Rapid Classification of Clostridioides difficile Strains Using MALDI-TOF MS Peak-Based Assay in Comparison with PCR-Ribotyping
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Study Design
2.3. Typing of C. difficile Isolates
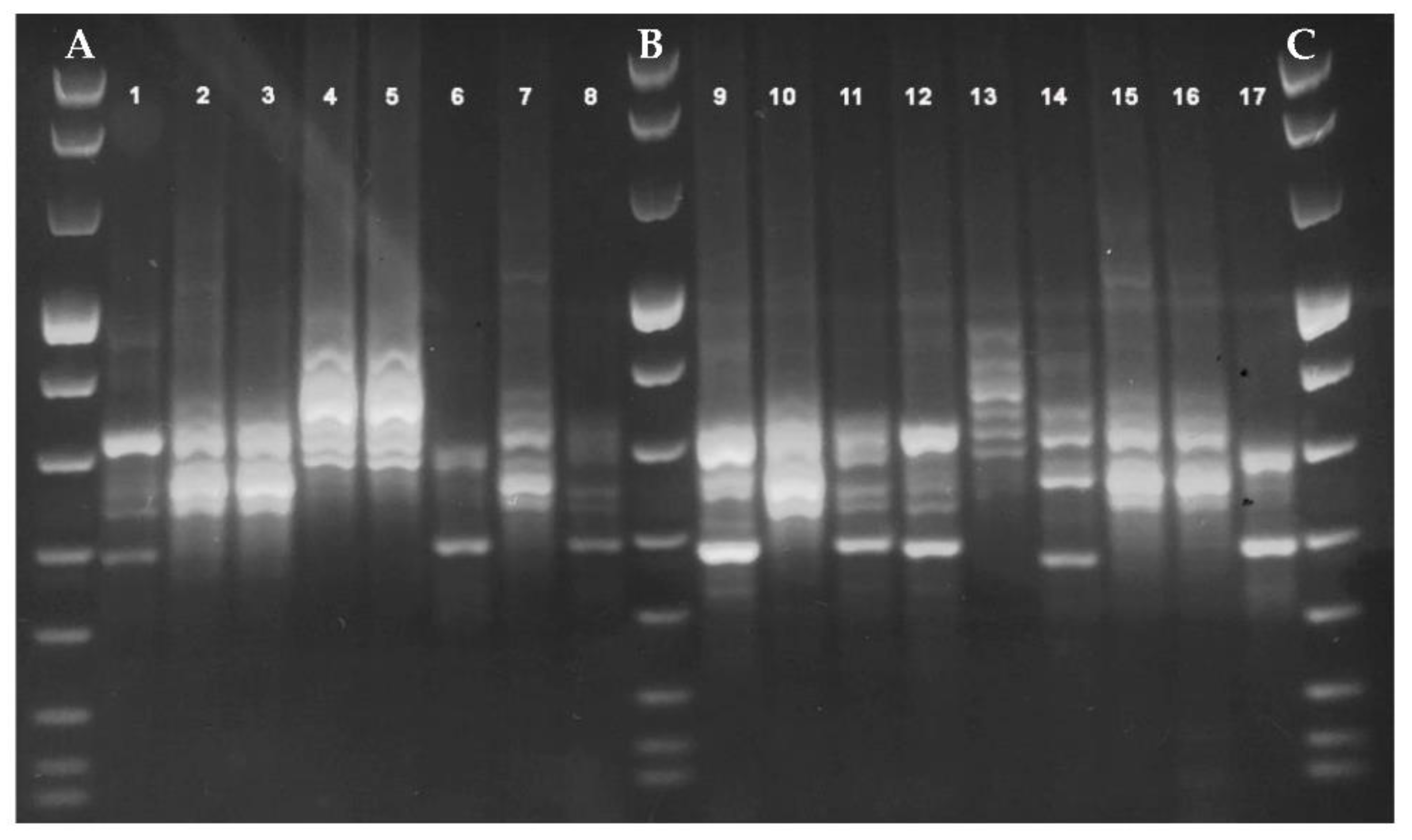
2.3.1. PCR-Ribotyping
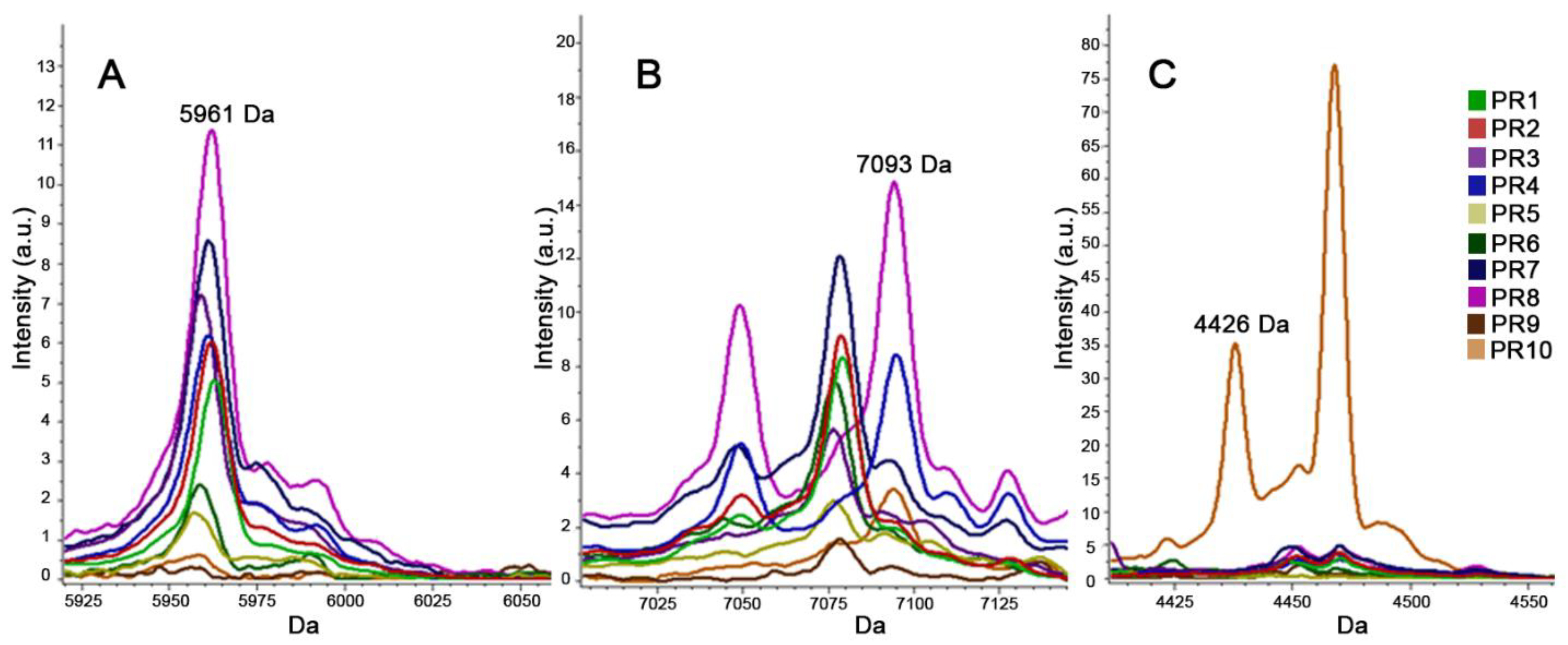
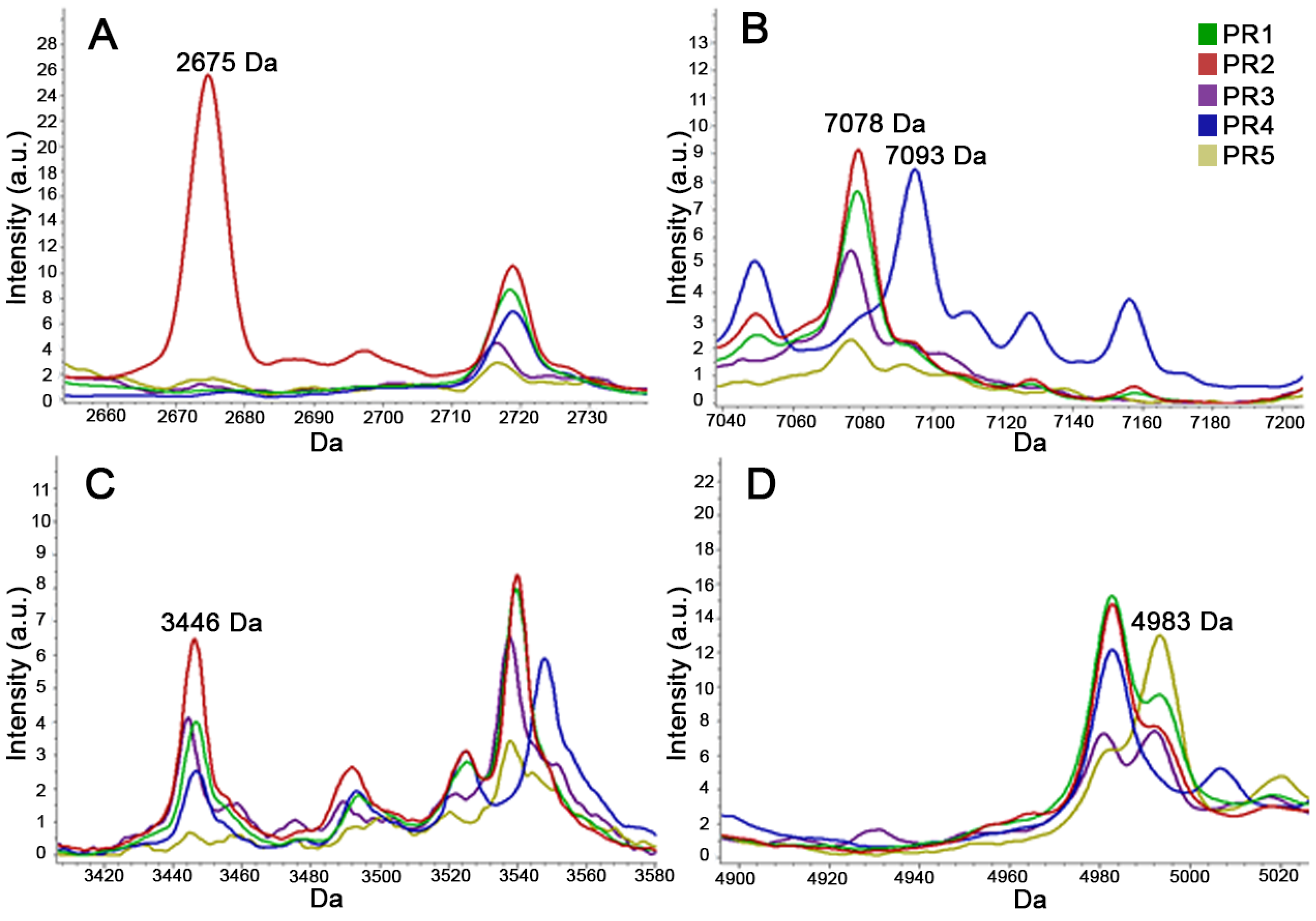
2.3.2. MALDI-TOF MS for Typing (T-MALDI)
2.3.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bartlett, J.G.; Moon, N.; Chang, T.W.; Taylor, N.; Onderdonk, A.B. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 1978, 75, 778–782. [Google Scholar] [CrossRef]
- Freeman, J.; Bauer, M.P.; Baines, S.D.; Corver, J.; Fawley, W.N.; Goorhuis, B.; Kuijper, E.J.; Wilcox, M.H. The Changing Epidemiology of Clostridium difficile Infections. Clin. Microbiol. Rev. 2010, 23, 529–549. [Google Scholar] [CrossRef]
- Khanna, S.; Gupta, A. Community-acquired Clostridium difficile infection: An increasing public health threat. Infect. Drug Resist. 2014, 7, 63–72. [Google Scholar] [CrossRef]
- Khanna, S.; Pardi, D.S.; Aronson, S.L.; Kammer, P.P.; Orenstein, R.; Sauver, J.L.S.; Harmsen, S.W.; Zinsmeister, A.R. The Epidemiology of Community-Acquired Clostridium difficile Infection: A Population-Based Study. Am. J. Gastroenterol. 2012, 107, 89–95. [Google Scholar] [CrossRef]
- Elliott, B.; Androga, G.O.; Knight, D.R.; Riley, T.V. Clostridium difficile infection: Evolution, phylogeny and molecular epidemiology. Infect. Genet. Evol. 2017, 49, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Furuya-Kanamori, L.; Marquess, J.; Yakob, L.; Riley, T.V.; Paterson, D.L.; Foster, N.F.; Huber, C.A.; Clements, A.C.A. Asymptomatic Clostridium difficile colonization: Epidemiology and clinical implications. BMC Infect. Dis. 2015, 15, 516. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Severe Clostridium difficile-associated disease in populations previously at low risk--four states, 2005. MMWR Morb. Mortal. Wkly. Rep. 2005, 54, 1201–1205. [Google Scholar]
- Del Prete, R.; Ronga, L.; Addati, G.; Magrone, R.; Abbasciano, A.; Decimo, M.; Miragliotta, G. Clostridium difficile. A review on an emerging infection. Clin. Ter. 2019, 170, e41–e47. [Google Scholar] [CrossRef]
- Clements, A.C.; Magalhães, R.J.S.; Tatem, A.J.; Paterson, D.L.; Riley, T.V. Clostridium difficile PCR ribotype 027: Assessing the risks of further worldwide spread. Lancet Infect. Dis. 2010, 10, 395–404. [Google Scholar] [CrossRef]
- Buffet-Bataillon, S.; Tattevin, P.; Senechal, H.; Cormier, M.; Vincent, P. Clostridium difficile O27 colitis: Hospital-onset but community-acquired. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2263–2267. [Google Scholar] [CrossRef] [PubMed]
- Barbut, F.; Jones, G.; Eckert, C. Epidemiology and control of Clostridium difficile infections in healthcare settings. Curr. Opin. Infect. Dis. 2011, 24, 370–376. [Google Scholar] [CrossRef]
- Couturier, J.; Davies, K.; Gateau, C.; Barbut, F. Ribotypes and New Virulent Strains Across Europe. Adv. Exp. Med. Biol. 2018, 1050, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Prim. 2016, 2, 1–20. [Google Scholar] [CrossRef]
- Goorhuis, A.; Bakker, D.; Corver, J.; Debast, S.B.; Harmanus, C.; Notermans, D.W.; Bergwerff, A.A.; Dekker, F.W.; Kuijper, E.J. Emergence ofClostridium difficileInfection Due to a New Hypervirulent Strain, Polymerase Chain Reaction Ribotype 078. Clin. Infect. Dis. 2008, 47, 1162–1170. [Google Scholar] [CrossRef]
- Warny, M.; Pepin, J.; Fang, A.; Killgore, G.; Thompson, A.; Brazier, J.; Frost, E.; McDonald, L.C. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 2005, 366, 1079–1084. [Google Scholar] [CrossRef]
- Stanley, J.; Bartlett, J.; Dart, B.; Ashcraft, J.; Iv, B.D. Clostridium difficile infection. Curr. Probl. Surg. 2013, 50, 302–337. [Google Scholar] [CrossRef]
- He, M.; Miyajima, F.; Roberts, P.; Ellison, L.; Pickard, D.J.; Martin, M.J.; Connor, T.R.; Harris, S.R.; Fairley, D.; Bamford, K.B.; et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 2013, 45, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.A.; Ashwin, H.; Longshaw, C.M.; Burns, D.A.; Davis, G.L.; Wilcox, M.H. Diversity of Clostridium difficile PCR ribotypes in Europe: Results from the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID), 2012 and 2013. Eurosurveillance 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, S.E.; Tanner, H.E.; Marriott, C.L.; Brazier, J.S.; Hardy, K.J.; Platt, S.; Hawkey, P.M. Extended Multilocus Variable-Number Tandem-Repeat Analysis of Clostridium difficile Correlates Exactly with Ribotyping and Enables Identification of Hospital Transmission. J. Clin. Microbiol. 2011, 49, 3523–3530. [Google Scholar] [CrossRef][Green Version]
- Knetsch, C.W.; Lawley, T.D.; Hensgens, M.P.; Corver, J.; Wilcox, M.W.; Kuijper, E.J. Current application and future perspectives of molecular typing methods to study Clostridium difficile infections. Eurosurveillance 2013, 18, 20381. [Google Scholar] [CrossRef] [PubMed]
- Emele, M.F.; Joppe, F.M.; Riedel, T.; Overmann, J.; Rupnik, M.; Cooper, P.; Kusumawati, R.L.; Berger, F.K.; Laukien, F.; Zimmermann, O.; et al. Proteotyping of Clostridioides difficile as Alternate Typing Method to Ribotyping Is Able to Distinguish the Ribotypes RT027 and RT176 From Other Ribotypes. Front. Microbiol. 2019, 10, 2087. [Google Scholar] [CrossRef]
- Sadeghifard, N.; Gürtler, V.; Beer, M.; Seviour, R.J. The Mosaic Nature of Intergenic 16S-23S rRNA Spacer Regions Suggests rRNA Operon Copy Number Variation in Clostridium difficile Strains. Appl. Environ. Microbiol. 2006, 72, 7311–7323. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Ling, L.; Stratton, C.W.; Li, C.; Polage, C.R.; Wu, B.; Tang, Y.-W. Advances in the diagnosis and treatment of Clostridium difficile infections. Emerg. Microbes Infect. 2018, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Burnham, C.-A.D.; Carroll, K.C. Diagnosis of Clostridium difficile Infection: An Ongoing Conundrum for Clinicians and for Clinical Laboratories. Clin. Microbiol. Rev. 2013, 26, 604–630. [Google Scholar] [CrossRef]
- Calderaro, A.; Arcangeletti, M.C.; Rodighiero, I.; Buttrini, M.; Montecchini, S.; Simone, R.V.; Medici, M.C.; Chezzi, C.; De Conto, F. Identification of different respiratory viruses, after a cell culture step, by matrix assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). Sci. Rep. 2016, 6, 36082. [Google Scholar] [CrossRef]
- Calderaro, A.; Arcangeletti, M.-C.; Rodighiero, I.; Buttrini, M.; Gorrini, C.; Motta, F.J.N.; Germini, D.; Medici, M.-C.; Chezzi, C.; De Conto, F. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry applied to virus identification. Sci. Rep. 2015, 4, 6803. [Google Scholar] [CrossRef]
- Florio, W.; Baldeschi, L.; Rizzato, C.; Tavanti, A.; Ghelardi, E.; Lupetti, A. Detection of Antibiotic-Resistance by MALDI-TOF Mass Spectrometry: An Expanding Area. Front. Cell. Infect. Microbiol. 2020, 10, 572909. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Buttrini, M.; Piergianni, M.; Montecchini, S.; Martinelli, M.; Covan, S.; Piccolo, G.; Medici, M.C.; Arcangeletti, M.C.; Chezzi, C.; et al. Evaluation of a modified meropenem hydrolysis assay on a large cohort of KPC and VIM carbapenemase-producing Enterobacteriaceae. PLoS ONE 2017, 12, e0174908. [Google Scholar] [CrossRef][Green Version]
- Croxatto, A.; Prod’hom, G.; Greub, G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 2012, 36, 380–407. [Google Scholar] [CrossRef]
- Sandrin, T.R.; Goldstein, J.E.; Schumaker, S. MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom. Rev. 2013, 32, 188–217. [Google Scholar] [CrossRef]
- Reil, M.; Erhard, M.; Kuijper, E.J.; Kist, M.; Zaiss, H.; Witte, W.; Gruber, H.; Borgmann, S. Recognition of Clostridium difficile PCR-ribotypes 001, 027 and 126/078 using an extended MALDI-TOF MS system. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1431–1436. [Google Scholar] [CrossRef]
- Rizzardi, K.; Åkerlund, T. High Molecular Weight Typing with MALDI-TOF MS—A Novel Method for Rapid Typing of Clostridium difficile. PLoS ONE 2015, 10, e0122457. [Google Scholar] [CrossRef]
- Ortega, L.; Ryberg, A.; Johansson, Å. HMW-profiling using MALDI-TOF MS: A screening method for outbreaks of Clostridioides difficile. Anaerobe 2018, 54, 254–259. [Google Scholar] [CrossRef]
- Calderaro, A.; Buttrini, M.; Martinelli, M.; Gorrini, C.; Montecchini, S.; Medici, M.C.; Arcangeletti, M.C.; De Conto, F.; Covan, S.; Chezzi, C. Comparative analysis of different methods to detect Clostridium difficile infection. New Microbiol. 2013, 36, 57–63. [Google Scholar]
- Bidet, P.; Lalande, V.; Salauze, B.; Burghoffer, B.; Avesani, V.; Delmée, M.; Rossier, A.; Barbut, F.; Petit, J.-C. Comparison of PCR-Ribotyping, Arbitrarily Primed PCR, and Pulsed-Field Gel Electrophoresis for Typing Clostridium difficile. J. Clin. Microbiol. 2000, 38, 2484–2487. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Healthcare-Associated Infections: Clostridium Difficile Infections; European Centre for Disease Prevention and Control: Solna Kommun, Sweden, 2018. [Google Scholar]
- Di Bella, S.; Musso, M.; Cataldo, M.A.; Meledandri, M.; Bordi, E.; Capozzi, D.; Cava, M.C.; Chiaradonna, P.; Prignano, G.; Petrosillo, N. Clostridium difficile infection in Italian urban hospitals: Data from 2006 through 2011. BMC Infect. Dis. 2013, 13, 146. [Google Scholar] [CrossRef] [PubMed]
- Spigaglia, P.; Barbanti, F.; Morandi, M.; Moro, M.L.; Mastrantonio, P. Diagnostic testing for Clostridium difficile in Italian microbiological laboratories. Anaerobe 2016, 37, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Visconti, V.; Brunetti, G.; Cuomo, M.R.; Giordano, A.; Raponi, G. Nosocomial-acquired and community-onset Clostridium difficile infection at an academic hospital in Italy: Epidemiology, recurrences and toxin genes distribution. J. Infect. Chemother. 2017, 23, 763–768. [Google Scholar] [CrossRef]
- Corver, J.; Sen, J.; Hornung, B.V.; Mertens, B.J.; Berssenbrugge, E.K.; Harmanus, C.; Sanders, I.M.; Kumar, N.; Lawley, T.D.; Kuijper, E.J.; et al. Identification and validation of two peptide markers for the recognition of Clostridioides difficile MLST-1 and MLST-11 by MALDI-MS. Clin. Microbiol. Infect. 2019, 25, 904.e1–904.e7. [Google Scholar] [CrossRef] [PubMed]
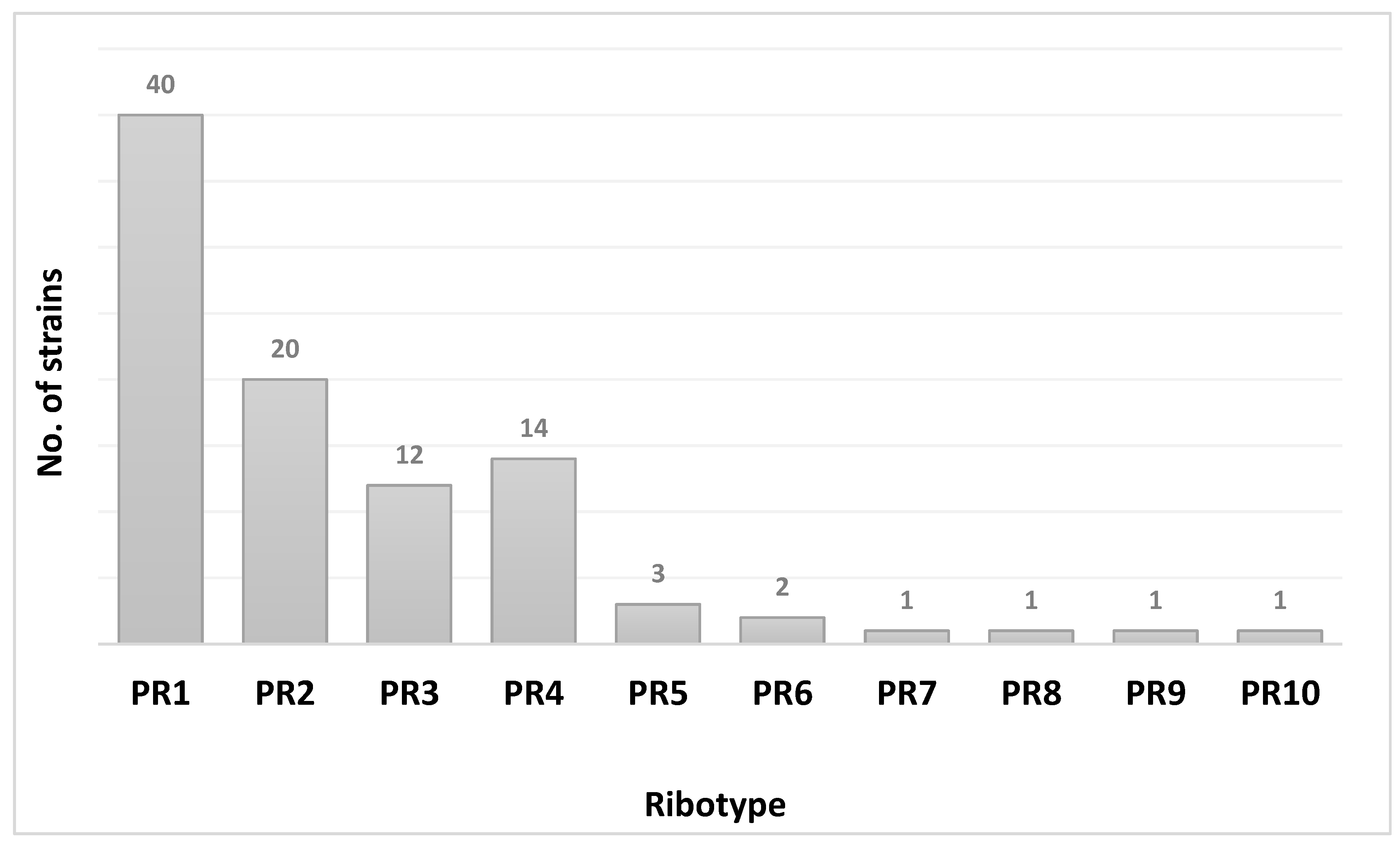
| Ribotype | PR1–PR10 CAM | PR1–PR5 CAM | ||
|---|---|---|---|---|
| RC (%) | CV (%) | RC (%) | CV (%) | |
| PR1 | 100 | 96.3 | 100 | 99.5 |
| PR2 | 98.7 | 95.1 | 100 | 92.5 |
| PR3 | 98.7 | 95.6 | 100 | 100 |
| PR4 | 100 | 91.5 | 100 | 95.5 |
| PR5 | 100 | 95.6 | 100 | 95.8 |
| PR6 | 100 | 78.3 | NA | NA |
| PR7 | 90 | 72.7 | NA | NA |
| PR8 | 100 | 95.8 | NA | NA |
| PR9 | 100 | 94.7 | NA | NA |
| PR10 | 100 | 100 | NA | NA |
| Overall | 98.7 | 91.6 | 100 | 96.6 |
| PCR-Ribotyping | T-MALDI | |||||
|---|---|---|---|---|---|---|
| Ribotype PR1 | Ribotype PR2 | Ribotype PR3 | Ribotype PR4 | Ribotype PR5 | No Classification | |
| Ribotype PR1 | 32 | |||||
| Ribotype PR2 | 12 | |||||
| Ribotype PR3 | 4 | |||||
| Ribotype PR4 | 5 | 1 | ||||
| Ribotype PR5 | 1 | |||||
| Ribotypes PR6-PR10 | 6 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calderaro, A.; Buttrini, M.; Martinelli, M.; Farina, B.; Moro, T.; Montecchini, S.; Arcangeletti, M.C.; Chezzi, C.; De Conto, F. Rapid Classification of Clostridioides difficile Strains Using MALDI-TOF MS Peak-Based Assay in Comparison with PCR-Ribotyping. Microorganisms 2021, 9, 661. https://doi.org/10.3390/microorganisms9030661
Calderaro A, Buttrini M, Martinelli M, Farina B, Moro T, Montecchini S, Arcangeletti MC, Chezzi C, De Conto F. Rapid Classification of Clostridioides difficile Strains Using MALDI-TOF MS Peak-Based Assay in Comparison with PCR-Ribotyping. Microorganisms. 2021; 9(3):661. https://doi.org/10.3390/microorganisms9030661
Chicago/Turabian StyleCalderaro, Adriana, Mirko Buttrini, Monica Martinelli, Benedetta Farina, Tiziano Moro, Sara Montecchini, Maria Cristina Arcangeletti, Carlo Chezzi, and Flora De Conto. 2021. "Rapid Classification of Clostridioides difficile Strains Using MALDI-TOF MS Peak-Based Assay in Comparison with PCR-Ribotyping" Microorganisms 9, no. 3: 661. https://doi.org/10.3390/microorganisms9030661
APA StyleCalderaro, A., Buttrini, M., Martinelli, M., Farina, B., Moro, T., Montecchini, S., Arcangeletti, M. C., Chezzi, C., & De Conto, F. (2021). Rapid Classification of Clostridioides difficile Strains Using MALDI-TOF MS Peak-Based Assay in Comparison with PCR-Ribotyping. Microorganisms, 9(3), 661. https://doi.org/10.3390/microorganisms9030661










