Bioinformatics Analysis of Oral, Vaginal, and Rectal Microbial Profiles during Pregnancy: A Pilot Study on the Bacterial Co-Residence in Pregnant Women
Abstract
1. Introduction
2. Patients and Methods
2.1. Ethics
2.2. Patients
2.3. Vaginal, Rectal, and Oral Sample Collection
2.4. DNA Extraction for Next-Generation Sequencing
2.5. 16S Ribosomal RNA (rRNA) Gene Amplification and Sequencing
2.6. Vaginal, Fecal, and Oral Microbiota Profiling
2.7. Statistics
3. Results
3.1. Vaginal Lactobacillus Classification by 16S rRNA Gene Sequencing Analysis
3.2. Bioinformatics Analysis of the Vaginal, Rectal, and Oral Microbial Profiles Using 16S rRNA Gene Amplicon Sequencing Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, L.; Johnson, H.L.; Cousens, S.; Perin, J.; Scott, S.; Lawn, J.E.; Rudan, I.; Campbell, H.; Cibulskis, R.; Li, M.; et al. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet 2012, 379, 2151–2161. [Google Scholar] [CrossRef]
- Goldenberg, R.L.; Culhane, J.F.; Iams, J.D.; Romero, R. Epidemiology and causes of preterm birth. Lancet 2008, 371, 75–84. [Google Scholar] [CrossRef]
- Villamor-Martinez, E.; Fumagalli, M.; Alomar, Y.I.; Passera, S.; Cavallaro, G.; Mosca, F.; Villamor, E. Cerebellar Hemorrhage in Preterm Infants: A Meta-Analysis on Risk Factors and Neurodevelopmental Outcome. Front. Physiol. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Bezold, K.Y.; Karjalainen, M.K.; Hallman, M.; Teramo, K.; Muglia, L.J. The genomics of preterm birth: From animal models to human studies. Genome Med. 2013, 5, 34. [Google Scholar] [CrossRef]
- Blencowe, H.; Cousens, S.; Chou, D.; Oestergaard, M.; Say, L.; Moller, A.B.; Kinney, M.; Lawn, J. Born Too Soon Preterm Birth Action Group Born too soon: The global epidemiology of 15 million preterm births. Reprod. Health 2013, 10 (Suppl. S1), S2. [Google Scholar] [CrossRef]
- Strauss, J.F., III; Romero, R.; Gomez-Lopez, N.; Haymond-Thornburg, H.; Modi, B.P.; Teves, M.E.; Pearson, L.N.; York, T.P.; Schenkein, H.A. Spontaneous preterm birth: Advances toward the discovery of genetic predisposition. Am. J. Obstet. Gynecol. 2018, 218, 294–314. [Google Scholar] [CrossRef]
- Tiensuu, H.; Haapalainen, A.M.; Karjalainen, M.K.; Pasanen, A.; Huusko, J.M.; Marttila, R.; Ojaniemi, M.; Muglia, L.J.; Hallman, M.; Ramet, M. Risk of spontaneous preterm birth and fetal growth associates with fetal SLIT2. PLoS Genet. 2019, 15, e1008107. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.; Wong, R.J.; Shaw, G.; et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef]
- Callahan, B.J.; DiGiulio, D.B.; Goltsman, D.S.A.; Sun, C.L.; Costello, E.K.; Jeganathan, P.; Biggio, J.R.; Wong, R.J.; Druzin, M.L.; Shaw, G.M.; et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc. Natl. Acad. Sci. USA 2017, 114, 9966–9971. [Google Scholar] [CrossRef]
- Koullali, B.; Oudijk, M.A.; Nijman, T.A.; Mol, B.W.; Pajkrt, E. Risk assessment and management to prevent preterm birth. Semin. Fetal Neonatal Med. 2016, 21, 80–88. [Google Scholar] [CrossRef]
- Kenyon, S.L.; Taylor, D.J.; Tarnow-Mordi, W.; ORACLE Collaborative Group. Broad-spectrum antibiotics for preterm, prelabour rupture of fetal membranes: The ORACLE I randomised trial. Lancet 2001, 357, 979–988. [Google Scholar] [CrossRef]
- Brocklehurst, P.; Gordon, A.; Heatley, E.; Milan, S.J. Antibiotics for treating bacterial vaginosis in pregnancy. Cochrane Database Syst. Rev. 2013, 1, CD000262. [Google Scholar] [CrossRef]
- Thinkhamrop, J.; Hofmeyr, G.J.; Adetoro, O.; Lumbiganon, P.; Ota, E. Antibiotic prophylaxis during the second and third trimester to reduce adverse pregnancy outcomes and morbidity. Cochrane Database Syst. Rev. 2015, 6, CD002250. [Google Scholar]
- Shimaoka, M.; Yo, Y.; Doh, K.; Kotani, Y.; Suzuki, A.; Tsuji, I.; Mandai, M.; Matsumura, N. Association between preterm delivery and bacterial vaginosis with or without treatment. Sci. Rep. 2019, 9, 509. [Google Scholar] [CrossRef]
- Javed, A.; Parvaiz, F.; Manzoor, S. Bacterial vaginosis: An insight into the prevalence, alternative treatments regimen and it’s associated resistance patterns. Microb. Pathog. 2019, 127, 21–30. [Google Scholar] [CrossRef]
- Sobel, J.D.; Kaur, N.; Woznicki, N.A.; Boikov, D.; Aguin, T.; Gill, G.; Akins, R.A. Prognostic Indicators of Recurrence of Bacterial Vaginosis. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Amabebe, E.; Anumba, D.O.C. The Vaginal Microenvironment: The Physiologic Role of Lactobacilli. Front. Med. 2018, 5, 181. [Google Scholar] [CrossRef]
- Srinivasan, S.; Fredricks, D.N. The human vaginal bacterial biota and bacterial vaginosis. Interdiscip. Perspect. Infect. Dis. 2008, 2008, 750479. [Google Scholar] [CrossRef]
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The vaginal microbiome and preterm birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef]
- Contini, C.; Rotondo, J.C.; Magagnoli, F.; Maritati, M.; Seraceni, S.; Graziano, A.; Poggi, A.; Capucci, R.; Vesce, F.; Tognon, M.; et al. Investigation on silent bacterial infections in specimens from pregnant women affected by spontaneous miscarriage. J. Cell. Physiol. 2018, 234, 100–107. [Google Scholar] [CrossRef]
- Lee, Y.H.; Cherkerzian, S.; Seidman, L.J.; Papandonatos, G.D.; Savitz, D.A.; Tsuang, M.T.; Goldstein, J.M.; Buka, S.L. Maternal Bacterial Infection During Pregnancy and Offspring Risk of Psychotic Disorders: Variation by Severity of Infection and Offspring Sex. Am. J. Psychiatry 2020, 177, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Brown, J.A.; Rager, S.L.; Sanidad, K.Z.; Ananthanarayanan, A.; Zeng, M.Y. Maternal Microbiome and Infections in Pregnancy. Microorganisms 2020, 8, 1996. [Google Scholar] [CrossRef]
- Shiozaki, A.; Yoneda, S.; Yoneda, N.; Yonezawa, R.; Matsubayashi, T.; Seo, G.; Saito, S. Intestinal microbiota is different in women with preterm birth: Results from terminal restriction fragment length polymorphism analysis. PLoS ONE 2014, 9, e111374. [Google Scholar] [CrossRef]
- Pelzer, E.; Gomez-Arango, L.F.; Barrett, H.L.; Nitert, M.D. Review: Maternal health and the placental microbiome. Placenta 2017, 54, 30–37. [Google Scholar] [CrossRef]
- Ide, M.; Papapanou, P.N. Epidemiology of association between maternal periodontal disease and adverse pregnancy outcomes—Systematic review. J. Periodontol. 2013, 84 (Suppl. S4), S181–S194. [Google Scholar] [CrossRef]
- Cobb, C.M.; Kelly, P.J.; Williams, K.B.; Babbar, S.; Angolkar, M.; Derman, R.J. The oral microbiome and adverse pregnancy outcomes. Int. J. Women’s Health 2017, 9, 551–559. [Google Scholar] [CrossRef]
- Fischer, L.A.; Demerath, E.; Bittner-Eddy, P.; Costalonga, M. Placental colonization with periodontal pathogens: The potential missing link. Am. J. Obstet. Gynecol. 2019, 221, 383–392. [Google Scholar] [CrossRef]
- De Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef]
- Theis, K.R.; Romero, R.; Winters, A.D.; Greenberg, J.M.; Gomez-Lopez, N.; Alhousseini, A.; Bieda, J.; Maymon, E.; Pacora, P.; Fettweis, J.M.; et al. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am. J. Obstet. Gynecol. 2019, 220, e1–e39. [Google Scholar] [CrossRef] [PubMed]
- Theis, K.R.; Romero, R.; Winters, A.D.; Jobe, A.H.; Gomez-Lopez, N. Lack of Evidence for Microbiota in the Placental and Fetal Tissues of Rhesus Macaques. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Holst, E. Reservoir of four organisms associated with bacterial vaginosis suggests lack of sexual transmission. J. Clin. Microbiol. 1990, 28, 2035–2039. [Google Scholar] [CrossRef] [PubMed]
- Antonio, M.A.; Rabe, L.K.; Hillier, S.L. Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J. Infect. Dis. 2005, 192, 394–398. [Google Scholar] [CrossRef]
- Sanchez-Ramos, L. The preterm labor index and fetal fibronectin for prediction of preterm delivery with intact membranes. Obstet. Gynecol. 2003, 102, 196. [Google Scholar] [PubMed]
- Yoneda, S.; Sakai, M.; Sasaki, Y.; Shiozaki, A.; Hidaka, T.; Saito, S. Interleukin-8 and glucose in amniotic fluid, fetal fibronectin in vaginal secretions and preterm labor index based on clinical variables are optimal predictive markers for preterm delivery in patients with intact membranes. J. Obstet. Gynaecol. Res. 2007, 33, 38–44. [Google Scholar] [CrossRef]
- Yoneda, S.; Shiozaki, A.; Yoneda, N.; Shima, T.; Ito, M.; Yamanaka, M.; Hidaka, T.; Sumi, S.; Saito, S. Prediction of exact delivery time in patients with preterm labor and intact membranes at admission by amniotic fluid interleukin-8 level and preterm labor index. J. Obstet. Gynaecol. Res. 2011, 37, 861–866. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef]
- MacIntyre, D.A.; Chandiramani, M.; Lee, Y.S.; Kindinger, L.; Smith, A.; Angelopoulos, N.; Lehne, B.; Arulkumaran, S.; Brown, R.; Teoh, T.G.; et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep. 2015, 5, 8988. [Google Scholar] [CrossRef]
- Freitas, A.C.; Chaban, B.; Bocking, A.; Rocco, M.; Yang, S.; Hill, J.E.; Money, D.M.; Group, V.R. The vaginal microbiome of pregnant women is less rich and diverse, with lower prevalence of Mollicutes, compared to non-pregnant women. Sci. Rep. 2017, 7, 9212. [Google Scholar] [CrossRef]
- Stafford, G.P.; Parker, J.L.; Amabebe, E.; Kistler, J.; Reynolds, S.; Stern, V.; Paley, M.; Anumba, D.O.C. Spontaneous Preterm Birth Is Associated with Differential Expression of Vaginal Metabolites by Lactobacilli-Dominated Microflora. Front. Physiol. 2017, 8, 615. [Google Scholar] [CrossRef]
- Coleman, J.S.; Gaydos, C.A. Molecular Diagnosis of Bacterial Vaginosis: An Update. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef]
- Hill, J.E.; Albert, A.Y.K.; Group, V.R. Resolution and Cooccurrence Patterns of Gardnerella leopoldii, G. swidsinskii, G. piotii, and G. vaginalis within the Vaginal Microbiome. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Onderdonk, A.B.; Delaney, M.L.; Fichorova, R.N. The Human Microbiome during Bacterial Vaginosis. Clin. Microbiol. Rev. 2016, 29, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.L.; Gottschick, C.; Bhuju, S.; Masur, C.; Abels, C.; Wagner-Dobler, I. Metatranscriptome Analysis of the Vaginal Microbiota Reveals Potential Mechanisms for Protection against Metronidazole in Bacterial Vaginosis. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Redelinghuys, M.J.; Geldenhuys, J.; Jung, H.; Kock, M.M. Bacterial Vaginosis: Current Diagnostic Avenues and Future Opportunities. Front. Cell. Infect. Microbiol. 2020, 10, 354. [Google Scholar] [CrossRef]
- Vicariotto, F.; Mogna, L.; Del Piano, M. Effectiveness of the two microorganisms Lactobacillus fermentum LF15 and Lactobacillus plantarum LP01, formulated in slow-release vaginal tablets, in women affected by bacterial vaginosis: A pilot study. J. Clin. Gastroenterol. 2014, 48 (Suppl. S1), S106–S112. [Google Scholar] [CrossRef]
- Cohen, C.R.; Wierzbicki, M.R.; French, A.L.; Morris, S.; Newmann, S.; Reno, H.; Green, L.; Miller, S.; Powell, J.; Parks, T.; et al. Randomized Trial of Lactin-V to Prevent Recurrence of Bacterial Vaginosis. N. Engl. J. Med. 2020, 382, 1906–1915. [Google Scholar] [CrossRef]
- Mastromarino, P.; Macchia, S.; Meggiorini, L.; Trinchieri, V.; Mosca, L.; Perluigi, M.; Midulla, C. Effectiveness of Lactobacillus-containing vaginal tablets in the treatment of symptomatic bacterial vaginosis. Clin. Microbiol. Infect. 2009, 15, 67–74. [Google Scholar] [CrossRef]
- Vujic, G.; Jajac Knez, A.; Despot Stefanovic, V.; Kuzmic Vrbanovic, V. Efficacy of orally applied probiotic capsules for bacterial vaginosis and other vaginal infections: A double-blind, randomized, placebo-controlled study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013, 168, 75–79. [Google Scholar] [CrossRef]
- Farkash, E.; Weintraub, A.Y.; Sergienko, R.; Wiznitzer, A.; Zlotnik, A.; Sheiner, E. Acute antepartum pyelonephritis in pregnancy: A critical analysis of risk factors and outcomes. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012, 162, 24–27. [Google Scholar] [CrossRef]
- Verma, I.; Avasthi, K.; Berry, V. Urogenital infections as a risk factor for preterm labor: A hospital-based case-control study. J. Obstet. Gynaecol. India 2014, 64, 274–278. [Google Scholar] [CrossRef][Green Version]
- Lewis, A.L.; Gilbert, N.M. Roles of the vagina and the vaginal microbiota in urinary tract infection: Evidence from clinical correlations and experimental models. GMS Infect. Dis. 2020, 8, Doc02. [Google Scholar]
- Roberts, C.L.; Algert, C.S.; Rickard, K.L.; Morris, J.M. Treatment of vaginal candidiasis for the prevention of preterm birth: A systematic review and meta-analysis. Syst. Rev. 2015, 4, 31. [Google Scholar] [CrossRef]
- Holzer, I.; Farr, A.; Kiss, H.; Hagmann, M.; Petricevic, L. The colonization with Candida species is more harmful in the second trimester of pregnancy. Arch. Gynecol. Obstet. 2017, 295, 891–895. [Google Scholar] [CrossRef]
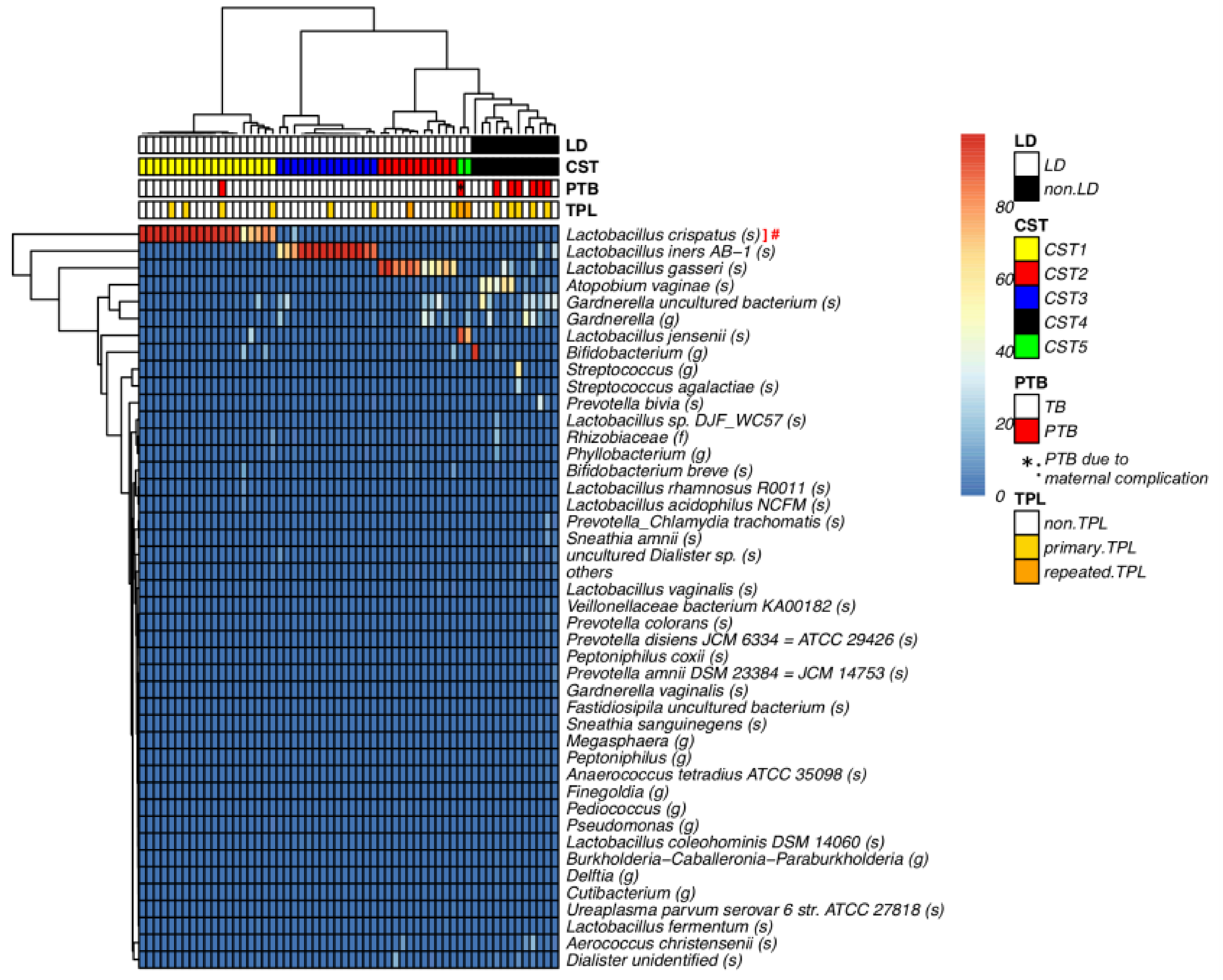
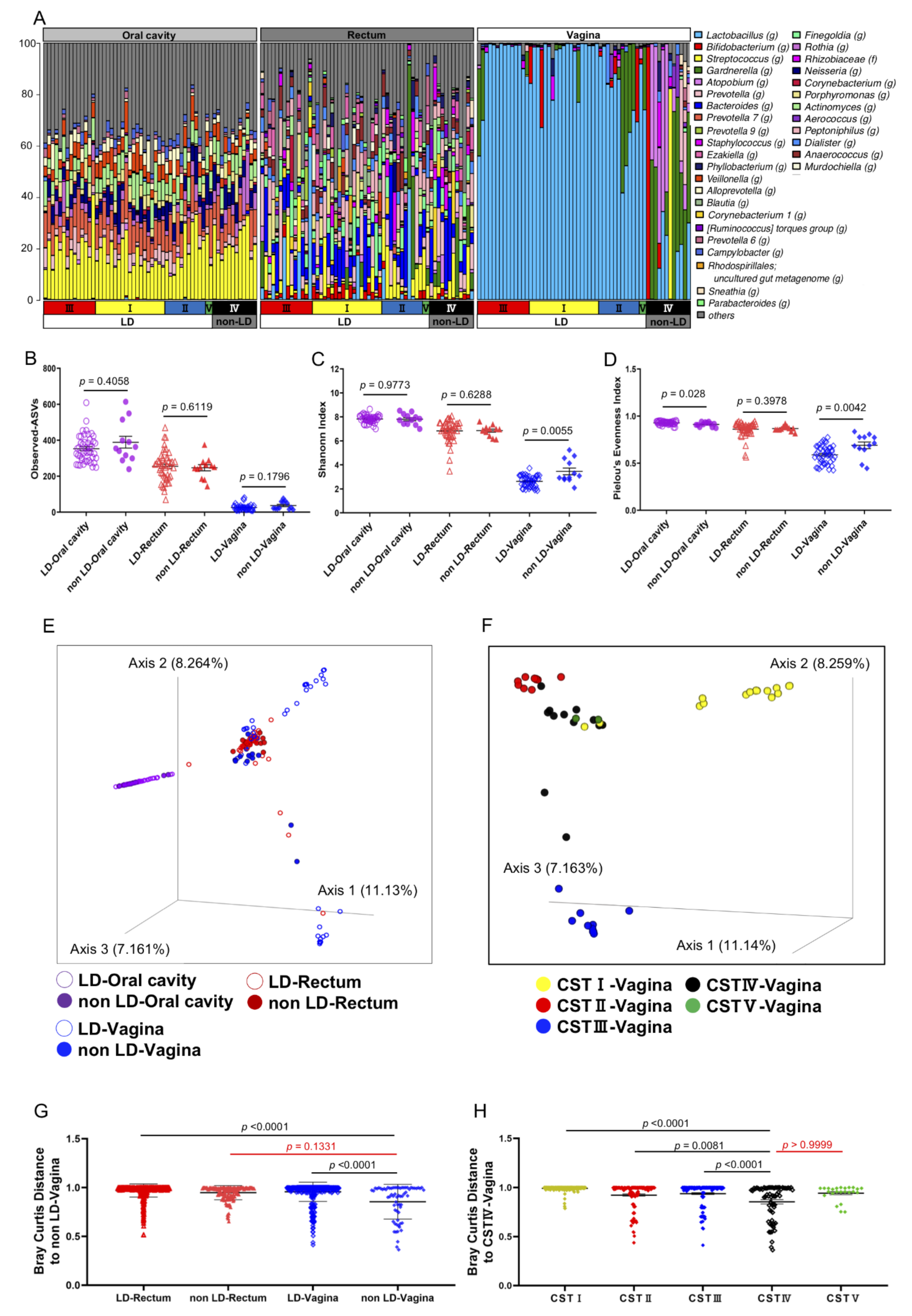
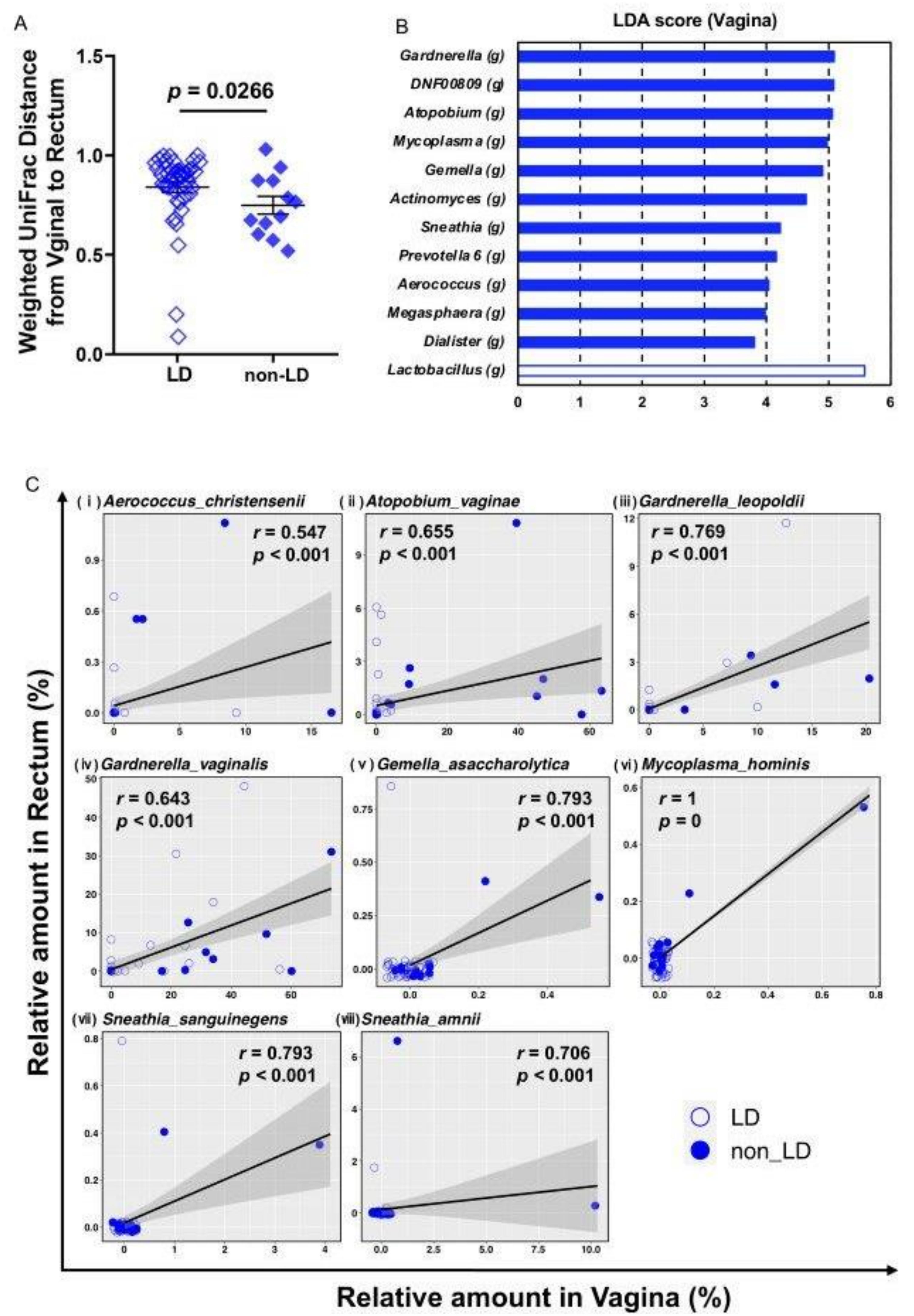
| Characteristics (n = 58 in total) | LD (n = 46) | non-LD (n = 12) | p Value |
|---|---|---|---|
| Age (median) | 33 (21–41) | 33 (30–40) | 0.87 |
| BMI (kg/m2) at pre-pregnancy (median) | 21.9 (16.1–32.9) | 21.6 (18.1–28.4) | 0.806 |
| BMI (kg/m2) at delivery (median) | 26.0 (19.1–34.0) | 24.9 (21.0–32.4) | 0.602 |
| Body weight gain during pregnancy (median) | 9.2 (−5–21.1) | 9.7 (2.8–15) | 0.719 |
| Parity | 18 (39.1%) | 4 (33.3%) | 0.493 |
| Uterine Leiomyoma | 6 (13.0%) | 1 (8.3%) | 1 |
| Diabetes Mellitus | 2 (4.3%) | 1 (8.3%) | 0.508 |
| Gestational Diabetes Mellitus | 8 (17.4%) | 0 | 0.185 |
| Hypertensive Disorders of Pregnancy | 3 (6.5%) | 1 (8.3%) | 1 |
| Threatened preterm labor | 10 (21.7%) | 5 (41.7%) | 0.265 |
| Premature rapture of membrane | 1 (2.2%) | 1 (8.3%) | 0.374 |
| Gestational age at delivery (median) | 38w5d (35w6d–41w4d) | 38w0d (34w0d–41w1d) | 0.076 |
| Spontaneous preterm birth | 1 (2.2%) | 5 (41.7%) | 0.001 |
| Birth weight of neonate (median) | 2990 (2285–4035) | 2772.5 (2056–3524) | 0.230 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fudaba, M.; Kamiya, T.; Tachibana, D.; Koyama, M.; Ohtani, N. Bioinformatics Analysis of Oral, Vaginal, and Rectal Microbial Profiles during Pregnancy: A Pilot Study on the Bacterial Co-Residence in Pregnant Women. Microorganisms 2021, 9, 1027. https://doi.org/10.3390/microorganisms9051027
Fudaba M, Kamiya T, Tachibana D, Koyama M, Ohtani N. Bioinformatics Analysis of Oral, Vaginal, and Rectal Microbial Profiles during Pregnancy: A Pilot Study on the Bacterial Co-Residence in Pregnant Women. Microorganisms. 2021; 9(5):1027. https://doi.org/10.3390/microorganisms9051027
Chicago/Turabian StyleFudaba, Megumi, Tomonori Kamiya, Daisuke Tachibana, Masayasu Koyama, and Naoko Ohtani. 2021. "Bioinformatics Analysis of Oral, Vaginal, and Rectal Microbial Profiles during Pregnancy: A Pilot Study on the Bacterial Co-Residence in Pregnant Women" Microorganisms 9, no. 5: 1027. https://doi.org/10.3390/microorganisms9051027
APA StyleFudaba, M., Kamiya, T., Tachibana, D., Koyama, M., & Ohtani, N. (2021). Bioinformatics Analysis of Oral, Vaginal, and Rectal Microbial Profiles during Pregnancy: A Pilot Study on the Bacterial Co-Residence in Pregnant Women. Microorganisms, 9(5), 1027. https://doi.org/10.3390/microorganisms9051027






