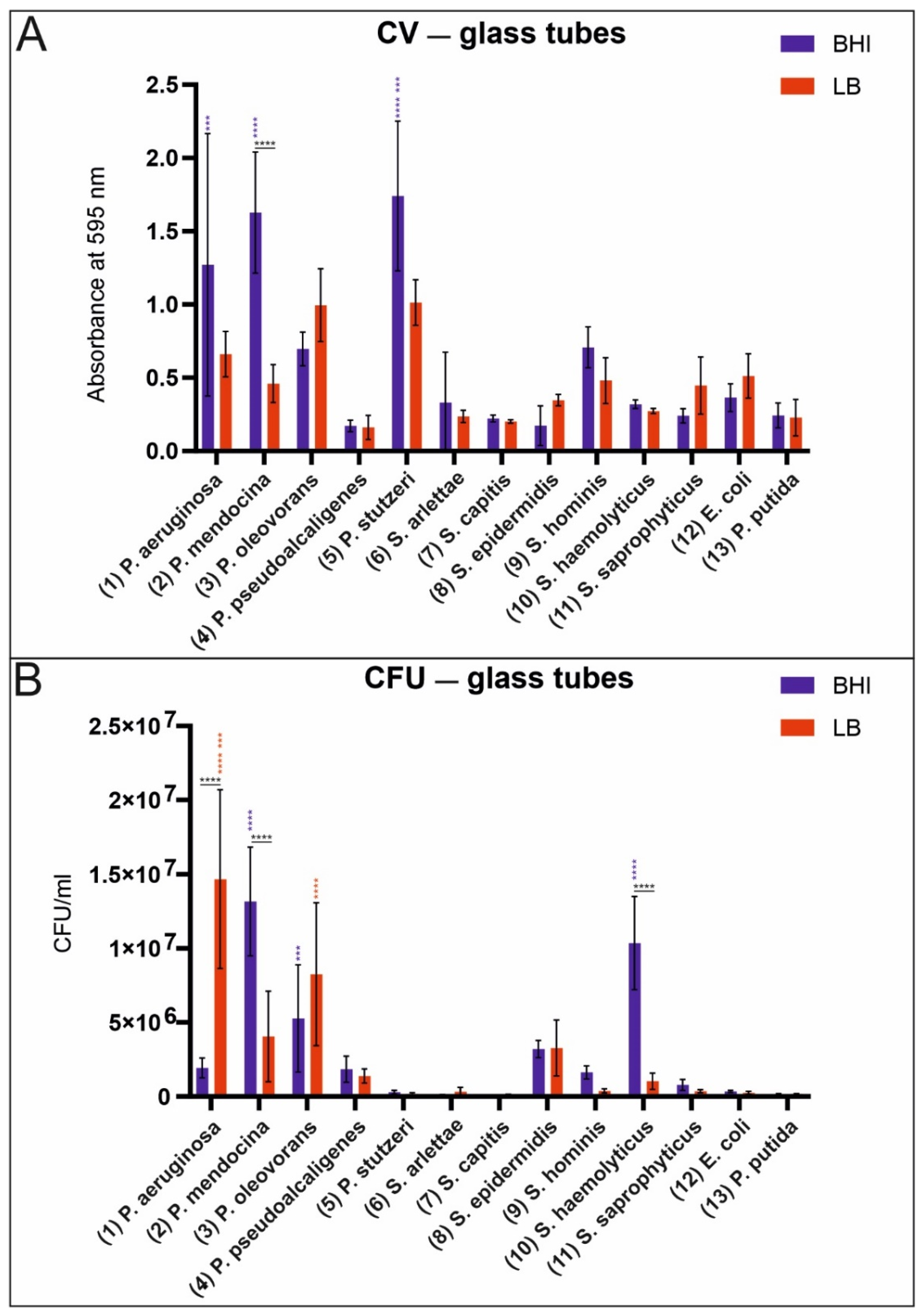
3.2. Analysis of Biofilm Formation
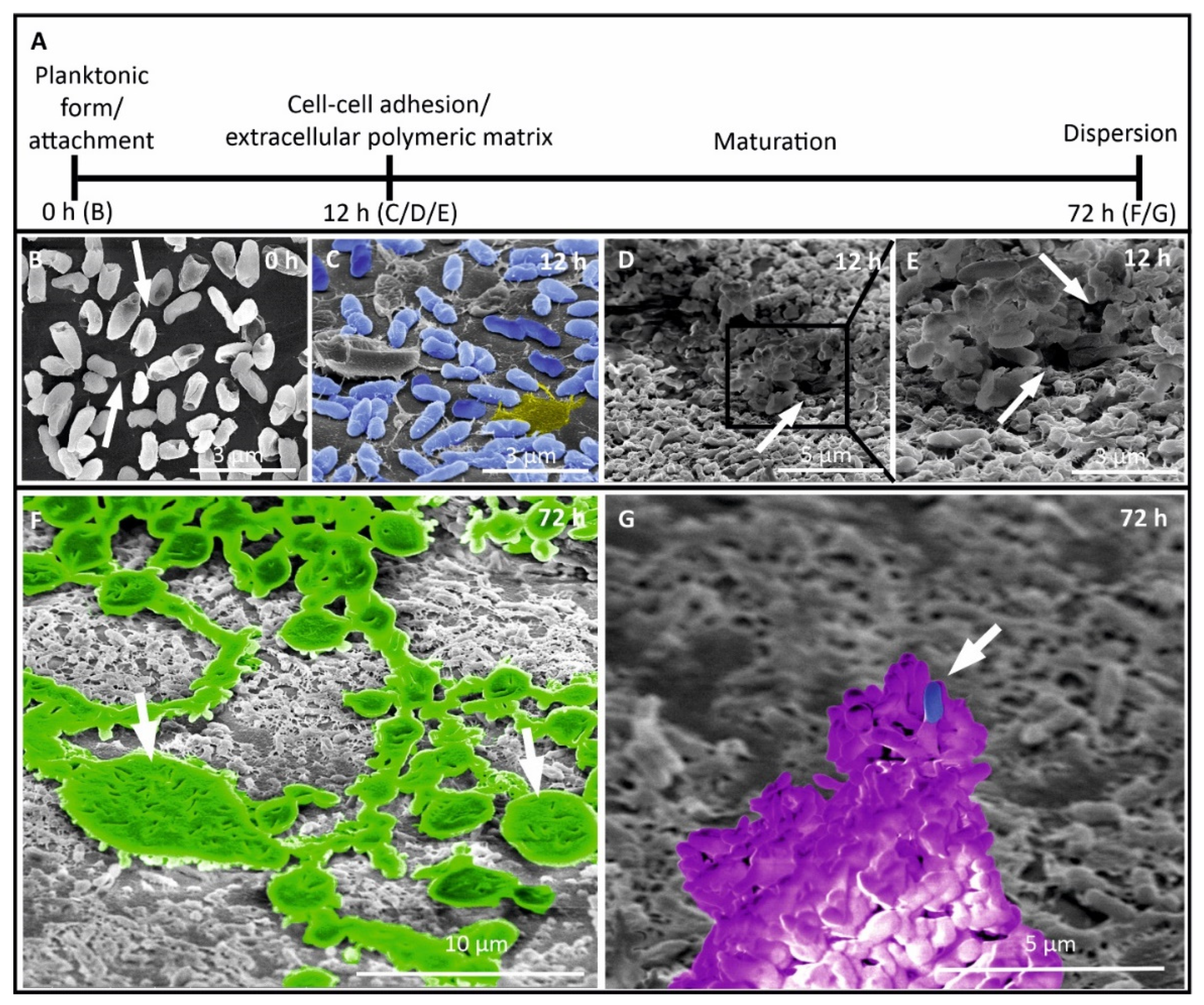
3.3. Ultrastructural Investigation of Biofilm Formation by Scanning Electron Microscopy (SEM)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bockmühl, D.P. Laundry Hygiene—How to Get More than Clean. J. Appl. Microbiol. 2017, 122, 1124–1133. [Google Scholar] [CrossRef]
- Laitala, K.; Jensen, H. Cleaning Effect of Household Laundry Detergents at Low Temperatures. Tenside Surfactants Deterg. 2010, 47, 413–420. [Google Scholar] [CrossRef]
- Terpstra, P.M.J. Domestic and Institutional Hygiene in Relation to Sustainability. Historical, Social and Environmental Implications. Int. Biodeterior. Biodegrad. 1998, 41, 169–175. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Y.; Li, X.; Liu, Y.; Zhao, L. Evaluation of Biofilm Development on Various Pipelines in the Domestic Hot Water System. Water Supply 2018, 18, 638–647. [Google Scholar] [CrossRef]
- Callewaert, C.; Van Nevel, S.; Kerckhof, F.-M.; Granitsiotis, M.S.; Boon, N. Bacterial Exchange in Household Washing Machines. Front. Microbiol. 2015, 6, 1381. [Google Scholar] [CrossRef]
- Gattlen, J.; Amberg, C.; Zinn, M.; Mauclaire, L. Biofilms Isolated from Washing Machines from Three Continents and Their Tolerance to a Standard Detergent. Biofouling 2010, 26, 873–882. [Google Scholar] [CrossRef]
- Raghupathi, P.K.; Zupančič, J.; Brejnrod, A.D.; Jacquiod, S.; Houf, K.; Burmølle, M.; Gunde-Cimerman, N.; Sørensen, S.J. Microbial Diversity and Putative Opportunistic Pathogens in Dishwasher Biofilm Communities. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Vilanova, C.; Iglesias, A.; Porcar, M. The Coffee-Machine Bacteriome: Biodiversity and Colonisation of the Wasted Coffee Tray Leach. Sci. Rep. 2015, 5, 17163. [Google Scholar] [CrossRef]
- Zupančič, J.; Raghupathi, P.K.; Houf, K.; Burmølle, M.; Sørensen, S.J.; Gunde-Cimerman, N. Synergistic Interactions in Microbial Biofilms Facilitate the Establishment of Opportunistic Pathogenic Fungi in Household Dishwashers. Front. Microbiol. 2018, 9, 21. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Characklis, W.G.; Trulear, M.G.; Bryers, J.D.; Zelver, N. Dynamics of Biofilm Processes: Methods. Water Res. 1982, 16, 1207–1216. [Google Scholar] [CrossRef]
- Munk, S.; Johansen, C.; Stahnke, L.H.; Adler-Nissen, J. Microbial Survival and Odor in Laundry. J. Surfact. Deterg. 2001, 4, 385–394. [Google Scholar] [CrossRef]
- Chan, S.; Pullerits, K.; Keucken, A.; Persson, K.M.; Paul, C.J.; Rådström, P. Bacterial Release from Pipe Biofilm in a Full-Scale Drinking Water Distribution System. npj Biofilms Microbiomes 2019, 5, 1–8. [Google Scholar] [CrossRef]
- Kerr, C.J.; Osborn, K.S.; Robson, G.D.; Handley, P.S. The Relationship between Pipe Material and Biofilm Formation in a Laboratory Model System. J. Appl. Microbiol. 1998, 85 (Suppl. 1), 29S–38S. [Google Scholar] [CrossRef]
- Mahapatra, A.; Padhi, N.; Mahapatra, D.; Bhatt, M.; Sahoo, D.; Jena, S.; Dash, D.; Chayani, N. Study of Biofilm in Bacteria from Water Pipelines. J. Clin. Diagn. Res. 2015, 9, DC09–DC11. [Google Scholar] [CrossRef] [PubMed]
- De Beer, D.; Stoodley, P.; Lewandowski, Z. Liquid Flow in Heterogeneous Biofilms. Biotechnol. Bioeng. 1994, 44, 636–641. [Google Scholar] [CrossRef]
- Costerton, J.W.; Cheng, K.J.; Geesey, G.G.; Ladd, T.I.; Nickel, J.C.; Dasgupta, M.; Marrie, T.J. Bacterial Biofilms in Nature and Disease. Annu. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef]
- Simões, M.; Simões, L.C.; Machado, I.; Pereira, M.O.; Vieira, M.J. Control of Flow-Generated Biofilms with Surfactants: Evidence of Resistance and Recovery. Food Bioprod. Process. 2006, 84, 338–345. [Google Scholar] [CrossRef]
- Stewart, P.S. Theoretical Aspects of Antibiotic Diffusion into Microbial Biofilms. Antimicrob. Agents Chemother. 1996, 40, 2517–2522. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- López, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010, 2, a000398. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The Connections between Quorum Sensing and Biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as Complex Differentiated Communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Jacksch, S.; Kaiser, D.; Weis, S.; Weide, M.; Ratering, S.; Schnell, S.; Egert, M. Influence of Sampling Site and Other Environmental Factors on the Bacterial Community Composition of Domestic Washing Machines. Microorganisms 2019, 8, 30. [Google Scholar] [CrossRef]
- Ranganathan, V. Biofilms: Microbial Cities of Scientific Significance. J. Microbiol. Exp. 2014, 1. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Biofilms and Their Consequences, with Particular Reference to Hygiene in the Food Industry. J. Appl. Bacteriol. 1993, 75, 499–511. [Google Scholar] [CrossRef]
- Gilbert, P.; Evans, D.J.; Evans, E.; Duguid, I.G.; Brown, M.R.W. Surface Characteristics and Adhesion of Escherichia Coli and Staphylococcus Epidermidis. J. Appl. Bacteriol. 1991, 71, 72–77. [Google Scholar] [CrossRef]
- Loosdrecht, M.C.v.; Lyklema, J.; Norde, W.; Schraa, G.; Zehnder, A.J. Electrophoretic Mobility and Hydrophobicity as a Measured to Predict the Initial Steps of Bacterial Adhesion. Appl. Environ. Microbiol. 1987, 53, 1898–1901. [Google Scholar]
- Palmer, J.; Flint, S.; Brooks, J. Bacterial Cell Attachment, the Beginning of a Biofilm. J. Ind. Microbiol. Biotechnol. 2007, 34, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Verran, J.; Whitehead, K.A. Assessment of Organic Materials and Microbial Components on Hygienic Surfaces. Food Bioprod. Process. 2006, 84, 260–264. [Google Scholar] [CrossRef][Green Version]
- Page, K.; Wilson, M.P.; Parkin, I. Antimicrobial Surfaces and Their Potential in Reducing the Role of the Inanimate Environment in the Incidence of Hospital-Acquired Infections. J. Mater. Chem. 2009, 19, 3819–3831. [Google Scholar] [CrossRef]
- Bixler, G.D.; Bhushan, B. Biofouling: Lessons from Nature. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2012, 370, 2381–2417. [Google Scholar] [CrossRef]
- Hou, T.-Y.; Chiang-Ni, C.; Teng, S.-H. Current Status of MALDI-TOF Mass Spectrometry in Clinical Microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Carbonnelle, E.; Mesquita, C.; Bille, E.; Day, N.; Dauphin, B.; Beretti, J.-L.; Ferroni, A.; Gutmann, L.; Nassif, X. MALDI-TOF Mass Spectrometry Tools for Bacterial Identification in Clinical Microbiology Laboratory. Clin. Biochem. 2011, 44, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.A.; Young, S.; Timm, K.; Novak-Weekley, S.; Marlowe, E.M.; Madisen, N.; Lillie, J.L.; Ledeboer, N.A.; Smith, R.; Hyke, J.; et al. Multicenter Evaluation of the Bruker MALDI Biotyper CA System for the Identification of Clinically Important Bacteria and Yeasts. Am. J. Clin. Pathol. 2017, 147, 623–631. [Google Scholar] [CrossRef]
- Colwell, R.R.; Grigorova, R. Current Methods for Classification and Identification of Microorganisms; Methods in Microbiology; Academic Press: Orlando, FL, USA, 1987; ISBN 978-0-08-086048-0. [Google Scholar]
- Förch, R.; Schönherr, H.; Jenkins, A.T.A. Surface Design: Applications in Bioscience and Nanotechnology; John Wiley & Sons: Hoboken, NJ, USA, 2009; ISBN 978-3-527-40789-7. [Google Scholar]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New Method for Detecting Slime Production by Coagulase Negative Staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [PubMed]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of Coagulase-Negative Staphylococci to Plastic Tissue Culture Plates: A Quantitative Model for the Adherence of Staphylococci to Medical Devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar]
- Christensen, G.D.; Simpson, W.A.; Bisno, A.L.; Beachey, E.H. Adherence of Slime-Producing Strains of Staphylococcus Epidermidis to Smooth Surfaces. Infect. Immun. 1982, 37, 318–326. [Google Scholar] [CrossRef]
- Krause, M.L.; Sohail, M.R.; Patel, R.; Wittich, C.M. Achromobacter Piechaudii Bloodstream Infection in an Immunocompetent Host. Am. J. Case Rep. 2012, 13, 265–267. [Google Scholar] [CrossRef]
- Coenye, T.; Vancanneyt, M.; Falsen, E.; Swings, J.; Vandamme, P. Achromobacter Insolitus Sp. Nov. and Achromobacter Spanius Sp. Nov., from Human Clinical Samples. Int. J. Syst. Evol. Microbiol. 2003, 53, 1819–1824. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Regalado, N.G.; Martin, G.; Antony, S.J. Acinetobacter Lwoffii: Bacteremia Associated with Acute Gastroenteritis. Travel Med. Infect. Dis. 2009, 7, 316–317. [Google Scholar] [CrossRef] [PubMed]
- Carr, E.; Ward, A.; Gurtler, V.; Seviour, R.J. Pyrolysis Mass Spectrometry (PyMS) and 16S-23S RDNA Spacer Region Fingerprinting Suggests the Presence of Novel Acinetobacters in Activated Sludge. Syst. Appl. Microbiol. 2001, 24, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Carr, E.L.; Kämpfer, P.; Patel, B.K.C.; Gürtler, V.; Seviour, R.J. Seven Novel Species of Acinetobacter Isolated from Activated Sludge. Int. J. Syst. Evol. Microbiol. 2003, 53, 953–963. [Google Scholar] [CrossRef]
- Parrey, A.H.; Sofi, F.; Ahmad, M.; Kuchay, A. Aerococcus Viridans Infection Presenting as Cutaneous Vasculitis in an Immunocompetent Patient. Reumatologia 2016, 54, 318–320. [Google Scholar] [CrossRef]
- Logan, N.A.; Vos, P.D. Bacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; American Cancer Society: Atlanta, GA, USA, 2015; pp. 1–163. ISBN 978-1-118-96060-8. [Google Scholar]
- Sonenshein, A.L.; Hoch, J.A.; Losick, R. Bacillus Subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics; American Society for Microbiology: Washington, DC, USA, 1993; ISBN 978-1-55581-053-5. [Google Scholar]
- Ivanova, E.P.; Christen, R.; Alexeeva, Y.V.; Zhukova, N.V.; Gorshkova, N.M.; Lysenko, A.M.; Mikhailov, V.V.; Nicolau, D.V. Brevibacterium Celere Sp. Nov., Isolated from Degraded Thallus of a Brown Alga. Int. J. Syst. Evol. Microbiol. 2004, 54, 2107–2111. [Google Scholar] [CrossRef]
- Yang, F.; Liu, H.; Zhang, R.; Chen, D.; Wang, X.; Li, S.; Hong, Q. Chryseobacterium Shandongense Sp. Nov., Isolated from Soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 1860–1865. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, N.O.G.; Brandt, K.K.; Nybroe, O.; Hansen, M. Delftia Lacustris Sp. Nov., a Peptidoglycan-Degrading Bacterium from Fresh Water, and Emended Description of Delftia Tsuruhatensis as a Peptidoglycan-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 2009, 59, 2195–2199. [Google Scholar] [CrossRef] [PubMed]
- Takarada, H.; Sekine, M.; Kosugi, H.; Matsuo, Y.; Fujisawa, T.; Omata, S.; Kishi, E.; Shimizu, A.; Tsukatani, N.; Tanikawa, S.; et al. Complete Genome Sequence of the Soil Actinomycete Kocuria Rhizophila. J. Bacteriol. 2008, 190, 4139–4146. [Google Scholar] [CrossRef]
- Kovács, G.; Burghardt, J.; Pradella, S.; Schumann, P.; Stackebrandt, E.; Màrialigeti, K. Kocuria Palustris Sp. Nov. and Kocuria Rhizophila Sp. Nov., Isolated from the Rhizoplane of the Narrow-Leaved Cattail (Typha Angustifolia). Int. J. Syst. Bacteriol. 1999, 49 Pt 1, 167–173. [Google Scholar] [CrossRef]
- Kloos, W.E.; TORNABENE, T.G.; SCHLEIFER, K.H. Isolation and Characterization of Micrococci From Human Skin, Including Two New Species: Micrococcus Lylae and Micrococcus Kristinae1. Int. J. Syst. Evol. Microbiol. 1974, 24, 79–101. [Google Scholar] [CrossRef]
- Kooken, J.M.; Fox, K.F.; Fox, A. Characterization of Micrococcus Strains Isolated from Indoor Air. Mol. Cell Probes 2012, 26, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hadano, Y.; Ito, K.; Suzuki, J.; Kawamura, I.; Kurai, H.; Ohkusu, K. Moraxella Osloensis: An Unusual Cause of Central Venous Catheter Infection in a Cancer Patient. Int. J. Gen. Med. 2012, 5, 875–877. [Google Scholar] [CrossRef]
- Lo, C.I.; Padhmanabhan, R.; Mediannikov, O.; Nguyen, T.T.; Raoult, D.; Fournier, P.-E.; Fenollar, F. Genome Sequence and Description of Pantoea Septica Strain FF5. Stand Genom. Sci. 2015, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Long, S.S.; Prober, C.G.; Fischer, M. Principles and Practice of Pediatric Infectious Diseases E-Book; Elsevier Health Sciences: Philadelphia, PA, USA, 2017; ISBN 978-0-323-46132-0. [Google Scholar]
- Gani, M.; Rao, S.; Miller, M.; Scoular, S. Pseudomonas Mendocina Bacteremia: A Case Study and Review of Literature. Am. J. Case Rep. 2019, 20, 453–458. [Google Scholar] [CrossRef]
- Lee, M.; Chandler, A.C. A Study of the Nature, Growth and Control of Bacteria in Cutting Compounds. J. Bacteriol. 1941, 41, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Saha, R.; Spröer, C.; Beck, B.; Bagley, S. Pseudomonas Oleovorans Subsp. Lubricantis Subsp. Nov., and Reclassification of Pseudomonas Pseudoalcaligenes ATCC 17440T as Later Synonym of Pseudomonas Oleovorans ATCC 8062 T. Curr. Microbiol. 2010, 60, 294–300. [Google Scholar] [CrossRef]
- Lalucat, J.; Bennasar, A.; Bosch, R.; García-Valdés, E.; Palleroni, N.J. Biology of Pseudomonas Stutzeri. Microbiol. Mol. Biol. Rev. 2006, 70, 510–547. [Google Scholar] [CrossRef] [PubMed]
- Bagley, S.T.; Seidler, R.J.; Brenner, D.J. Klebsiella Planticola Sp. Nov.: A New Species of Enterobacteriaceae Found Primarily in Nonclinical Environments. Curr. Microbiol. 1981, 6, 105–109. [Google Scholar] [CrossRef]
- Drancourt, M.; Bollet, C.; Carta, A.; Rousselier, P. Phylogenetic Analyses of Klebsiella Species Delineate Klebsiella and Raoultella Gen. Nov., with Description of Raoultella Ornithinolytica Comb. Nov., Raoultella Terrigena Comb. Nov. and Raoultella Planticola Comb. Nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Seidler, R.J.; Knittel, M.D.; Brown, C. Potential Pathogens in the Environment: Cultural Reactions and Nucleic Acid Studies on Klebsiella Pneumoniae from Clinical and Environmental Sources. Appl. Microbiol. 1975, 29, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Labbate, M.; Queck, S.Y.; Koh, K.S.; Rice, S.A.; Givskov, M.; Kjelleberg, S. Quorum Sensing-Controlled Biofilm Development in Serratia Liquefaciens MG1. J. Bacteriol. 2004, 186, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Khanna, A.; Khanna, M.; Aggarwal, A. Serratia Marcescens-A Rare Opportunistic Nosocomial Pathogen and Measures to Limit Its Spread in Hospitalized Patients. J. Clin. Diagn. Res. 2013, 7, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Schleifer, K.H.; Kilpper-Bälz, R.; Devriese, L.A. Staphylococcus Arlettae Sp. Nov., S. Equorum Sp. Nov. and S. K1oosii Sp. Nov.: Three New Coagulase-Negative, Novobiocin-Resistant Species from Animals. Syst. Appl. Microbiol. 1984, 5, 501–509. [Google Scholar] [CrossRef]
- Kloos, W.E.; Schleifer, K.H. Isolation and Characterization of Staphylococci from Human Skin II. Descriptions of Four New Species: Staphylococcus Warneri, Staphylococcus Capitis, Staphylococcus Hominis, and Staphylococcus Simulans1. Int. J. Syst. Evol. Microbiol. 1975, 25, 62–79. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus Epidermidis–the “Accidental” Pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef]
- Mendoza-Olazarán, S.; Morfin-Otero, R.; Rodríguez-Noriega, E.; Llaca-Díaz, J.; Flores-Treviño, S.; González-González, G.M.; Villarreal-Treviño, L.; Garza-González, E. Microbiological and Molecular Characterization of Staphylococcus Hominis Isolates from Blood. PLoS ONE 2013, 8, e61161. [Google Scholar] [CrossRef]
- Renaud, F.; Etienne, J.; Bertrand, A.; Brun, Y.; Greenland, T.B.; Freney, J.; Fleurette, J. Molecular Epidemiology of Staphylococcus Haemolyticus Strains Isolated in an Albanian Hospital. J. Clin. Microbiol. 1991, 29, 1493–1497. [Google Scholar] [CrossRef]
- Ehlers, S.; Merrill, S.A. Staphylococcus Saprophyticus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Kaci, G.; Goudercourt, D.; Dennin, V.; Pot, B.; Doré, J.; Ehrlich, S.D.; Renault, P.; Blottière, H.M.; Daniel, C.; Delorme, C. Anti-Inflammatory Properties of Streptococcus Salivarius, a Commensal Bacterium of the Oral Cavity and Digestive Tract. Appl. Environ. Microbiol. 2014, 80, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Silva-Jiménez, H.; Araujo-Palomares, C.L.; Macías-Zamora, J.V.; Ramírez-Álvarez, N.; García-Lara, B.; Corrales-Escobosa, A.R.; Silva-Jiménez, H.; Araujo-Palomares, C.L.; Macías-Zamora, J.V.; Ramírez-Álvarez, N.; et al. Identification by MALDI-TOF MS of Environmental Bacteria with High Potential to Degrade Pyrene. J. Mex. Chem. Soc. 2018, 62, 214–225. [Google Scholar] [CrossRef]
- Bononi, I.; Balatti, V.; Gaeta, S.; Tognon, M. Gram-Negative Bacterial Lipopolysaccharide Retention by a Positively Charged New-Generation Filter. Appl. Environ. Microbiol. 2008, 74, 6470–6472. [Google Scholar] [CrossRef] [PubMed]
- Thewes, N.; Thewes, A.; Loskill, P.; Peisker, H.; Bischoff, M.; Herrmann, M.; Santen, L.; Jacobs, K. Stochastic Binding of Staphylococcus Aureus to Hydrophobic Surfaces. Soft Matter 2015, 11, 8913–8919. [Google Scholar] [CrossRef]
- Maikranz, E.; Spengler, C.; Thewes, N.; Thewes, A.; Nolle, F.; Jung, P.; Bischoff, M.; Santen, L.; Jacobs, K. Different Binding Mechanisms of Staphylococcus Aureus to Hydrophobic and Hydrophilic Surfaces. Nanoscale 2020, 12, 19267–19275. [Google Scholar] [CrossRef] [PubMed]
- Spengler, C.; Thewes, N.; Jung, P.; Bischoff, M.; Jacobs, K. Determination of the Nano-Scaled Contact Area of Staphylococcal Cells. Nanoscale 2017, 9, 10084–10093. [Google Scholar] [CrossRef]
- Priester, J.H.; Horst, A.M.; Van De Werfhorst, L.C.; Saleta, J.L.; Mertes, L.A.K.; Holden, P.A. Enhanced Visualization of Microbial Biofilms by Staining and Environmental Scanning Electron Microscopy. J. Microbiol. Methods 2007, 68, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Miquel Guennoc, C.; Rose, C.; Guinnet, F.; Miquel, I.; Labbé, J.; Deveau, A. A New Method for Qualitative Multi-Scale Analysis of Bacterial Biofilms on Filamentous Fungal Colonies Using Confocal and Electron Microscopy. J. Vis. Exp. 2017. [Google Scholar] [CrossRef]
- Henry, V.A.; Jessop, J.L.P.; Peeples, T.L. Differentiating Pseudomonas Sp. Strain ADP Cells in Suspensions and Biofilms Using Raman Spectroscopy and Scanning Electron Microscopy. Anal. Bioanal. Chem. 2017, 409, 1441–1449. [Google Scholar] [CrossRef]
- Zhou, J.-W.; Luo, H.-Z.; Jiang, H.; Jian, T.-K.; Chen, Z.-Q.; Jia, A.-Q. Hordenine: A Novel Quorum Sensing Inhibitor and Antibiofilm Agent against Pseudomonas Aeruginosa. J. Agric. Food Chem. 2018, 66, 1620–1628. [Google Scholar] [CrossRef]
- Pugazhendhi, A.; Prabakar, D.; Jacob, J.M.; Karuppusamy, I.; Saratale, R.G. Synthesis and Characterization of Silver Nanoparticles Using Gelidium Amansii and Its Antimicrobial Property against Various Pathogenic Bacteria. Microb. Pathog. 2018, 114, 41–45. [Google Scholar] [CrossRef]
- Carette, J.; Nachtergael, A.; Duez, P.; Jaziri, M.E.; Rasamiravaka, T. Natural Compounds Inhibiting Pseudomonas Aeruginosa Biofilm Formation by Targeting Quorum Sensing Circuitry. Bact. Biofilms 2020. [Google Scholar] [CrossRef][Green Version]
| Bacteria | Substrate Material | Gram Status | Occurrence/Source | Characterization Method | Score |
|---|---|---|---|---|---|
| Achromobacter piechaudii | Plastic | - | Soil and water [41] | MALDI-TOF-MS | 2.09 |
| Achromobacter spanius | Plastic | - | Human blood [42] | MALDI-TOF-MS | 2.03 |
| Acinetobacter lwoffii | Glass | - | Normal flora of the oropharynx and skin [43] | MALDI-TOF-MS | 2.28 |
| Acinetobacter towneri | Elastomer | - | Activated sludge, Australia [44,45] | MALDI-TOF-MS | 2.29 |
| Aerococcus viridans | Plastic, metal, elastomer | + | Hospital environments and room air [46] | MALDI-TOF-MS | 2.06 |
| Bacillus circulans | Plastic | + | Soil, marine water, plants, animals [47] | MALDI-TOF-MS | 2.11 |
| Bacillus pumilus | Glass | + | Soil [48]; | Classical microbiology | - |
| Brevibacterium celere | Glass | + | Alga Fucus evanescens [49] | Classical microbiology | - |
| Chryseobacterium shandongense | Elastomer | - | Soil [50] | MALDI-TOF-MS | 2.05 |
| Delftia lacustris | Plastic | - | Mesotrophic lake water [51] | MALDI-TOF-MS | 2.41 |
| Kocuria rhizophila | Elastomer | + | Soil [52], rhizosphere of Typha angustifolia [53] | MALDI-TOF-MS | 2.31 |
| Micrococcus luteus | Elastomer | + | Human skin [54], air [55] | MALDI-TOF-MS | 2.45 |
| Moraxella osloensis | Elastomer | - | Environmental sources in hospitals and normal human respiratory tract [56] | MALDI-TOF-MS | 2.12 |
| Pantoea septica | Metal | - | Environment, plants, seeds, vegetables, human skin [57] | MALDI-TOF-MS | 2.17 |
| Pseudomonas aeruginosa | Plastic, elastomer | - | Water and soil [58] | MALDI-TOF-MS | 2.51 |
| Pseudomonas mendocina | Plastic, elastomer | - | Water and soil [59] | MALDI-TOF-MS | 2.23 |
| Pseudomonas oleovorans | Plastic, metal, glass, elastomer | - | Cutting fluid [60] | MALDI-TOF-MS | 2.22 |
| Pseudomonas pseudoalcaligenes | Plastic | - | Cutting fluid [60,61] | MALDI-TOF-MS | 2.09 |
| Pseudomonas stutzeri | Glass, elastomer | - | Denitrifying bacteria widely distributed in the environment [62] | MALDI-TOF-MS | 2.30 |
| Raoultella planticola (Synonym: Klebsiella planticola) | Plastic | - | Radishroot, water, [63,64,65] | MALDI-TOF-MS | 2.29 |
| Serratia liquefaciens | Plastic | - | River water, domestic sewage, fish [66] | MALDI-TOF-MS | 2.03 |
| Serratia marcescens | Plastic | - | Water [67] | MALDI-TOF-MS | 2.17 |
| Staphylococcus arlettae | Metal | + | Skin of mammals and birds [68] | Classical microbiology | - |
| Staphylococcus capitis | Metal | + | Human skin [69] | MALDI-TOF-MS | 2.09 |
| Staphylococcus epidermidis | Plastic, glass | + | Skin [70] | MALDI-TOF-MS | 2.14 |
| Staphylococcus hominis | Metal, glass, elastomer | + | Skin [71] | MALDI-TOF-MS | 2.26 |
| Staphylococcus haemolyticus | Metal, glass, elastomer | + | Skin [72] | MALDI-TOF-MS | 2.23 |
| Staphylococcus saprophyticus | Elastomer | + | Perineum, rectum urethra, cervix, and gastrointestinal tract of humans, pigs and cows [73] | MALDI-TOF-MS | 2.17 |
| Streptococcus salivarius | Glass | + | Human oral cavity [74] | MALDI-TOF-MS | 2.24 |
| Bacteria | CV Absorbance, F-Bottom Polystyrene Plates (Hydrophilic) | CV Absorbance, U-Bottom Polystyrene Plates (Hydrophobic) | CFU, Glass Tubes (CFU/mL) | CV Absorbance, Glass Tubes | ||||
|---|---|---|---|---|---|---|---|---|
| BHI | LB | BHI | LB | BHI | LB | BHI | LB | |
| (1) P. aeruginosa | 4.30 | 4.82 | 4.56 | 4.92 | 1.93 × 106 | 1.47 × 107 | 1.27 | 0.66 |
| (2) P. mendocina | 3.07 | 4.02 | 0.28 | 4.22 | 1.32 × 107 | 4.05 × 106 | 1.63 | 0.46 |
| (3) P. oleovorans | 0.92 | 0.36 | 1.20 | 0.19 | 5.27 × 106 | 8.27 × 106 | 0.70 | 1.00 |
| (4) P. pseudoalcaligenes | 0.18 | 0.16 | 0.20 | 0.26 | 1.85 × 106 | 1.39 × 106 | 0.17 | 0.16 |
| (5) P. stutzeri | 0.29 | 2.94 | 0.11 | 0.13 | 2.88 × 105 | 1.67 × 105 | 1.74 | 1.01 |
| (6) S. arlettae | 0.14 | 0.22 | 0.19 | 3.59 | 8.33 × 104 | 3.35 × 105 | 0.33 | 0.24 |
| (7) S. capitis | 0.37 | 0.29 | 0.13 | 0.15 | 4.86 × 104 | 8.80 × 104 | 0.22 | 0.20 |
| (8) S. epidermidis | 0.67 | 1.07 | 0.23 | 0.23 | 3.20 × 106 | 3.27 × 106 | 0.17 | 0.35 |
| (9) S. hominis | 0.67 | 0.263 | 1.32 | 0.51 | 1.64 × 106 | 3.78 × 105 | 0.71 | 0.48 |
| (10) S. haemolyticus | 0.44 | 0.20 | 0.46 | 0.57 | 1.04 × 107 | 1.03 × 106 | 0.32 | 0.27 |
| (11) S. saprophyticus | 0.44 | 0.15 | 0.18 | 0.13 | 7.85 × 105 | 3.63 × 105 | 0.24 | 0.45 |
| Comparative Bacteria | ||||||||
| (12) E. coli XL1-blue | 1.40 | 0.15 | 2.64 | 0.85 | 3.53 × 105 | 2.50 × 105 | 0.36 | 0.51 |
| (13) P. putida | 0.36 | 0.22 | 0.30 | 0.81 | 1.38 × 105 | 1.72× 105 | 0.24 | 0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asghari, E.; Kiel, A.; Kaltschmidt, B.P.; Wortmann, M.; Schmidt, N.; Hüsgen, B.; Hütten, A.; Knabbe, C.; Kaltschmidt, C.; Kaltschmidt, B. Identification of Microorganisms from Several Surfaces by MALDI-TOF MS: P. aeruginosa Is Leading in Biofilm Formation. Microorganisms 2021, 9, 992. https://doi.org/10.3390/microorganisms9050992
Asghari E, Kiel A, Kaltschmidt BP, Wortmann M, Schmidt N, Hüsgen B, Hütten A, Knabbe C, Kaltschmidt C, Kaltschmidt B. Identification of Microorganisms from Several Surfaces by MALDI-TOF MS: P. aeruginosa Is Leading in Biofilm Formation. Microorganisms. 2021; 9(5):992. https://doi.org/10.3390/microorganisms9050992
Chicago/Turabian StyleAsghari, Ehsan, Annika Kiel, Bernhard Peter Kaltschmidt, Martin Wortmann, Nadine Schmidt, Bruno Hüsgen, Andreas Hütten, Cornelius Knabbe, Christian Kaltschmidt, and Barbara Kaltschmidt. 2021. "Identification of Microorganisms from Several Surfaces by MALDI-TOF MS: P. aeruginosa Is Leading in Biofilm Formation" Microorganisms 9, no. 5: 992. https://doi.org/10.3390/microorganisms9050992
APA StyleAsghari, E., Kiel, A., Kaltschmidt, B. P., Wortmann, M., Schmidt, N., Hüsgen, B., Hütten, A., Knabbe, C., Kaltschmidt, C., & Kaltschmidt, B. (2021). Identification of Microorganisms from Several Surfaces by MALDI-TOF MS: P. aeruginosa Is Leading in Biofilm Formation. Microorganisms, 9(5), 992. https://doi.org/10.3390/microorganisms9050992








