Evaluation of a New Culture-Based AtbFinder Test-System Employing a Novel Nutrient Medium for the Selection of Optimal Antibiotics for Critically Ill Patients with Polymicrobial Infections within 4 h
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Samples and Laboratory Settings
2.2. Antibiotics
2.3. Bacterial Strains and Growth Conditions
2.4. Biological Specimen Processing and Bacterial Isolation
2.5. Identification of Bacteria in Biological Specimens
2.6. The AtbFinder System and Interpretation of Results
2.7. Gold Standard Definition
2.8. Data Analysis
2.9. Bacterial Diversity Analysis
2.10. Statistics
3. Results
3.1. Comparison of Bacterial Growth Rate on TGV Medium to That on Other Media
3.2. Estimation of the Diversity of Bacteria Grown from Patient Samples on TGV Medium
3.3. Antibiotic Selection in Monomicrobial Cultures Using AtbFinder
3.4. Antibiotic Selection in Polymicrobial Cultures Using AtbFinder
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Leekha, S.; Terrell, C.L.; Edson, R.S. General principles of antimicrobial therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Postma, D.F.; Van Werkhoven, C.H.; Van Elden, L.J.; Thijsen, S.F.; Hoepelman, A.I.; Kluytmans, J.A.; Boersma, W.G.; Compaijen, C.J.; Van Der Wall, E.; Prins, J.M.; et al. Antibiotic treatment strategies for community-acquired pneumonia in adults. N. Engl. J. Med. 2015, 372, 1312–1323. [Google Scholar] [CrossRef]
- Mettler, J.; Simcock, M.; Sendi, P.; Widmer, A.F.; Bingisser, R.; Battegay, M.; Fluckiger, U.; Bassetti, S. Empirical use of antibiotics and adjustment of empirical antibiotic therapies in a university hospital: A prospective observational study. BMC Infect. Dis. 2007, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Braykov, N.P.; Morgan, D.J.; Schweizer, M.L.; Uslan, D.Z.; Kelesidis, T.; Weisenberg, S.A.; Johannsson, B.; Young, H.; Cantey, J.; Srinivasan, A.; et al. Assessment of empirical antibiotic therapy optimisation in six hospitals: An observational cohort study. Lancet Infect. Dis. 2014, 14, 1220–1227. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.; Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar]
- Burnham, C.-A.D.; Leeds, J.; Nordmann, P.; O’Grady, J.; Patel, J. Diagnosing antimicrobial resistance. Nat. Rev. Genet. 2017, 15, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Fu, J. Rapid laboratory diagnosis for respiratory infectious diseases by using MALDI-TOF mass spectrometry. J. Thorac. Dis. 2014, 6, 507–511. [Google Scholar]
- Burns, J.L.; Rolain, J.-M. Culture-based diagnostic microbiology in cystic fibrosis: Can we simplify the complexity? J. Cyst. Fibros. 2014, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cummings, L.A.; Kurosawa, K.; Hoogestraat, D.R.; SenGupta, D.J.; Candra, F.; Doyle, M.; Thielges, S.; Land, T.A.; Rosenthal, C.A.; Hoffman, N.G.; et al. Clinical next generation sequencing outperforms standard microbiological culture for characterizing polymicrobial samples. Clin. Chem. 2016, 62, 1465–1473. [Google Scholar] [CrossRef]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef]
- Domann, E.; Hong, G.; Imirzalioglu, C.; Turschner, S.; Kühle, J.; Watzel, C.; Hain, T.; Hossain, H.; Chakraborty, T. Culture-independent identification of pathogenic bacteria and polymicrobial infections in the genitourinary tract of renal transplant recipients. J. Clin. Microbiol. 2003, 41, 5500–5510. [Google Scholar] [CrossRef] [PubMed]
- Cillóniz, C.; Ewig, S.; Ferrer, M.; Polverino, E.; Gabarrús, A.; De La Bellacasa, J.P.; Mensa, J.; Torres, A. Community-acquired polymicrobial pneumonia in the intensive care unit: Aetiology and prognosis. Crit. Care 2011, 15, R209. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Difrancesco, L.F.; Liapikou, A.; Rinaudo, M.; Carbonara, M.; Bassi, G.L.; Gabarrus, A.; Torres, A. Polymicrobial intensive care unit-acquired pneumonia: Prevalence, microbiology and outcome. Crit. Care 2015, 19, 450. [Google Scholar] [CrossRef]
- Shanmugam, P.; Jeya, M. The bacteriology of diabetic foot ulcers, with a special reference to multidrug resistant strains. J. Clin. Diagn. Res. 2013, 7, 441. [Google Scholar] [CrossRef]
- Sorg, R.A.; Lin, L.; Van Doorn, G.S.; Sorg, M.; Olson, J.; Nizet, V.; Veening, J.-W. Collective resistance in microbial communities by intracellular antibiotic deactivation. PLoS Biol. 2016, 14, e2000631. [Google Scholar] [CrossRef] [PubMed]
- Croxall, G.; Weston, V.; Joseph, S.; Manning, G.; Cheetham, P.; McNally, A. Increased human pathogenic potential of Escherichia coli from polymicrobial urinary tract infections in comparison to isolates from monomicrobial culture samples. J. Med. Microbiol. 2011, 60, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Combes, A.; Figliolini, C.; Trouillet, J.-L.; Kassis, N.; Wolff, M.; Gibert, C.; Chastre, J. Incidence and outcome of polymicrobial ventilator-associated pneumonia. Chest 2002, 121, 1618–1623. [Google Scholar] [CrossRef]
- Mayrand, D. Virulence promotion by mixed bacterial infections. In The Pathogenesis of Bacterial Infections; Springer: Berlin/Heidelberg, Germany, 1985; pp. 281–292. [Google Scholar]
- Salsgiver, E.L.; Fink, A.K.; Knapp, E.A.; LiPuma, J.J.; Olivier, K.N.; Marshall, B.C.; Saiman, L. Changing epidemiology of the respiratory bacteriology of patients with cystic fibrosis. Chest 2016, 149, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Fishman, J.A. Infections in immunocompromised hosts and organ transplant recipients: Essentials. Liver Transplant. 2011, 17, S34–S37. [Google Scholar] [CrossRef]
- Jørgensen, I.M.; Johansen, H.K.; Frederiksen, B.; Pressler, T.; Hansen, A.; Vandamme, P.; Høiby, N.; Koch, C. Epidemic spread of Pandoraea apista, a new pathogen causing severe lung disease in cystic fibrosis patients. Pediatr. Pulmonol. 2003, 36, 439–446. [Google Scholar] [CrossRef]
- Del Castillo, E.; Meier, R.; Chung, M.; Koestler, D.C.; Chen, T.; Paster, B.J.; Charpentier, K.; Kelsey, K.T.; Izard, J.; Michaud, D.S. The Microbiomes of Pancreatic and Duodenum Tissue Overlap and are Highly Subject Specific but Differ between Pancreatic Cancer and Non-Cancer Subjects. Cancer Epidemiol. Prev. Biomark. 2019, 28, 370–383. [Google Scholar] [CrossRef]
- Tetz, V.; Tetz, G. Draft genome sequence of the uropathogenic Herbaspirillum frisingense strain ureolyticus. VT-16-41. Genome Announc. 2017, 5, e00279-17. [Google Scholar] [CrossRef]
- Wang, J.T.; Fang, C.T.; Hsueh, P.R.; Chang, S.C.; Luh, K.T. Spontaneous bacterial empyema caused by Aeromonas veronii biotype sobria. Diagn. Microbiol. Infect. Dis. 2000, 37, 271–273. [Google Scholar] [CrossRef]
- Liu, W.; Røder, H.L.; Madsen, J.S.; Bjarnsholt, T.; Sørensen, S.J.; Burmølle, M. Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front. Microbiol. 2016, 7, 1366. [Google Scholar] [CrossRef]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of ‘unculturable’bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ‘unculturable’human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543. [Google Scholar] [CrossRef]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef]
- Bertelli, C.; Greub, G. Rapid bacterial genome sequencing: Methods and applications in clinical microbiology. Clin. Microbiol. Infect. 2013, 19, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Liu, C.M.; Melendez, J.H.; Frankel, Y.M.; Engelthaler, D.; Aziz, M.; Bowers, J.; Rattray, R.; Ravel, J.; Kingsley, C.; et al. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: Impact of diabetes and antibiotics on chronic wound microbiota. PLoS ONE 2009, 4, e6462. [Google Scholar] [CrossRef] [PubMed]
- Pulido, M.R.; García-Quintanilla, M.; Martín-Peña, R.; Cisneros, J.M.; McConnell, M.J. Progress on the development of rapid methods for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 2710–2717. [Google Scholar] [CrossRef] [PubMed]
- Charnot-Katsikas, A.; Tesic, V.; Love, N.; Hil, B.; Bethel, C.; Boonlayangoor, S.; Beavis, K.G. Use of Accelerate Pheno™ system for identification and antimicrobial susceptibility testing (ID/AST) of pathogens in positive blood cultures and impact on time to results and workflow. J. Clin. Microbiol. 2017, 56, e01166-17. [Google Scholar] [CrossRef]
- Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef]
- Davenport, M.; Mach, K.E.; Shortliffe, L.M.D.; Banaei, N.; Wang, T.H.; Liao, J.C. New and developing diagnostic technologies for urinary tract infections. Nat. Rev. Urol. 2017, 14, 296–310. [Google Scholar] [CrossRef]
- Ellington, M.J.; Ekelund, O.; Aarestrup, F.M.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.T.G.; Hopkins, K.L.; et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Maurer, F.P.; Christner, M.; Hentschke, M.; Rohde, H. Advances in rapid identification and susceptibility testing of bacteria in the clinical microbiology laboratory: Implications for patient care and antimicrobial stewardship programs. Infect. Dis. Rep. 2017, 9, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.V.; Tetz, V.V. Complete genome sequence of Bacilli bacterium strain VT-13-104 isolated from the intestine of a patient with duodenal cancer. Genome Announc. 2015, 3, e00705-15. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lee, F.; Tai, C.; Kasai, H. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA–DNA hybridization in the Bacillus subtilis group. Int. J. Syst. Evol. Microbiol. 2007, 57, 1846–1850. [Google Scholar] [CrossRef]
- Su, M.; Satola, S.W.; Read, T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2018, 57. [Google Scholar] [CrossRef]
- Sundsfjord, A.; Simonsen, G.S.; Haldorsen, B.C.; Haaheim, H.; Hjelmevoll, S.O.; Littauer, P.; Dahl, K.H. Genetic methods for detection of antimicrobial resistance. Apmis 2004, 112, 815–837. [Google Scholar] [CrossRef]
- Martineau, F.; Picard, F.J.; Lansac, N.; Ménard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Correlation between the Resistance Genotype Determined by Multiplex PCR Assays and the Antibiotic Susceptibility Patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2000, 44, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Schön, T.; Miotto, P.; Köser, C.; Viveiros, M.; Böttger, E.; Cambau, E. Mycobacterium tuberculosis drug-resistance testing: Challenges, recent developments and perspectives. Clin. Microbiol. Infect. 2017, 23, 154–160. [Google Scholar] [CrossRef]
- American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 388. [Google Scholar] [CrossRef]
- Mandell, L.A.; Wunderink, R.G.; Anzueto, A.; Bartlett, J.G.; Campbell, G.D.; Dean, N.C.; Dowell, S.F.; File, T.M., Jr.; Musher, D.M.; Niederman, M.S.; et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 2007, 44, S27–S72. [Google Scholar] [CrossRef]
- Kalil, A.C.; Metersky, M.L.; Klompas, M.; Muscedere, J.; Sweeney, D.A.; Palmer, L.B.; Napolitano, L.M.; O’grady, N.P.; Bartlett, J.G.; Carratalà, J.; et al. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin. Infect. Dis. 2016, 63, e61–e111. [Google Scholar] [CrossRef] [PubMed]
- Bergogne-Bérézin, E. Pharmacodynamics of antibiotics in the respiratory tree. Expert Opin. Pharmacother. 2004, 5, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Foulds, G.; Shepard, R.M.; Johnson, R.B. The pharmacokinetics of azithromycin in human serum and tissues. J. Antimicrob. Chemother. 1990, 25, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, C.; MacGowan, A.P. A review of the pharmacokinetics and pharmacodynamics of aztreonam. J. Antimicrob. Chemother. 2016, 71, 2704–2712. [Google Scholar] [CrossRef]
- Beam, T.R.; Galask, R.P.; Friedhoff, L.T.; Platt, T.B.; A Leitz, M. Aztreonam concentrations in human tissues obtained during thoracic and gynecologic surgery. Antimicrob. Agents Chemother. 1986, 30, 505–507. [Google Scholar] [CrossRef] [PubMed]
- Boselli, E.; Breilh, D.; Duflo, F.; Saux, M.-C.; DeBon, R.; Chassard, D.; Allaouchiche, B. Steady-state plasma and intrapulmonary concentrations of cefepime administered in continuous infusion in critically ill patients with severe nosocomial pneumonia. Crit. Care Med. 2003, 31, 2102–2106. [Google Scholar] [CrossRef]
- Panidis, D.; Markantonis, S.L.; Boutzouka, E.; Karatzas, S.; Baltopoulos, G. Penetration of gentamicin into the alveolar lining fluid of critically ill patients with ventilator-associated pneumonia. Chest 2005, 128, 545–552. [Google Scholar] [CrossRef]
- Rodvold, K.A.; Danziger, L.H.; Gotfried, M.H. Steady-state plasma and bronchopulmonary concentrations of intravenous levofloxacin and azithromycin in healthy adults. Antimicrob. Agents Chemother. 2003, 47, 2450–2457. [Google Scholar] [CrossRef] [PubMed]
- Honeybourne, D.; Tobin, C.; Jevons, G.; Andrews, J.; Wise, R. Intrapulmonary penetration of linezolid. J. Antimicrob. Chemother. 2003, 51, 1431–1434. [Google Scholar] [CrossRef]
- Conte, J.E.; Golden, J.A.; Kipps, J.; Zurlinden, E. Intrapulmonary pharmacokinetics of linezolid. Antimicrob. Agents Chemother. 2002, 46, 1475–1480. [Google Scholar] [CrossRef] [PubMed]
- Byl, B.; Jacobs, F.; Roucloux, I.; De Franquen, P.; Cappello, M.; Thys, J.-P. Penetration of meropenem in lung, bronchial mucosa, and pleural tissues. Antimicrob. Agents Chemother. 1999, 43, 681–682. [Google Scholar] [CrossRef] [PubMed]
- Boselli, E.; Breilh, D.; Cannesson, M.; Xuereb, F.; Rimmelé, T.; Chassard, D.; Saux, M.-C.; Allaouchiche, B. Steady-state plasma and intrapulmonary concentrations of piperacillin/tazobactam 4 g/0.5 g administered to critically ill patients with severe nosocomial pneumonia. Intensiv. Care Med. 2004, 30, 976–979. [Google Scholar] [CrossRef]
- Cruciani, M.; Gatti, G.; Lazzarini, L.; Furlan, G.; Broccali, G.; Malena, M.; Franchini, C.; Concia, E. Penetration of vancomycin into human lung tissue. J. Antimicrob. Chemother. 1996, 38, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Kullberg, B.J.; Vasquez, J.; Mootsikapun, P.; Nucci, M.; Paiva, J.-A.; Garbino, J.; Yan, J.L.; Aram, J.; Capparella, M.R.; Conte, U.; et al. Efficacy of anidulafungin in 539 patients with invasive candidiasis: A patient-level pooled analysis of six clinical trials. J. Antimicrob. Chemother. 2017, 72, 2368–2377. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, T.; Hieda, J.; Bratescu, M.A.; Saito, N.; Takai, O. Biomimetic materials processing. Nanostructured Thin Films II 2009, 7404, 74040M. [Google Scholar] [CrossRef]
- Kageyama, A.; Benno, Y. Rapid detection of human fecal Eubacterium species and related genera by nested PCR method. Microbiol. Immunol. 2001, 45, 315–318. [Google Scholar] [CrossRef]
- Wilson, D.A.; Reischl, U.; Hall, G.S.; Procop, G.W. Use of partial 16S rRNA gene sequencing for identification of Legionella pneumophila and non-pneumophila Legionella spp. J. Clin. Microbiol. 2006, 45, 257–258. [Google Scholar] [CrossRef][Green Version]
- Chun, J.; Lee, J.H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K.; Lim, Y.W. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar] [CrossRef]
- National Committee for Clinical Laboratory Standards. Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline M26-A; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 1999. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 9th ed.; Approved Standard M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI supplement M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Parikh, R.; Mathai, A.; Parikh, S.; Sekhar, G.C.; Thomas, R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008, 56, 45–50. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Delhaes, L.; Monchy, S.; Fréalle, E.; Hubans, C.; Salleron, J.; Leroy, S.; Prevotat, A.; Wallet, F.; Wallaert, B.; Dei-Cas, E.; et al. The airway microbiota in cystic fibrosis: A complex fungal and bacterial community—Implications for therapeutic management. PLoS ONE 2012, 7, e36313. [Google Scholar] [CrossRef]
- Michalopoulos, A.; Kasiakou, S.K.; Mastora, Z.; Rellos, K.; Kapaskelis, A.M.; E Falagas, M. Aerosolized colistin for the treatment of nosocomial pneumonia due to multidrug-resistant Gram-negative bacteria in patients without cystic fibrosis. Crit. Care 2005, 9, R53–R59. [Google Scholar] [CrossRef] [PubMed]
- Firmida, M.C.; Pereira, R.H.; Silva, E.A.; Marques, E.A.; Lopes, A.J. Clinical impact of Achromobacter xylosoxidans colonization/infection in patients with cystic fibrosis. Braz. J. Med. Biol. Res. 2016, 49, e5097. [Google Scholar] [CrossRef]
- Glass, M.B.; Gee, J.E.; Steigerwalt, A.G.; Cavuoti, D.; Barton, T.; Hardy, R.D.; Godoy, D.; Spratt, B.G.; Clark, T.A.; Wilkins, P.P. Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J. Clin. Microbiol. 2006, 44, 4601–4604. [Google Scholar] [CrossRef]
- Han, J.K.; Kerschner, J.E. Streptococcus milleri: An organism for head and neck infections and abscess. Arch. Otolaryngol. Head Neck Surg. 2001, 127, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Parkins, M.D.; Sibley, C.D.; Surette, M.G.; Rabin, H.R. The Streptococcus milleri group—An unrecognized cause of disease in cystic fibrosis: A case series and literature review. Pediatr. Pulmonol. 2008, 43, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Yakut, N.; Kepenekli, E.K.; Karaaslan, A.; Atici, S.; Akkoc, G.; Demir, S.O.; Soysal, A.; Bakir, M. Bacteremia due to Acinetobacter ursingii in infants: Reports of two cases. Pan Afr. Med. J. 2016, 23. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Tetz, V. Draft Genome Sequence of Chryseobacterium mucoviscidosis sp. nov. Strain VT16-26, Isolated from the Bronchoalveolar Lavage Fluid of a Patient with Cystic Fibrosis. Genome Announc. 2018, 6, e01473-17. [Google Scholar] [CrossRef]
- Tetz, V.; Tetz, G. Draft genome sequence of Bacillus obstructivus VT-16-70 isolated from the bronchoalveolar lavage fluid of a patient with chronic obstructive pulmonary disease. Genome Announc. 2017, 5, e01754-16. [Google Scholar] [CrossRef] [PubMed]
- Galera-Laporta, L.; Garcia-Ojalvo, J. Antithetic population response to antibiotics in a polybacterial community. Sci. Adv. 2020, 6, eaaz5108. [Google Scholar] [CrossRef]
- Van Belkum, A.; Bachmann, T.T.; Lüdke, G.; Lisby, J.G.; Kahlmeter, G.; Mohess, A.; Becker, K.; Hays, J.P.; Woodford, N.; Mitsakakis, K.; et al. Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2019, 17, 51–62. [Google Scholar] [CrossRef]
- Battleman, D.S.; Callahan, M.; Thaler, H.T. Rapid antibiotic delivery and appropriate antibiotic selection reduce length of hospital stay of patients with community-acquired pneumonia: Link between quality of care and resource utilization. Arch. Intern. Med. 2002, 162, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.J.; Belitsos, N.J.; Hamilton, S.R. Bacterial esophagitis in immunocompromised patients. Arch. Intern. Med. 1986, 146, 1345–1348. [Google Scholar] [CrossRef] [PubMed]
- Torre, D.; Crespi, E.; Bernasconi, M.; Rapazzini, P. Urinary tract infection caused by Kluyvera ascorbata in an immunocompromised patient: Case report and review. Scand. J. Infect. Dis. 2005, 37, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Montero, J.G.; Paiva, J.A. When antibiotic treatment fails. Intensiv. Care Med. 2017, 44, 73–75. [Google Scholar] [CrossRef]
- Samanta, S.; Poddar, B.; Azim, A.; Singh, R.K.; Gurjar, M.; Baronia, A.K. Significance of Mini Bronchoalveolar Lavage Fluid Amylase Level in Ventilator-Associated Pneumonia: A Prospective Observational Study. Crit. Care Med. 2018, 46, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Tetz, V. Introducing the sporobiota and sporobiome. Gut Pathog. 2017, 9, 1–6. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef]
- Le Dorze, M.; Gault, N.; Foucrier, A.; Ruppé, E.; Mourvillier, B.; Woerther, P.L.; Birgand, G.; Montravers, P.; Dilly, M.P.; Tubach, F.; et al. Performance and impact of a rapid method combining mass spectrometry and direct antimicrobial susceptibility testing on treatment adequacy of patients with ventilator-associated pneumonia. Clin. Microbiol. Infect. 2015, 21, 468.e1–468.e6. [Google Scholar] [CrossRef]
- Toosky, M.N.; Grunwald, J.T.; Pala, D.; Shen, B.; Zhao, W.; D’Agostini, C.; Coghe, F.; Angioni, G.; Motolese, G.; Abram, T.J.; et al. A rapid, point-of-care antibiotic susceptibility test for urinary tract infections. J. Med. Microbiol. 2020, 69, 52–62. [Google Scholar] [PubMed]
- Kasas, S.; Malovichko, A.; Villalba, M.; Vela, M.; Yantorno, O.; Willaert, R. Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing. Antibiotics 2021, 10, 287. [Google Scholar] [CrossRef]
- Blot, S.I.; Pea, F.; Lipman, J. The effect of pathophysiology on pharmacokinetics in the critically ill patient—Concepts appraised by the example of antimicrobial agents. Adv. Drug Deliv. Rev. 2014, 77, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Büdingen, F.V.; Gonzalez, D.; Tucker, A.N.; Derendorf, H. Relevance of liver failure for anti-infective agents: From pharmacokinetic alterations to dosage adjustments. Ther. Adv. Infect. Dis. 2014, 2, 17–42. [Google Scholar] [CrossRef] [PubMed]
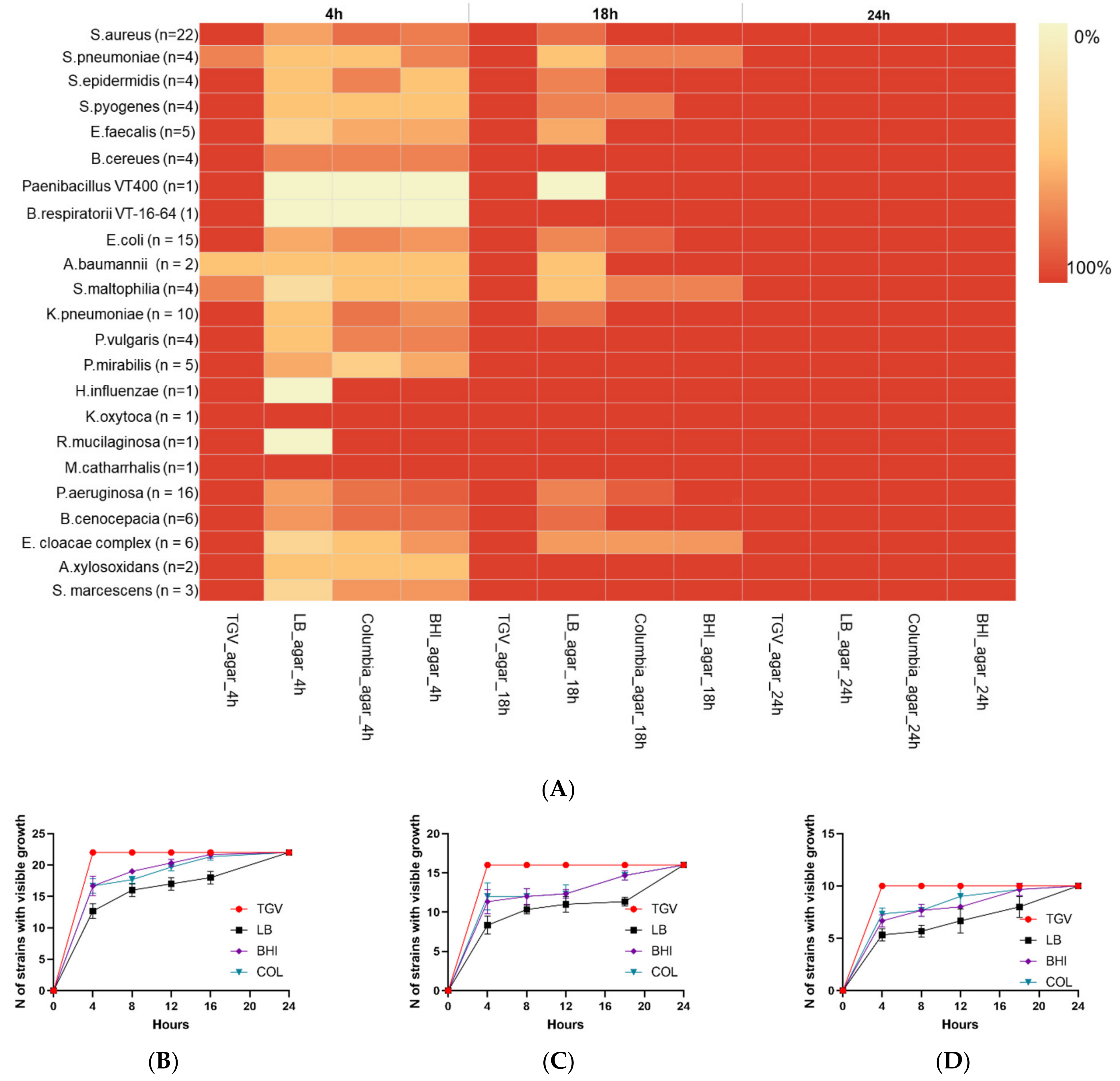
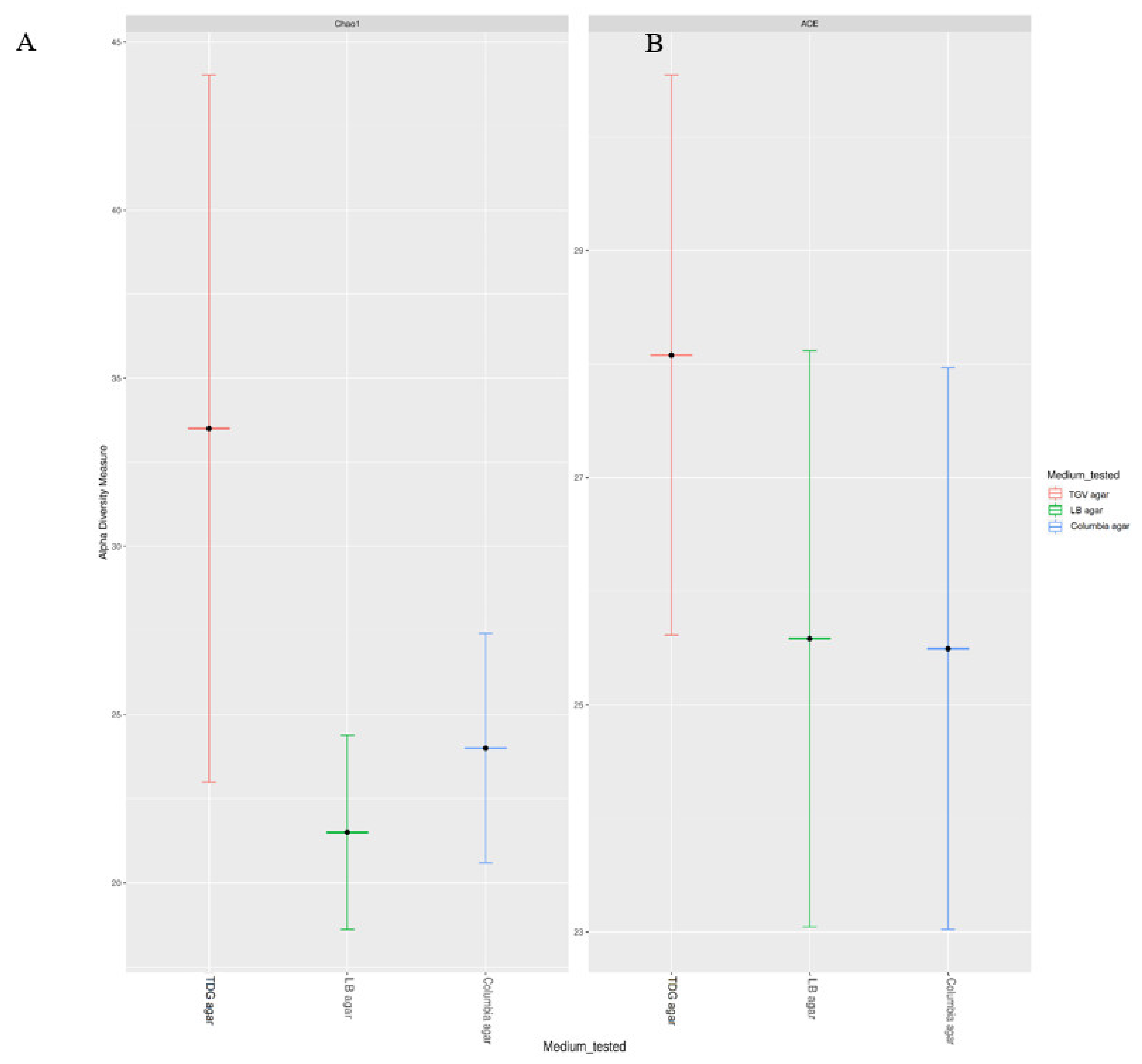
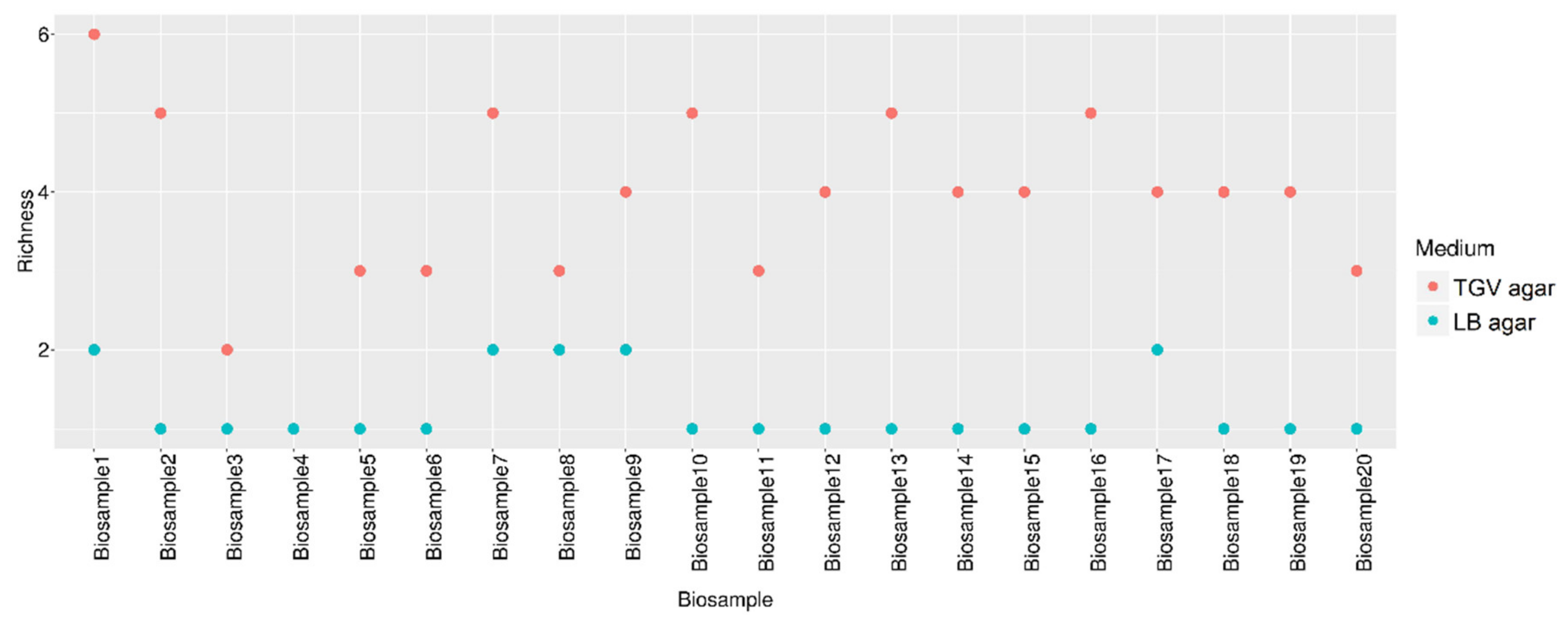
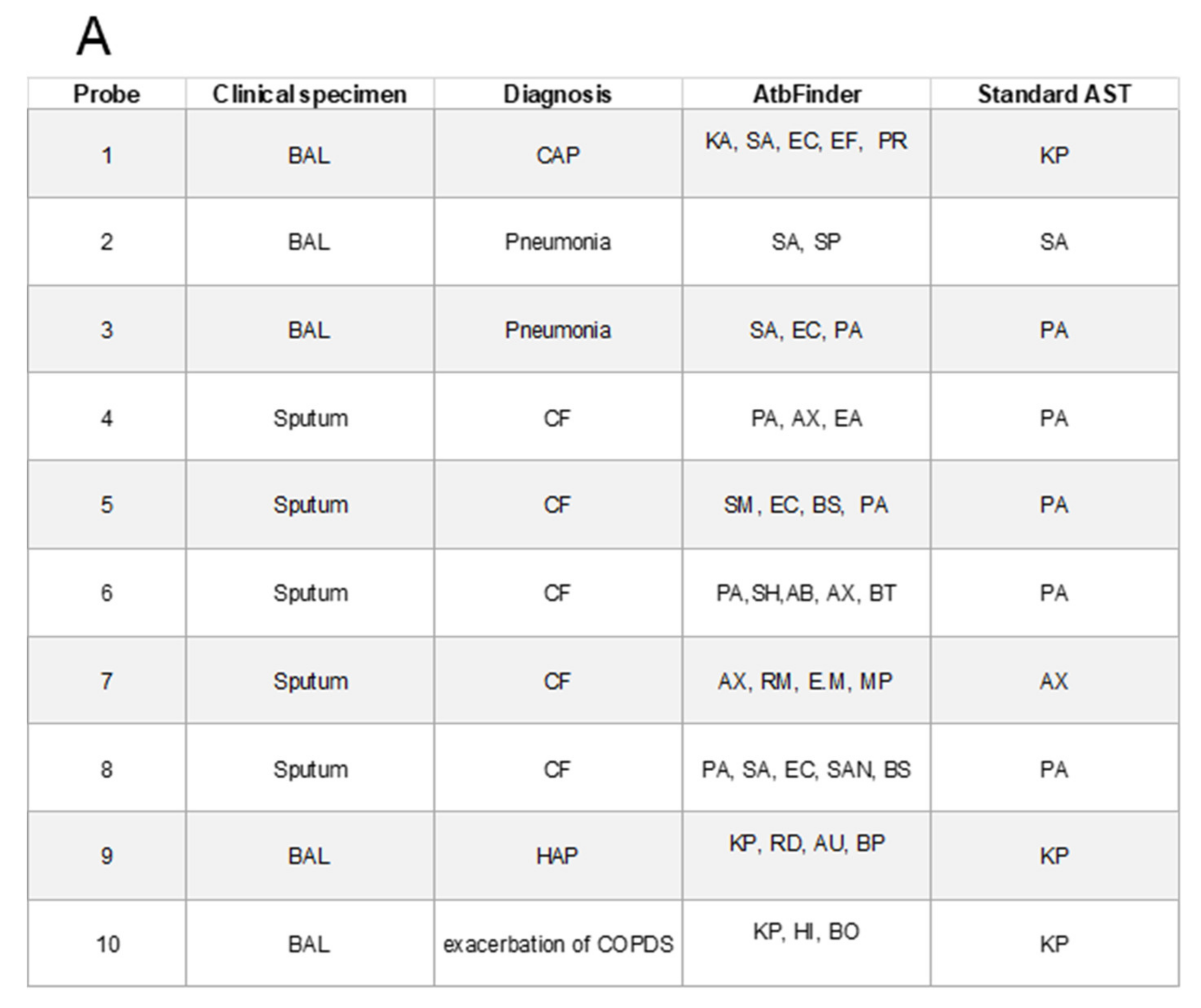


| # | Clinical Specimen | Diagnosis | Bacteria that Showed Growth on TGV Medium | Bacteria Identified with Standard Media * | ||
|---|---|---|---|---|---|---|
| 4 h | 24 h | 4 h | 24 h | |||
| 1 | BAL | COPD | Burkholderia gladioli Neisseria spp. Haemophilus parainfluenzae S. mitis R. mucilaginosa Streptomyces violaceruber | B. gladioli Neisseria spp. H. parainfluenzae S. mitis R. mucilaginosa S. violaceruber | - | S. mitis R. mucilaginosa |
| 2 | BAL | COPD | S. aureus K. pneumoniae E. coli E. faecalis Proteus spp. | S. aureus K. pneumoniae E. coli E. faecalis Proteus spp. | K. pneumoniae | K. pneumoniae |
| 3 | BAL | CAP | S. aureus S. pneumoniae | S. aureus S. pneumoniae | - | S. aureus |
| 4 | BAL | CAP | S. aureus | S. aureus | S. aureus | S. aureus |
| 5 | BAL | CAP | P. aeruginosa H. influenza M. catarrhalis | P. aeruginosa H. influenza M. catarrhalis | P. aeruginosa | P. aeruginosa |
| 6 | BAL | VAP | S. aureus E. cloacae P. aeruginosa | S. aureus E. cloacae P. aeruginosa | - | P. aeruginosa |
| 7 | BAL | VAP | S. aureus K. pneumoniae E. coli C. mucoviscidosis S. aureus | S. aureus K. pneumoniae E. coli C. mucoviscidosis S. aureus | - | S. aureus E. coli |
| 8 | Sputum | CF | P. aeruginosa S. aureus R. mucilaginosa | P. aeruginosa S. aureus R. mucilaginosa | P. aeruginosa | P. aeruginosa S. aureus |
| 9 | Sputum | CF | P. aeruginosa S. aureus S. maltophilia B. cepacia | P. aeruginosa S. aureus S. maltophilia B. cepacia | - | P. aeruginosa B. cepacia |
| 10 | Sputum | CF | P. aeruginosa Micrococcus luteus S. aureus S. oralis S. milleri | P. aeruginosa M. luteus S. aureus S. oralis S. milleri | - | P. aeruginosa |
| 11 | Sputum | CF | P. aeruginosa A. xylosoxidans E. coli | P. aeruginosa A. xylosoxidans E. coli | P. aeruginosa | P. aeruginosa |
| 12 | Sputum | CF | S. maltophilia E. coli Bacillaceae spp. P. aeruginosa | S. maltophilia E. coli Bacillaceae spp. P. aeruginosa | P. aeruginosa | P. aeruginosa |
| 13 | Sputum | CF | P. aeruginosa S. haemoliticus A. baumannii A. xylosoxidans B. thailandensis | P. aeruginosa S. haemoliticus A. baumannii A. xylosoxidans B. thailandensis | - | P. aeruginosa |
| 14 | Sputum | CF | B. multivorans Corynebacterium pseudodiphtheriticum Moraxella catharrhalis Paenibacillus pabuli | B. multivorans C. pseudodiphtheriticum M. catharrhalis P. pabuli | - | C. pseudodiphtheriticum |
| 15 | Sputum | CF | A. xylosoxidans R. mucilaginosa Elizabethkingia miricola Microbacterium paraoxydans | A. xylosoxidans R. mucilaginosa E. miricola M.paraoxydans | - | A. xylosoxidans |
| 16 | Sputum | CF | P. aeruginosa S. aureus E. coli S. anginosus B. sonorensis | P. aeruginosa S. aureus E. coli S. anginosus B. sonorensis | P. aeruginosa | P. aeruginosa |
| 17 | Sputum | CF | P. aeruginosa S. aureus E. coli B. cereus | P. aeruginosa S. aureus E. coli B. cereus | P. aeruginosa | P. aeruginosa S. aureus |
| 18 | BAL | HAP | K. pneumoniae Rothia dentocariosa A.ursingii Bacillus pumilus | K. pneumoniae R. dentocariosa A. ursingii B. pumilus | - | K. pneumoniae |
| 19 | BAL | CAP | H. influenzae Actinobacillus suis Eikenella corrodens Actinomyces oris | H. influenzae A. suis E. corrodens A.oris | - | H. influenzae |
| 20 | BAL | COPD | K. pneumoniae H. influenza Bacillus obstructivus | K. pneumoniae H. influenza Bacillus obstructivus | - | K. pneumoniaee |
| Species | N True Positive | N True Negative | N False Positive | N False Negative | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | |
| S. aureus | 134 | 134 | 84 | 84 | 1 | 1 | 1 | 1 | 99.3 | 99.3 | 98.8 | 98.8 | 99.3 | 99.3 | 98.8 | 98.8 |
| S. pneumoniae | 12 | 16 | 18 | 24 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| S. epidermidis | 16 | 17 | 23 | 23 | 1 | 0 | 0 | 0 | 100 | 100 | 95.8 | 100 | 94.1 | 94.1 | 100 | 100 |
| S. pyogenes | 10 | 10 | 30 | 30 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| E. faecalis | 27 | 27 | 22 | 22 | 1 | 1 | 0 | 0 | 100 | 100 | 95.6 | 95.6 | 96.4 | 96.4 | 100 | 100 |
| B. cereues | 17 | 17 | 23 | 23 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Paenibacillus VT400 | 5 | 5 | 4 | 4 | 1 | 1 | 0 | 0 | 100 | 100 | 80 | 80 | 83.3 | 83.3 | 100 | 100 |
| B. respiratorii VT1664 | 3 | 3 | 7 | 7 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| E. coli | 70 | 71 | 78 | 78 | 2 | 1 | 0 | 0 | 100 | 100 | 97.5 | 98.8 | 97.2 | 97.2 | 100 | 100 |
| A. baumannii | 8 | 16 | 1 | 4 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| S. maltophilia | 19 | 28 | 10 | 11 | 1 | 1 | 0 | 0 | 100 | 100 | 90.1 | 91.6 | 95 | 95 | 100 | 100 |
| K. pneumoniae | 51 | 51 | 47 | 47 | 1 | 1 | 1 | 1 | 98.1 | 98.1 | 97.9 | 97.9 | 98.1 | 98.1 | 97.9 | 97.9 |
| P. vulgaris | 18 | 18 | 22 | 22 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| P. mirabilis | 22 | 22 | 28 | 28 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| H. influenzae | 2 | 2 | 8 | 8 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| K. oxytoca | 6 | 6 | 4 | 4 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| R. mucilaginosa | 4 | 4 | 6 | 6 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| M. catharrhalis | 4 | 4 | 6 | 6 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| P. aeruginosa | 122 | 122 | 35 | 35 | 2 | 2 | 1 | 1 | 99.2 | 99.2 | 94.6 | 94.6 | 98.4 | 98.4 | 97.2 | 97.2 |
| B. cenocepacia | 53 | 53 | 7 | 7 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| E. cloacae complex | 31 | 31 | 29 | 29 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| A. xylosoxidans | 16 | 16 | 4 | 4 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| S. marcescens | 14 | 14 | 16 | 16 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Gram-positive bacteria | 403 | 408 | 259 | 265 | 6 | 5 | 2 | 2 | 99.9 | 99.9 | 96.8 | 97.2 | 97.4 | 97.4 | 99.6 | 99.6 |
| Gram-negative bacteria | 437 | 455 | 295 | 298 | 14 | 14 | 2 | 2 | 99.8 | 99.8 | 98.6 | 98.8 | 99.2 | 99.2 | 99.7 | 99.7 |
| Antibiotic | Category Agreement: No of Correct Identifications/Total No of Observations (%) | No of Errors | ||||
|---|---|---|---|---|---|---|
| Very Major (False-Positive) | Major (False-Negative) | |||||
| 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | |
| Piperacillin-tazobactam | 118/119 (99.2) | 122/122 (100) | 1 | 0 | 0 | 0 |
| Meropenem | 116/119 (97.4) | 119/122 (97.5) | 2 | 2 | 1 | 1 |
| Levofloxacin | 119/119 (100) | 122/122 (100) | 0 | 0 | 0 | 0 |
| Aztreonam | 118/119 (99.2) | 121/122 (99.2) | 1 | 1 | 0 | 0 |
| Gentamicin | 118/119 (99.2) | 121/122 (99.2) | 0 | 0 | 1 | 1 |
| Amikacin | 117/119 (98.3) | 120/122 (98.3) | 2 | 2 | 0 | 0 |
| Azithromycin | 117/119 (98.3) | 121/122 (99.2) | 2 | 1 | 0 | 0 |
| Vancomycin | 119/119 (100) | 122/122 (100) | 0 | 0 | 0 | 0 |
| Cefepime | 117/119 (98.3) | 120/122 (98.3) | 2 | 2 | 0 | 0 |
| Linezolid | 118/119 (99.2) | 121/122 (99.2) | 0 | 0 | 1 | 1 |
| Overall performance | 1177/1190 (98.9) | 1209/1220 (99.1) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tetz, G.; Tetz, V. Evaluation of a New Culture-Based AtbFinder Test-System Employing a Novel Nutrient Medium for the Selection of Optimal Antibiotics for Critically Ill Patients with Polymicrobial Infections within 4 h. Microorganisms 2021, 9, 990. https://doi.org/10.3390/microorganisms9050990
Tetz G, Tetz V. Evaluation of a New Culture-Based AtbFinder Test-System Employing a Novel Nutrient Medium for the Selection of Optimal Antibiotics for Critically Ill Patients with Polymicrobial Infections within 4 h. Microorganisms. 2021; 9(5):990. https://doi.org/10.3390/microorganisms9050990
Chicago/Turabian StyleTetz, George, and Victor Tetz. 2021. "Evaluation of a New Culture-Based AtbFinder Test-System Employing a Novel Nutrient Medium for the Selection of Optimal Antibiotics for Critically Ill Patients with Polymicrobial Infections within 4 h" Microorganisms 9, no. 5: 990. https://doi.org/10.3390/microorganisms9050990
APA StyleTetz, G., & Tetz, V. (2021). Evaluation of a New Culture-Based AtbFinder Test-System Employing a Novel Nutrient Medium for the Selection of Optimal Antibiotics for Critically Ill Patients with Polymicrobial Infections within 4 h. Microorganisms, 9(5), 990. https://doi.org/10.3390/microorganisms9050990






