Abstract
The respiratory chain is very important for bacterial survival and pathogenicity, yet the roles of the respiratory chain in P. aeruginosa remain to be fully elucidated. Here, we not only proved experimentally that the operon PA4427-PA4431 of Pseudomonas aeruginosa PAO1 encodes respiratory chain complex III (cytobc1), but also found that it played important roles in virulence and pathogenicity. PA4429–31 deletion reduced the production of the virulence factors, including pyocyanin, rhamnolipids, elastase, and extracellular polysaccharides, and it resulted in a remarkable decrease in pathogenicity, as demonstrated in the cabbage and Drosophila melanogaster infection models. Furthermore, RNA-seq analysis showed that PA4429–31 deletion affected the expression levels of the genes related to quorum-sensing systems and the transport of iron ions, and the iron content was also reduced in the mutant strain. Taken together, we comprehensively illustrated the function of the operon PA4427–31 and its application potential as a treatment target in P. aeruginosa infection.
1. Introduction
The respiratory chain, including the aerobic and anaerobic respiratory branches, is one of the most key supplying energy pathways for cellular metabolism. The shared main respiratory equipment of the aerobic and anaerobic respiration pathways contains NADH dehydrogenase (complexes I), coenzyme Q, cytochrome bc1 complex (complex III), and cytochrome c. In addition, in Pseudomonas aeruginosa, the aerobic chain contains terminal oxidases (complex IV, e.g., panthenol and cytochrome c oxidases), and the anaerobic respiratory chain possesses a series of NOx reductases (e.g., nitrate, nitrite, nitric oxide, and nitrous oxide reductases) [1]. Protons are pumped out across the bacterial membrane during electron transfer through complexes I, III, and IV, which form the proton gradient for ATP synthesis [2]. The bc1 complex in the center of the respiratory chain is responsible for the electrons transferring from coenzyme Q to cytochrome c [3]. All bc1 complexes contain three redox subunits: cyt b (two b-type hemes: bL and bH), cyt c1 (a c-type heme), and ISP (two Fe-S clusters) [4].
Since the respiratory chain is indispensable in the energy metabolism of all living organisms, mutation of the respiratory chain complexes can lead to various severe physiological defects [5]. For example, damage to the mitochondrial respiratory chain complex [6] results in membrane potential loss [7], a reduction of ATP production [8], age-related neurodegenerative and metabolic syndrome, and even cancer [6]. The respiratory chain has also been reported to play a crucial role in the viability and virulence of bacteria. Functionality impairment of the respiratory chain causes a rapid loss of cell viability in Mycobacterium tuberculosis [9]. Inactivation of complex I of the respiratory chain disrupts the aerobic respiration and virulence of some microbes, like Streptococcus agalactiae [10], S. sanguinis, and S. pneumoniae [11,12].
The importance of the respiratory chain’s role makes it the main target of various natural and synthetic antimicrobial drugs for both eukaryotic and prokaryotic cells [13]. Several compounds have been reported to be complex III inhibitors in some eukaryotes, like atovaquone against Toxoplasma gondii and Pneumocystis sp. and 2-hydroxy-naphthoquinone, pyridones, quinolones, acridones, acridine diones, and rhinacanthin against malaria parasites [14]. Amilorides, the well-known inhibitors of Na+/H+ and Na+/Ca2+ antiporters and Na+ channels, have also been reported to be able to inhibit bacterial and mitochondrial NADH-quinone oxidoreductase (complex I) [15]. Cytochrome bc1 of M. tuberculosis is inhibited by a new small molecule substance Q203, which has become a clinical drug target for M. tuberculosis infection treatment [16].
P. aeruginosa, a ubiquitous Gram-negative bacterium, is able to survive in various environments, such as soils, waters, or medical device surfaces [17]. It rarely affects healthy individuals but causes high morbidity and mortality in cystic fibrosis (CF) patients and immunocompromised individuals [18]. Treatment of P. aeruginosa infections has become a great challenge, due to its high innate and acquired resistance to many available antibiotics [19]. Discovery of new specific drug targets is helpful for P. aeruginosa infection treatment. The multibranched respiratory chain could be a potential target of P. aeruginosa infection treatment because it not only produces more than 90% of cellular ATP but also enhances the adaptability of this bacterium [2,14]. According to genomic sequence analysis, there are at least 17 kinds of dehydrogenases [20], 3 kinds of cytochrome c, and 5 kinds of terminal oxidase in its genome [21]. Among them, the genes PA4429-PA4431 have been predicted to code cytochrome bc1, according to the KEGG pathway database (https://www.kegg.jp/kegg/pathway.html) (accessed on 1 May 2019) [22] and the P. aeruginosa genome database (http://www.pseudomonas.com) (accessed on 1 May 2019) [23]. However, very limited experimental evidence is available regarding the identification of cytbc1 in P. aeruginosa PAO1, as well as its roles in pathogenicity.
In our previous work, a mutant with a threefold increased resistance to aminoglycoside antibiotics was screened out from a random transposon mutant library [24]. In this report, the transposon insertion site was found to be located in the gene PA4431. PA4429–31 were experimentally demonstrated to encode cytbc1. Next, the effects of PA4427–31 on virulence and pathogenicity were investigated by cabbage and Drosophila melanogaster infection models, and the reasons for the reduction of pathogenicity were illustrated by RNA-Seq analysis and molecular experiments. Taken together, all the genetic and biochemical evidence proved the important contributions of cytbc1 to the virulence and pathogenicity of P. aeruginosa PAO1, indicating that it might be used as a potential therapeutic target for P. aeruginosa infection.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
The P. aeruginosa and Escherichia coli strains listed in the Table 1 were routinely cultured using Luria–Bertani (LB) agar plates or broth at 37 °C. In addition, Pseudomonas isolate agar (PIA) plates were also used for P. aeruginosa growth. When necessary, tetracycline (Tc, 15 μg/mL), kanamycin (Kan, 50 μg/mL), or gentamicin (Gm, 15 μg/mL) were used for E. coli, and tetracycline (Tc, 300 μg/mL), gentamicin (Gm, 150 μg/mL) or carbenicillin (Cb, 250 μg/mL) was used for P. aeruginosa. All antibiotics used were purchased from Amresco (Solon, PA, USA).

Table 1.
The strains and plasmids used in this study.
2.2. RT-PCR
The various mutants were grown in LB broth, and the total RNA was extracted using an RNA extraction kit (RNA prep Pure Cell/Bacteria Kit, TIANGEN, Beijing, China). Then, 1 μg of RNA and random PCR primers (Table 2) were used to synthesize cDNA with a Prime Script TM RT reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. To determine the operon profile, different primer sets were designed and used for PCR (Table 2), and the non-retrotranscribed mRNA was used as a negative control.

Table 2.
The primers used in this study.
2.3. Construction of Mutants
A sacB-based method was used to construct the in-frame deletion mutants [25]. Taking PAO1(ΔPA4429) as an example, the 0.78 kb upstream fragment of PA4429 was amplified using a forward primer PA4429-UP-S and a reverse primer PA4429-UP-A containing EcoR I and Sac I restriction sites, respectively (Table 2). The 0.75 kb downstream region of PA4429 was generated with the forward primer PA4429-D-S and the reverse primer PA4429-D-A containing the Sac I and Kpn I sites, respectively. The upstream and downstream fragments of PA4429 were digested and then successively ligated into pEX18Tc to generate the plasmid pEX18Tc-PA4429. PAO1(ΔPA4429) was obtained by triparental mating as reported previously [26]. In brief, PAO1, E. coli DH10B containing pEX18Tc-PA4429, and E. coli DH10B containing pRK2013 were cultured overnight, collected, washed with PBS, and finally suspended in SOC medium. The bacteria were mixed at a ratio of 1:1:1 in a microcentrifuge tube and then spotted onto a LB agar plate. After 12 h of growth at 37 °C, the bacteria were resuspended in 500 mL of LB medium and then plated onto two PIA plates with a tetracycline addition. After being cultured on 10% sucrose-containing PIA agar, the resultant mutants were selected and verified by PCR.
2.4. Complementation of the In-Frame Deletion Mutants
To complement the PA4429 deletion, PA4429, PA4429–31, and PA4427–31 were respectively PCR amplified and ligated into the pAK1900 vector (Biovector NTCC Inc. Beijing, China). Taking PAO1(ΔPA4429)C1 as an example, PA4429 was PCR amplified by the forward primer and the reverse primer with the Hind Ⅲ and Bam HⅠ restriction sites, respectively. The fragment was ligated into pAK1900 to form a new plasmid pAK4429, which was next transferred into E. coli DH10B. The transfer of pAK4429 into the PAO1(ΔPA4429) was performed by electroporation. The integrants were selected on PIA plates containing 250 μg/mL Cb. The resultant strain was named PAO1(ΔPA4429)C1. Similarly, PAO1(ΔPA4430)C1, PAO1(ΔPA4430)C2, PAO1(ΔPA4430)C3, PAO1(ΔPA4431)C1, PAO1(ΔPA4431)C2, PAO1(ΔPA4431)C3, PAO1(ΔPA4429–31)C1, PAO1(ΔPA4429–31)C2, and PAO1(ΔPA4429–31)C3 were respectively constructed (Table 1).
2.5. The Disk Diffusion and Antibiotics Susceptibility Tests
For disk diffusion test, 10 μL of the 50 μg/mL antibiotic was spotted on filter disks with a 6-mm diameter, which were placed on LB agar plates with 40 μL of bacterial cells with 0.5 OD600. The plates were incubated at 37 °C for 24 h. The diameters of the inhibition zones around the antimicrobial agents were observed [27].
The minimum inhibitory concentrations (MICs) of the antibiotics were determined by twofold dilutions of the bacterial culture in 96-well plates. In brief, overnight bacterial cultures were diluted at 1:150 in LB, added to the wells containing different concentrations of antibiotics and incubated at 37 °C for 24 h. The MICs were recorded as the lowest concentrations of the antibiotics by measuring the growth after 24 h of incubation at 37 °C.
2.6. Biofilm Assay
The biofilm formation of the various mutant strains was compared with the wild-type strain, as previously described in [28]. In brief, after the various mutant strains were grown in LB overnight, 1 μL of culture was inoculated into 99 μL of LB broth in a 96-well polyvinyl chloride plate (Corning-Costar, NY, USA) and kept at 37 °C for 12 h. After the 96-well plate was stained by using 0.1% crystal violet for 10 min and washed 5 times with water, it was visualized and compared.
2.7. ATP Measurement
The amount of ATP was measured by the luciferin luciferase method, following the protocol of the ATP detection kit (Beyotime, Beijing, China) [29]. After being cultured in LB medium for 10 h, PAO1, PAO1(Δ4429–31) and PAO1(Δ4429–31)C were adjusted to an OD600 value of 1.0. Then, 2 mL of each cell culture was collected and centrifuged at 12,000 rpm for 5 min. The pellets were treated with 200 μL of a lysis buffer from the ATP detection kit. After being centrifuged at 13,000 rpm for 5 min at 4 °C, the supernatant was transferred to a new centrifuge tube for ATP testing. The luminescence from a 100 μL sample was assayed in a Victor2 multilabel counter (Wallac model 1450, Perkin-Elmer, Waltham, MA, USA) together with 100 μL of an ATP detection buffer from the ATP detection kit [30].
2.8. Assays of Pyocyanin, Rhamnolipid, Extracellular Polysaccharide, Elastase, and Motility
Pyocyanin was extracted from the culture supernatants of PAO1, PAO1(Δ4429–31), and PAO1(Δ4429–31)C and measured as previously described in [31]. Briefly, 3 mL of chloroform was added to 5 mL of the culture supernatant, vigorously vortexed for 2 min, and then centrifuged for 10 min at 8,000 rpm. After extraction, the chloroform layer was transferred to a fresh tube and mixed with 1 mL 0.2 M HCI. Then, the top layer was removed, and the OD520 was measured. The amount of pyocyanin, in μg/mL, was calculated using the following formula: OD520/OD600 × 17.072 =μg of pyocyanin per mL.
The rhamnolipid was analyzed according to the previous report [32]. For rhamnolipid extraction, P. aeruginosa PAO1, PAO1(Δ4429–31), and PAO1(Δ4429–31)C were grown at 37 °C for 24 h in LB medium. After incubation, the bacterial culture was centrifuged at 10,000 rpm for 5 min. The cell-free supernatant was mixed with an equal volume of ethyl acetate and vigorously vortexed to obtain rhamnolipid in its organic phase. The rhamnolipid extract was dissolved in 4 mL chloroform and put in 200 µL of freshly prepared 1 g/L methylene blue solution (pH 8.6) and 4.9 mL of distilled water. After vortexing for 4 min and standing for 15 min, the absorbance of the solution at 638 nm was measured, and the values were converted to rhamnolipid concentrations using a calibration curve.
An extracellular polysaccharide assay was performed by examining the colony color in the Congo Red agar plates (1% tryptone, 1% agar, 4% Congo Red, and 2% Coomassie brilliant blue) [28]. After being grown overnight, 2 µL of culture was inoculated in the plates and kept at 37 °C overnight. The plates were visualized and analyzed.
Elastase examination was carried out according to the previous method [33]. The overnight culture in LB was centrifugated, and 1 mL of supernatant was added to a new tube containing 5 mg Elastin-Congo Red and 2 mL of a 0.1 M phosphate buffer (pH 7.0) (Sigma, Saint Louis, MO, USA). After being kept at 37 °C for 3 h, the absorbance at 495 nm was measured.
The motility was assayed in different media [28]. Briefly, for the swarming and swimming test, 2 µL of overnight culture in LB broth was inoculated in the swarming (0.8% BD nutrient broth, 0.5% glucose, and 0.5% agar) and swimming (1% tryptone, 0.5% yeast extract, 1% NaCl, and 0.3% agar) plates, respectively. After keeping the overnight culture at 37 °C, the swarming and swimming zones were examined. The twitching motility was examined by stab-inoculating strains through a thin LB (1% tryptone, 0.5% yeast extract, 1% NaCl, and 1% agar) plate. After being kept at 30 °C for 24–48 h, the twitching zones were visualized at the agar plate interface by using 0.1% crystal violet.
2.9. In Vivo Virulence Assays
The virulence of PAO1 and PAO1(Δ4429–31) was investigated with cabbage (Brassica pekinensis) and fruitfly (Drosophila melanogaster Canton S) infection models, as reported previously in [34]. In brief, for the cabbage infection model, bacterial strains were cultured overnight in M9 broth (22 mM Na2HPO4, 22 mM KH2PO4, 85 mM NaCl, 0.1% NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, and 0.4% glucose) at 37 °C. Cells were collected and rinsed with 10 mM MgSO4. After adjusting the OD600 to 0.5, 10 mL of the dilution was injected into the stems with a micropipette [26]. The cabbage leaves were incubated at room temperature and assessed daily for visual signs of infection. For the fruit fly feeding model [35], the overnight cultures’ OD600 values were adjusted to 2.0 using M9 medium. The pellets from the 1.5 mL cultures were suspended in 100 µL of 5% sucrose. The suspensions were spotted on the sterile filter on the bottom of the culture flask. After cultivation at 37 °C for 30 min, 20 male flies anesthetized with carbon dioxide were sorted and placed in each flask. A PBS buffer was used as a negative control. The number of live fruit flies was counted at 24 h intervals.
2.10. RNA-Seq and Differentially Expressed Genes (DEGs) Analysis
The overnight culture of PAO1 and PAO1(Δ4429–31) was transferred to fresh LB broth until reaching an OD600 of 0.6–0.8, respectively. After being washed once with sterilized fresh PBS, the bacterial cells were quickly frozen with liquid nitrogen for sequencing. The RNA-Seq was carried out on an Illumina HiSeq TM 2500 (Gene Denovo Biotechnology, Guangzhou, China). Three independent experiments were performed. The DEGs of PAO1 and PAO1(Δ4429–31) were identified using the edgeR package (http://www.r-project.org/) (accessed on 1 June 2020) in R. Genes with a fold change ≥2 and a false discovery rate (FDR) <0.05 were treated as significant DEGs.
2.11. The Expression Level Measurement of the Genes Related to Quorum Sensing and Iron Absorption
The vector pMS402 was used to detect gene expression, as reported previously in [26]. The promoter region of the target genes was amplified and fused with the luxCDABE gene in the vector. In the assay, the overnight culture of the reporter strains was diluted at a 1:300 ratio in LB media, and 90 µL of these solutions were added to the wells of a 96-well black plate. The luminescence and OD595 values were measured every 0.5 h for a total of 24 h using a Victor2 multilabel counter (Wallac model 1450, Perkin-Elmer, MA, USA).
2.12. Measurement of Intracellular Iron Concentration
PAO1 and PAO1(Δ4429–31) were inoculated and cultured in 100 mL of LB liquid medium overnight. After centrifugation, the cell sediment was washed with a PBS solution 2–3 times and digested with 65% nitric acid for 30 min. The treated solution was diluted to 3%, and the iron concentration was measured through ICP-MS (Agilent, Santa Clara, CA, USA).
2.13. Statistical Analysis
Statistical analysis was performed using the computer software program SPSS version 18 for descriptive statistics including frequencies and the cross-tabulation of microbiological data. P < 0.05 was considered to be statistically significant.
3. Results
3.1. Deletion of PA4429–31 Increased Aminoglycoside Antibiotics Resistance
A transposon mutant with a threefold increase of aminoglycoside antibiotics resistance was screened from our transposition mutant library [24]. The transposition insertion site was determined to be located in the PA4431 gene by arbitrary PCR and sequencing. The genes PA4427, PA4428, PA4429, PA4430, and PA4431 were predicted to constitute an operon according to the P. aeruginosa genome database (http://www.pseudomonas.com) (accessed on 1 May 2019), and it was consistent with the RT-PCR results (Figure 1A). To further investigate the contribution to the aminoglycoside antibiotics resistance of the individual genes, the in-frame deletion mutants of PA4429, PA4430, PA4431, and PA4429–31 were constructed and named as PAO1 (ΔPA4429), PAO1 (ΔPA4430), PAO1 (ΔPA4431), and PAO1 (ΔPA4429–31), respectively. The disk diffusion test showed that, compared with the wild-type strain PAO1, all the mutants exhibited smaller inhibition zones under the treatment condition of four kinds of the tested aminoglycoside antibiotics, including kanamycin, gentamycin, tobramycin, and amikacin (Figure 1B). Furthermore, their MIC values were determined, and the results showed that the MIC values of the single-gene deletion or the three-gene deletion were all increased threefold for the four antibiotics, indicating a resistance increase for the aminoglycoside antibiotics of these mutants (Table 3). In addition, the single-gene deletion and the three-gene deletion both exhibited an increase in the biofilm formation ability (Figure 1C). Therefore, we focused on the function of the operon PA4429–31 in the study.

Figure 1.
The transcription profiles of PA4427-PA4431 and the antibiotic susceptibility change after their mutation. (A) The products of PCR and RT-PCR using different primers and templates. The gray box between PA4428 and PA4429 is the intergenic region. Lane 1: r1 and r2 primers and genomic DNA template; Lane 2: r1 and r2 primers and cDNA template; Lane 5: r5 and r6 primers and genomic DNA template; Lane 6: r5 and r6 primers and cDNA template; Lane 9: r3 and r4 primers and genomic DNA template; Lane 10: r3 and r4 primers and cDNA template; Lanes 3, 7, and 11: non-retro-transcribed mRNA template as a negative control; and Lanes 4 and 8: 1 kb ladder marker. (B) Antibiotic susceptibility of PAO1, PAO1(Δ4429), PAO1(Δ4430), and PAO1(Δ4431) in LB plates with different antibiotics. Kan: kanamycin; Amk: amikacin; Gm: gentamicin; Tob: tobramycin. (C) The biofilm was examined in the PAO1, PAO1(ΔPA4428), PAO1(ΔPA4429), PAO1(ΔPA4430), PAO1(ΔPA4431), and PAO1(ΔPA4429–31) strains. The error bars represent standard errors. **** significantly different, p < 0.001.

Table 3.
MIC values of the mutants against four aminoglycoside antibiotics.
3.2. The Function Analysis of PA4429–31
Consistent with the predicted result of the P. aeruginosa genome database (http://www.pseudomonas.com) (accessed on 1 May 2019), the phylogenetic tree analysis showed that that the three genes (PA4429–PA4431) were presumed to encode the cytochrome c1 precursor, cytochrome b, and iron–sulfur protein and were believed to synthesize the cytochrome bc1 complex (complex III) [36]. If the cytochrome bc1 complex is defective, PAO1 is able not to carry out aerobic respiration but can perform anaerobic respiration using nitrate and not nitrite [37]. Therefore, we measured the growth of ΔPA4429–31 under anaerobic conditions with nitrate or nitrite addition. After 18 h of incubation under anaerobic conditions, ΔPA4429–31 displayed growth when nitrate was added, although the growth was weaker compared with the wild-type PAO1 (Figure 2A). Regarding nitrite, no growth was observed for ΔPA4429–31 (Figure 2B).
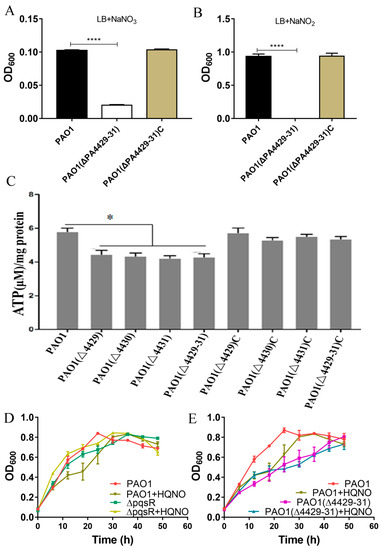
Figure 2.
The physiological effects after PA4427–PA4431 mutation. (A,B) The effect of nitrate and nitrite on the bacterial growth. (C) ATP detection in the various strains. (D,E) The effect of the gene pqsR and the molecule HQNQ on the bacterial growth in the LB broth. The error bars represent standard errors. * p < 0.05. **** p < 0.001.
During the electron transport process, cytochrome complexes I, III, and IV pump intracellular protons out of the membrane, which forms the proton driving force and catalyzes the synthesis of ATP by ATP synthase [38]. Therefore, we tested the intracellular ATP levels of ΔPA4429, ΔPA4430, ΔPA4431, and ΔPA4429–31. The results indicated that the ATP contents of the mutants were significantly lower than that of the wild type (Figure 2C).
It has been reported that HQNO (2-n-heptyl-4-hydroxyquinoline-N-oxide) is a quorum-sensing signaling molecule that specifically inhibits cytochrome bc1 and induces programmed cell death [39]. PqsR is a transcriptional regulator of the gene cluster pqsABCD that is responsible for HQNO synthesis, and ΔpqsR cannot produce HQNO. ΔpqsR did not show autolysis when cultured for 48 h, but the autolysis phenomenon was recovered after exogenous HQNO addition (Figure 2D). However, we found that the PA4429–31 mutant did not exhibit autolysis within 48 h, and the autolysis was not able to recover after HQNO addition, indicating that PA4429–31 was the target cytochrome bc1 of HQNO (Figure 2E). Taken together, all the results indicated that PA4429–31 encoded cytochrome bc1.
3.3. The Effects of PA4429–31 Deficiency on the Phenotypic Traits Related to Virulence of P. aeruginosa PAO1
Pyocyanin, a class of phenazine compounds, is a critical virulence factor secreted by P. aeruginosa, and phzA1 and phzA2 are the key genes responsible for its synthesis [40]. Therefore, the transcription of phzA1 and phzA2 and the pyocyanin production were examined in wild-type and ΔPA4429–31 strains. The results showed a strain-dependent difference for the phzA1 transcription profiles. Before 11 h, the transcription levels of phzA1 for the single- or three-mutant strains all significantly reduced, but those of the single mutants ΔPA4429 and ΔPA4430 were higher compared with the wild-type strain after 15 h (Figure 3A). For phzA2, the transcription levels of all the mutants of single- or three-mutant strains significantly reduced (Figure 3A). These results indicated that the gene PA4431 might be more important for pyocyanin regulation, especially for phzA1. The mutant produced remarkably less pyocyanin compared with the wild-type strain (Figure 3B).
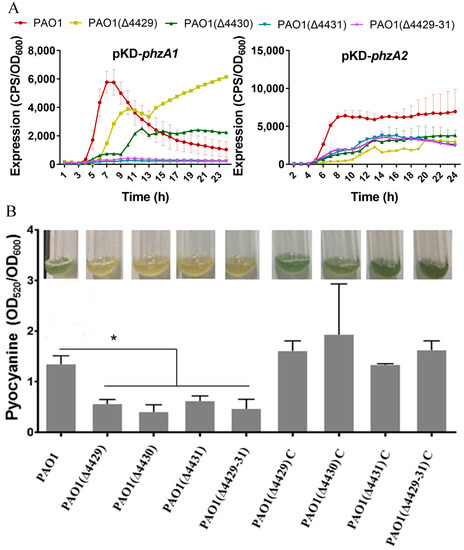
Figure 3.
The effects of PA4429–31 deficiency on pyocyanin production. (A,B) The transcription levels of phzA1 and phzA2 in the wild-type strain PAO1 and the mutants. (B) The content of pyocyanin in the wild-type strain PAO1 and the mutants. The error bars represent standard errors. * p < 0.05.
In addition to pyocyanin, P. aeruginosa also produces some other virulence factors, such as rhamnolipids, extracellular polysaccharide, and elastase [33,41,42]. These virulence factors were examined in this study. The results showed that the mutants produced much less rhamnolipid compared with the wild-type and complemented strains (Figure 4A). Extracellular polysaccharides are carbohydrate compounds, which affect bacteria attachment and biofilm formation. To explore extracellular polysaccharide synthesis, Congo Red was used in the LB agar plates. The results showed that no colonies were red in the mutants, indicating that all the mutants produced less extracellular polysaccharides compared with the wild-type strain (Figure 4B). Elastase is another important virulence factor in local infection of P. aeruginosa. The results showed that the elastase production of the mutants was markedly reduced compared with wild-type PAO1 (Figure 5A).
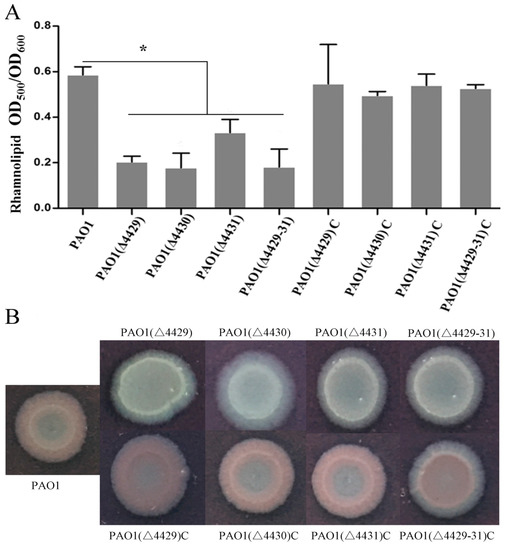
Figure 4.
The effects of PA4429–31 deficiency on rhamnolipids (A) and polysaccharides (B) in the wild-type strain PAO1 and the mutants. (A) Rhamnolipid contents were normalized by the OD600 of the bacteria. (B) The strains were cultured in the plates with Congo Red. The error bars represent standard errors. * p < 0.05.
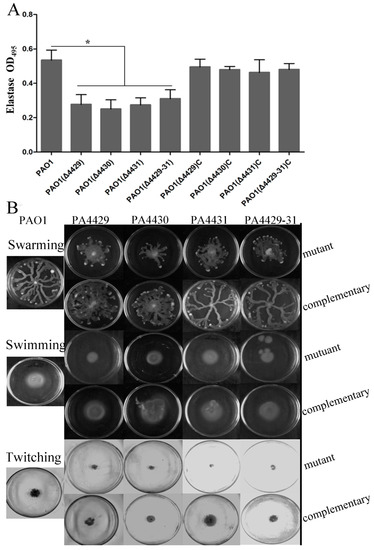
Figure 5.
The effects of PA4429–31 deficiency on virulence and motility. (A) Expression levels of elastase in the wild-type strain PAO1 and the mutants. (B) Swarming, swimming, and twitching of the wild-type strain PAO1 and the mutants were detected. The error bars represent standard errors. * p < 0.05.
Since motility is also involved in P. aeruginosa infection, besides the virulence mentioned above, we further examined the effect of PA4429–31 deletion on P. aeruginosa motility, including swarming, swimming, and twitching. The results showed that the three kinds of motility zones in all the mutant strains were reduced by about half compared with those of the wild-type strain (Figure 5B), indicating that the motility ability for the mutant strains was impaired, but not totally inhibited.
3.4. The Effects of PA4429–31 on the Pathogenicity of P. aeruginosa PAO1
The reduction of virulence may affect bacterial pathogenicity. Therefore, we further explored the effects of PA4429–31 deletion on the pathogenicity of P. aeruginosa PAO1. The cabbage and Drosophila melanogaster infection models were used. The results showed that in the cabbage infection model, PA4429–31deletion obviously reduced the decay area compared with that of the wild-type strain (Figure 6A). In the Drosophila melanogaster infection model, the mortality rate with the infection of the PA4429–31 mutant was significantly lower compared with the wild-type strain (Figure 6B). All the results indicated that the complete pathogenicity of P. aeruginosa PAO1 required a functional cytbc1.
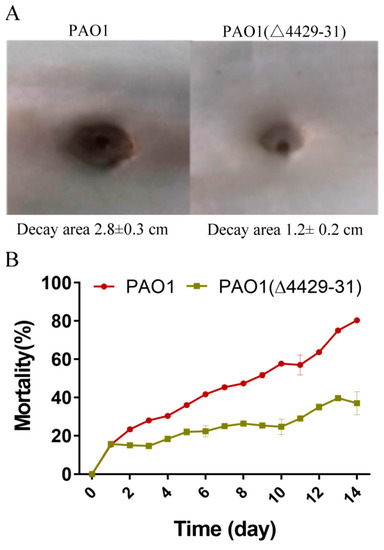
Figure 6.
The deficiency of PA4429–31 has influenced bacterial pathogenicity. (A) The decay area of PAO1 and PAO1(ΔPA4429–31) in the infection model of Chinese cabbage. (B) The mortality of PAO1 and PAO1(ΔPA4429–31) in the infection model of Drosophila melanogaster.
3.5. PA4429–31 Deletion Affected the Quorum Sensing and Iron Adsorption Systems of P. aeruginosa PAO1
To investigate the underlying mechanism of the virulence and pathogenicity reduction in the cytbc1 mutant, RNA-Seq analysis was performed. The results showed that the genes involved in the quorum sensing and iron adsorption systems remarkably changed (Table 4 and Table 5). These two systems have been reported to be related to the virulence and pathogenicity of P. aeruginosa [43,44,45], and therefore, we examined them in the next experiments. The expression of these related genes was measured using the lux-based reporter system. The results indicated that all the quorum sensing genes tested, including lasR, lasI, rhlR, and pqsH, were all downregulated in the PA4429–31 mutant (Figure 7A–D). The expression levels of the two key genes related to iron adsorption, fur and tonB1, remarkably increased (Figure 7E,F). In addition, the iron content of the PA4429–31mutant was obviously lower than that of the wild-type strain (Figure 6G). Taken together, it indicated that the quorum sensing and iron adsorption systems might be involved in the reduction of the virulence and pathogenicity of the cytbc1 mutant.

Table 4.
Differential expression genes involved in the quorum sensing system.

Table 5.
Differential expression genes involved in the iron metabolism.
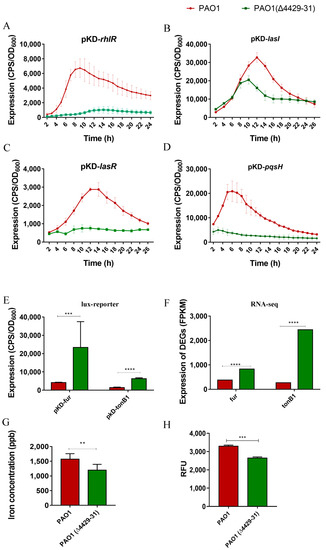
Figure 7.
The transcriptional profiles of the genes related to the quorum sensing system, iron metabolism and the iron concentration in bacterial cells. (A–D) The transcriptional levels of lasI, lasR, pqsH, and rhlR were detected through the CPS (count per second) values. (E,F) The transcriptional levels of tonB1 and fur were detected through the CPS values and RNA-Seq analysis. The FPKM (fragments per kilobase of exon model per million mapped fragments) is the normalized number of transcripts measured for each gene. (G) The content of iron was tested through ICP-MS in PAO1 and PAO1(Δ4429–31). (H) The ROS was detected by using 2’, 7’-dichlorofluorescein diacetate (carboxy-H2DCFDA) in PAO1 and PAO1(Δ4429–31). RFU: relative fluorescence units. The error bars represent standard errors. ** p < 0.01. *** p < 0.001. **** p < 0.0001.
4. Discussion
P. aeruginosa has a multibranched, complex respiratory chain, which allows it to adapt to a variety of living conditions [46]. The respiratory chain is not only important for the survival of bacteria, but also has an impact on the drug resistance [47] and pathogenicity [48] of bacteria. In this study, the mutation of the respiratory chain complex III, cytbc1, displayed increased resistance to aminoglycoside antibiotics and decreased contents of various virulence factors such as pyocyanin, rhamnolipid, and extracellular polysaccharides. The entry of aminoglycoside antibiotics into bacteria requires a potential difference between the inside and outside of the cell membrane [49]. The bacterial respiratory chain complex I or Fe-S cluster synthesis gene ISC mutation can increase the aminoglycoside resistance. In this study, the deletion of cytbc1 caused an increase in aminoglycoside resistance, which might be due to disturbing the complex III transmembrane transport proton or reducing the synthesis of complex I and complex II, resulting in the loss of iron–sulfur protein. In addition, many studies have found that antibiotics can induce the production of ROS and lead to bacterial death. In this study, it was found that the ROS level of the cytbc1 mutant was a bit lower than that of the wild-type strain (Figure 7H), and therefore, this situation might also lead to an increase in antibiotic resistance for the mutants.
The most obvious change in the P. aeruginosa cytbc1 mutant was the reduction of pyocyanin, rhamnolipid production and extracellular polysaccharides. P. aeruginosa has the ability to produce a pigmented secondary metabolite: phenazine. Pyocyanin, one kind of phenazine, is a critical virulence factor in P. aeruginosa during chronic infection [50]. It appeared that the reduction of pyocyanin production was mediated by the downregulation of the phzA1 and phzA2 operons, indicating that cytbc1 affected pyocyanin production by directly regulating the phz genes. Cytbc1 might also directly affect the synthesis of rhamnolipids and polysaccharides. However, it is well known that the quorum sensing system controls the production of virulence factors in P. aeruginosa. The reduced expression of quorum sensing-related genes, such as lasI, lasR, rhlR, and pqsH, in the cytbc1 mutant indicated that the cytbc1 indirectly affected virulence factor production through the quorum sensing systems. Exopolysaccharides, pyocyanin, and rhamnolipids all impact the formation of P. aeruginosa biofilm, besides virulence. Biofilm formation is an advantage in many infection situations and enhances the bacterial ability to resist antibiotics and harsh environmental conditions [50]. Therefore, cytbc1 might be a target for biofilm infection treatment.
Taken together, the work illustrated that the influence of cytbc1 deficiency was multifarious, which increased the antibiotic resistance but reduced the virulence and pathogenicity. It also affected quorum sensing and iron adsorption, and in return, both changed many phenotypes of this bacterium. These results help to comprehensively understand the function of the respiratory chain and develop new drug targets for P. aeruginosa infection.
Author Contributions
S.W. and L.S. conceived and designed the research; S.W., L.S., L.G., J.Z. and Y.W. conducted the experiments; M.Y., Y.F. and L.W. contributed new reagents or analytical tools; S.W., L.S., J.Z. and Y.W. analyzed the data; S.W. and L.S. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project was financially supported by grants from the Provincial Natural Science Foundation of Shaanxi Province (No. 2018ZDXM-SF-004) and National Natural Science Foundation of China (81770434, 31770152, and 31400094).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, J.; Strous, M. Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim. Biophys Acta 2013, 1827, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Arai, H. Regulation and Function of Versatile Aerobic and Anaerobic Respiratory Metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2011, 2, 103. [Google Scholar] [CrossRef] [PubMed]
- Mazat, J.P.; Ransac, S. The cytochrome bc1 complex in the mitochondrial respiratory chain functions according to the Q cycle hypothesis of Mitchell: The proof using a stochastic approach? Med. Sci. (Paris) 2010, 26, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Esser, L.; Tang, W.; Zhou, F.; Zhou, Y.; Yu, L.; Yu, C. Structural analysis of cytochrome bc1 complexes: Implications to the mechanism of function. Biochim. Biophys. Acta 2013, 1827, 1278–1294. [Google Scholar] [CrossRef]
- Vartak, R.; Porras, C.A.; Bai, Y. Respiratory supercomplexes: Structure, function and assembly. Protein Cell 2013, 4, 582–590. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, X.; Cui, G.; Chan, J.W.; Wang, L.; Li, C.; Shan, L.; Xu, C.; Zhang, Q.; Wang, Y.; et al. A novel agent exerts antitumor activity in breast cancer cells by targeting mitochondrial complex II. Oncotarget 2016, 7, 32054–32064. [Google Scholar] [CrossRef]
- Sharaf, M.S.; Stevens, D.; Kamunde, C. Mitochondrial transition ROS spike (mTRS) results from coordinated activities of complex I and nicotinamide nucleotide transhydrogenase. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 955–965. [Google Scholar] [CrossRef]
- Small, J.L.; Park, S.W.; Kana, B.D.; Ioerger, T.R.; Sacchettini, J.C.; Ehrt, S. Perturbation of cytochrome c maturation reveals adaptability of the respiratory chain in Mycobacterium tuberculosis. mBio 2013, 4, e00475-13. [Google Scholar] [CrossRef] [PubMed]
- Lencina, A.M.; Franza, T.; Sullivan, M.J.; Ulett, G.C.; Ipe, D.S.; Gaudu, P.; Gennis, R.B.; Schurig-Briccio, L.A. Type 2 NADH Dehydrogenase Is the Only Point of Entry for Electrons into the Streptococcus agalactiae Respiratory Chain and Is a Potential Drug Target. mBio 2018, 9, e01034-18. [Google Scholar] [CrossRef]
- Auzat, I.; Chapuy-Regaud, S.; Le Bras, G.; Dos Santos, G.; Ogunniyi, A.D.; Le Thomas, I.; Garel, J.R.; Paton, J.C.; Trombe, M.C. The NADH oxidase of Streptococcus pneumoniae: Its involvement in competence and virulence. Mol. Microbiol. 1999, 34, 1018–1028. [Google Scholar] [CrossRef]
- Ge, X.; Yu, Y.; Zhang, M.; Chen, L.; Chen, W.; Elrami, F.; Kong, F.; Kitten, T.; Xu, P. Involvement of NADH Oxidase in Competition and Endocarditis Virulence in Streptococcus sanguinis. Infect. Immun. 2016, 84, 1470–1477. [Google Scholar] [CrossRef]
- Xiao, Y.; Esser, L.; Zhou, F.; Li, C.; Zhou, Z.; Yu, C.; Qin, Z.; Xia, D. Studies on inhibition of respiratory cytochrome bc1 complex by the fungicide pyrimorph suggest a novel inhibitory mechanism. PLoS ONE 2014, 9, e93765. [Google Scholar] [CrossRef] [PubMed]
- Martins Vde, P.; Dinamarco, T.M.; Curti, C.; Uyemura, S.A. Classical and alternative components of the mitochondrial respiratory chain in pathogenic fungi as potential therapeutic targets. J. Bioenerg. Biomembr. 2011, 43, 81–88. [Google Scholar] [CrossRef]
- Murai, M.; Habu, S.; Murakami, S.; Ito, T.; Miyoshi, H. Production of new amilorides as potent inhibitors of mitochondrial respiratory complex I. Biosci. Biotechnol. Biochem. 2015, 79, 1061–1066. [Google Scholar] [CrossRef]
- Bald, D.; Villellas, C.; Lu, P.; Koul, A. Targeting Energy Metabolism in Mycobacterium tuberculosis, a New Paradigm in Antimycobacterial Drug Discovery. mBio 2017, 8, e00272-17. [Google Scholar] [CrossRef]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.J.; Orange, N.; Dufour, A.; Cornelis, P. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef]
- Karpeisky, M.; Ilyin, V.A. Analysis of non-polar regions in proteins. J. Mol. Biol. 1992, 224, 629–638. [Google Scholar] [CrossRef]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed]
- Raba, D.A.; Rosas-Lemus, M.; Menzer, W.M.; Li, C.; Fang, X.; Liang, P.; Tuz, K.; Minh, D.D.L.; Juárezt, O. Characterization of the Pseudomonas aeruginosa NQR complex, a bacterial proton pump with roles in autopoisoning resistance. J. Biol. Chem. 2018, 293, 15664–15677. [Google Scholar] [CrossRef]
- Williams, H.D.; Zlosnik, J.E.; Ryall, B. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 2007, 52, 1–71. [Google Scholar]
- Slizen, M.V.; Galzitskaya, O.V. Comparative Analysis of Proteomes of a Number of Nosocomial Pathogens by KEGG Modules and KEGG Pathways. Int. J. Mol. Sci. 2020, 21, 7839. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic. Acids. Res. 2016, 44, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, L.; Zhao, X.Y.; Shen, L.X.; Duan, K.M. Identification of Pseudomonas aeruginosa genes associated with antibiotic susceptibility. Sci. China Life Sci. 2010, 53, 1247–1251. [Google Scholar] [CrossRef]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
- Starkey, M.; Rahme, L.G. Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat. Protoc. 2009, 4, 117–124. [Google Scholar] [CrossRef]
- Poole, K.; Krebes, K.; McNally, C.; Neshatt, S. Multiple antibiotic resistance in Pseudomonas aeruginosa: Evidence for involvement of an efflux operon. J. Bacteriol. 1993, 175, 7363–7372. [Google Scholar] [CrossRef]
- Wang, S.W.; Yu, S.; Zhang, Z.; Wei, Q.; Yan, L.; Ai, G.; Liu, H.; Ma, L.Z. Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. App. Environ. Microbiol. 2014, 80, 6724–6732. [Google Scholar] [CrossRef]
- John, J.B. Determination of ATP in Chlorella with the luciferin-luciferase enzyme system. Anal. Biochem. 1970, 37, 409–416. [Google Scholar] [CrossRef]
- Peng, X.; Wang, S.; Yu, S.; Zhang, Z.; Wei, Q.; Yan, L.; Ai, G.; Liu, H.; Ma, L.Z. Expression of a mitochondrial gene orfH79 from the CMS-HongLian rice inhibits Saccharomyces cerevisiae growth and causes excessive ROS accumulation and decrease in ATP. Biotechnol. Lett. 2009, 31, 409–414. [Google Scholar] [CrossRef]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [PubMed]
- Pinzon, N.M.; Ju, L.K. Analysis of rhamnolipid biosurfactants by methylene blue complexation. App. Microbiol. Biotech. 2009, 82, 975–981. [Google Scholar] [CrossRef]
- Taha, M.N.; Saafan, A.E.; Ahmedy, A.; El Gebaly, E.; Khairalla, A.S. Two novel synthetic peptides inhibit quorum sensing-dependent biofilm formation and some virulence factors in Pseudomonas aeruginosa PAOJ. Microbiology 2019, 57, 618–625. [Google Scholar]
- Qi, J.; Wang, B.; Li, J.; Ning, H.; Wang, Y.; Kong, W.; Shen, L. Genetic determinants involved in the biodegradation of naphthalene and phenanthrene in Pseudomonas aeruginosa PAO1. Environ. Sci. Pollut. Res. Int. 2015, 22, 6743–6755. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Dammel, C.; Stein, J.; Rabin, H.; Surette, M.G. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 2003, 50, 1477–1491. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Toyofuku, M.; Miyano, T.; Nomura, N. cbb3-type cytochrome c oxidases, aerobic respiratory enzymes, impact the anaerobic life of Pseudomonas aeruginosa PAO1. J. Bacteriol. 2014, 196, 3881–3889. [Google Scholar] [CrossRef]
- Schreiber, K.; Krieger, R.; Benkert, B.; Eschbach, M.; Arai, H.; Schobert, M.; Jahn, D. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 2007, 189, 4310–4314. [Google Scholar] [CrossRef]
- Hazan, R.; Que, Y.A.; Maura, D.; Strobel, B.; Majcherczyk, P.A.; Hopper, L.R.; Wilbur, D.J.; Hreha, T.N.; Barquera, B.; Rahme, L.G. Auto Poisoning of the Respiratory Chain by a Quorum-Sensing-Regulated Molecule Favors Biofilm Formation and Antibiotic Tolerance. Curr. Biol. 2016, 26, 195–206. [Google Scholar] [CrossRef]
- Kawakami, T.; Kuroki, M.; Ishii, M.; Igarashi, Y.; Arai, H. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ. Microbiol. 2010, 12, 1399–1412. [Google Scholar]
- Cui, Q.; Lv, H.; Qi, Z.; Jiang, B.; Xiao, B.; Liu, L.; Ge, Y.; Hu, X. Cross-Regulation between the phz1 and phz2 Operons Maintain a Balanced Level of Phenazine Biosynthesis in Pseudomonas aeruginosa PAO1. PLoS ONE 2016, 11, e0144447. [Google Scholar] [CrossRef]
- Pearson, J.P.; Pesci, E.C.; Iglewski, B.H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997, 179, 5756–5767. [Google Scholar] [CrossRef] [PubMed]
- Colvin, K.M.; Irie, Z.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012, 14, 1913–1928. [Google Scholar] [CrossRef] [PubMed]
- Hnamte, S.; Parasuraman, P.; Ranganathan, S.; Ampasala, D.R.; Reddy, D.; Kumavath, R.N.; Suchiang, K.; Mohanty, S.K.; Busi, S. Mosloflavone attenuates the quorum sensing controlled virulence phenotypes and biofilm formation in Pseudomonas aeruginosa PAO1: In vitro, in vivo and in silico approach. Microb. Pathog. 2019, 131, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Minandri, F.; Imperi, F.; Frangipani, E.; Bonchi, C.; Visaggio, D.; Facchini, M.; Pasquali, P.; Bragonzi, A.; Visca, P. Role of Iron Uptake Systems in Pseudomonas aeruginosa Virulence and Airway Infection. Infect. Immun. 2016, 84, 2324–2335. [Google Scholar] [CrossRef] [PubMed]
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2002, 99, 7072–7077. [Google Scholar] [CrossRef] [PubMed]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef]
- Torres, A.; Kasturiarachi, N.; DuPont, M.; Cooper, V.S.; Bomberger, J.; Zemke, A. NADH Dehydrogenases in Pseudomonas aeruginosa Growth and Virulence. Front. Microbiol. 2019, 10, 75. [Google Scholar] [CrossRef]
- Taber, H.W.; Mueller, J.P.; Miller, P.F.; Arrow, A.S. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 1987, 51, 439–457. [Google Scholar] [CrossRef]
- Ezraty, B.; Vergnes, A.; Banzhaf, M.; Duverger, Y.; Huguenot, A.; Brochado, A.R.; Su, S.Y.; Espinosa, L.; Loiseau, L.; Py, B.; et al. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 2013, 340, 1583–1587. [Google Scholar] [CrossRef]
- Newman, J.W.; Floyd, R.V.; Fothergill, J.L. The contribution of Pseudomonas aeruginosa virulence factors and host factors in the establishment of urinary tract infections. FEMS. Microbiol. Lett. 2017, 364, fnx124. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).