Abstract
HAC1 encodes a key transcription factor that transmits the unfolded protein response (UPR) from the endoplasmic reticulum (ER) to the nucleus and regulates downstream UPR genes in Saccharomyces cerevisiae. In response to the accumulation of unfolded proteins in the ER, Ire1p oligomers splice HAC1 pre-mRNA (HAC1u) via a non-conventional process and allow the spliced HAC1 (HAC1i) to be translated efficiently. However, leaky splicing and translation of HAC1u may occur in non-UPR cells to induce undesirable UPR. To control accidental UPR activation, multiple fail-safe mechanisms have been proposed to prevent leaky HAC1 splicing and translation and to facilitate rapid degradation of translated Hac1up and Hac1ip. Among proposed regulatory mechanisms is a degron sequence encoded at the 5′ end of the HAC1 intron that silences Hac1up expression. To investigate the necessity of an intron-encoded degron sequence that specifically targets Hac1up for degradation, we employed publicly available transcriptomic data to quantify leaky HAC1 splicing and translation in UPR-induced and non-UPR cells. As expected, we found that HAC1u is only efficiently spliced into HAC1i and efficiently translated into Hac1ip in UPR-induced cells. However, our analysis of ribosome profiling data confirmed frequent occurrence of leaky translation of HAC1u regardless of UPR induction, demonstrating the inability of translation fail-safe to completely inhibit Hac1up production. Additionally, among 32 yeast HAC1 surveyed, the degron sequence is highly conserved by Saccharomyces yeast but is poorly conserved by all other yeast species. Nevertheless, the degron sequence is the most conserved HAC1 intron segment in yeasts. These results suggest that the degron sequence may indeed play an important role in mitigating the accumulation of Hac1up to prevent accidental UPR activation in the Saccharomyces yeast.
1. Introduction
Folding of nascent proteins in the endoplasmic reticulum (ER) is an error-prone process in eukaryotic cells, and the accumulation of unfolded/misfolded proteins can lead to lethal consequences if left unregulated [1]. In response to the increase of unfolded/misfolded proteins, yeasts have evolved a complex Ire1p+Hac1p-mediated signaling pathway to tightly control protein folding [2,3]. This unfolded protein response (UPR) increases ER folding capacity by increasing the production of specific chaperon proteins, reducing folding load by reducing global translation activity and triggers apoptosis when UPR fails to restore ER homeostasis.
During ER stress, the ER transmembrane kinase/endonuclease Ire1p [4] detects UPR via a core luminal domain [5,6] and responds by undergoing trans-autophosphorylation and congregation into discrete foci of Ire1p oligomers [7]. HAC1 pre-mRNA (HAC1u) is recruited, through recognition of its 3′ bipartite element (3′ BE) [8], to the Ire1p oligomers to undergo splicing. What is particular about HAC1u splicing is that it is not spliced through the conventional spliceosome mechanism.
To form mature HAC1 mRNAs (HAC1i) in Saccharomyces cerevisiae, oligomerized Ire1p cleaves out a single 252nt HAC1 intron (Figure 1a) [4,9] with its RNAse activity [5,7,9,10,11,12,13], and the exons are then ligated by the tRNA ligase Trl1p [14,15,16,17,18,19]. Aside from S. cerevisiae, Ire1p-mediated HAC1 splicing has been observed in Candida albicans [20,21], Candida glabrata [21], Kluyveromyces lactis [21], Pichia pastoris [22], Hansenula polymorpha [23], and Yarrowia lipolytica [24] under ER stress conditions. Furthermore, through a comparative gene study, Hooks and Griffiths-Jones [25] determined HAC1 homologs in 19 yeasts with intron lengths ranging from 19nt to 379nt, with most yeast lineages that diverged early having short HAC1 introns.
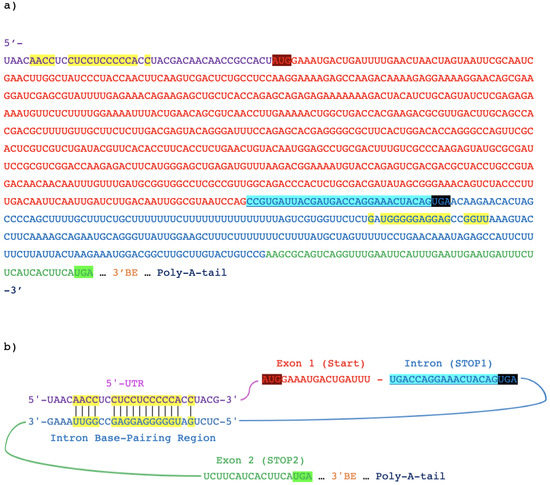
Figure 1.
The HAC1 gene in S. cerevisiae. (a) The annotated HAC1u mRNA illustrating its sequence elements: 5′ untranslated region (5′ UTR) in purple, first exon in red, the intron in blue, and the second exon in green. The start codon (START) is highlighted in dark red, and the in-frame stop codons in the intron (STOP1) and in the second exon (STOP2) are respectively highlighted in black and green. Nucleotides involved in the long-ranged base-pair interactions between the 5′ UTR and intron are highlighted in yellow, while the intronic sequence proposed to encode a C-terminal degron in Hac1up is shaded in blue. (b) An illustration showing the base-pairs formed between the 5′ UTR and intron in HAC1u to inhibit translation initiation of the pre-mRNA, with the same color scheme as in (a).
The HAC1i mRNA is efficiently translated when the HAC1 intron is removed through Ire1p-mediated splicing, and the translated protein (Hac1ip) is a key transcription activator that transmits UPR from the ER to the nucleus [4,9,26,27,28]. Hac1ip contains a nuclear localization signal that binds to UPR related genes such as KAR2, PDI1, EUG1, FKB2, and LHS1 that feature one of the two conserved UPR elements (UPREs): UPRE1 (GACAGCGTGTC) [9,29,30,31], and UPRE2 (TACGTGT) [31,32,33,34]. This signaling pathway increases the production of UPR related proteins to stimulate protein folding [4,9,32].
In absence of UPR, the unspliced HAC1u mRNA is still constitutively transcribed and stable, and it is thought to be slowly translated into Hac1up [4,9]. Hac1up is a truncated protein because of the presence of an in-frame stop codon (STOP1) in the intronic sequence (Figure 1a) [35]. In S. cerevisiae, the truncated Hac1up misses the 18 amino acids transcriptional activation domain [35] encoded by the second exon. Consequently, Hac1up has reduced transactivation activity [4] on UPRE promoter elements [36] relative to Hac1ip.
As a mechanism to block undesirable translation of HAC1u, part of the intron forms base pairs with the 5′ untranslated region (5′ UTR) in the unspliced form (Figure 1b). This interaction has been experimentally shown to inhibit the undesired translation of HAC1u pre-mRNAs [11,26,37,38,39,40] and may prevent unnecessary UPR activation in absence of ER stress [41]. Indeed, leaky translation of HAC1u was considered minimal given the well-documented intron-mediated translation inhibition (Figure 1b). Another fail-safe mechanism that prevents leaky splicing is handled by the Kar2p chaperon. When Kar2p is free from protein folding, it binds to Ire1p to hinder Ire1p oligomerization and prevent its RNase splicing activity [8,42,43]. A third fail-safe mechanism against undesired HAC1u splicing is the rapid removal of the 3′ BE in the 3′ UTR of HAC1u transcripts under non-UPR-induced (hereafter referred to as just non-UPR) condition. This prevents the translocation of HAC1u to Ire1 foci [11] and limits Ire1p-mediated splicing. Resultantly, HAC1i mRNAs are hardly detectable in non-UPR cells [44].
Nonetheless, leaky translation and leaky splicing of HAC1u may still occur during non-ER stress, and this is undesirable because it could lead to the accumulation of Hac1ip and accidental induction of UPR. While leaky translation of HAC1u may be rare [39], there is substantial evidence that Hac1up will accumulate without efficient protein degradation [38] and may lead to accidental induction of UPR [41]. Thus, rapid degradation of Hac1p may be a much-required fail-safe mechanism in non-UPR cells to counteract leaky translation of Hac1up and Hac1ip. It was previously proposed that this process requires a nuclear localization signal (29)RKRAKTK(35) that is encoded in the first exon [45] and is recognized by the ubiquitin-proteasome pathway [38,45]. Since the degradation signal is located in the first exon, it is shared by Hac1up and Hac1ip, which explains why both protein forms are highly unstable in the yeast [26,46] with half-life of about 1.5–2.0 min [26,45,46].
Recently, Di Santo et al. [38] suggest that S. cerevisiae contains an additional post-translational regulatory signal, encoded by a 10 amino acid “degron” at the C-terminus of Hac1up that is located just before STOP1 (Figure 1a). Their meticulous experimental design involves GFP-HAC1 intron constructs in a fluorescent measurement-based assay. To summarize, they found that the degron plays a functionally important role in the post-translational silencing of Hac1up. When the degron was present in the construct, fluorescence was barely detectable in cells expressing green fluorescent protein (GFP), regardless of the presence of the downstream intronic sequence. When the degron sequence was removed from the intron, fluorescence was restored to a level comparable with the intron-less constructs. Furthermore, GFP silencing was unaffected when the degron was recoded with synonymous codons, but silencing was abolished when the 10 amino acid sequence was altered or contained a premature stop codon. To further demonstrate that the degron acts by promoting Hac1up degradation and not by disrupting translation, the GFP-HAC1 intron construct were non-covalently linked by an upstream HA-mRuby reporter, which was expressed only if translation was not affected, and HA-mRuby gene expression was indeed not affected by the presence of the degron [38]. Together, these reporter constructs demonstrated that the translation of the C-terminus degron could effectively silence HAC1u expression by rapidly degrading Hac1up.
As mentioned previously, HAC1 may encode a nuclear localization sequence in the first exon that signals for the rapid degradation of both Hac1ip and Hac1up. In addition, translation of HAC1u is blocked by a 5′ UTR loop. Nonetheless, having an additional fail-safe degron that specifically targets Hac1up for degradation would be advantageous if the HAC1u translation block does not adequately eliminate leaky HAC1u translation. Furthermore, if this intron-encoded degron is an important post-translational control, then we expect this sequence to be evolutionarily conserved among closely related yeast species under similar selection pressure.
Our investigation considered the HAC1 gene sequence in 32 yeast species and the transcriptomics data from UPR-induced and non-UPR S. cerevisiae yeast. Transcript profiling analyses suggest that HAC1u is efficiently spliced into HAC1i, and HAC1i is efficiently translated into Hac1ip in UPR-induced cells. In contrast, HAC1u is constitutively translated into Hac1up in both UPR-induced and non-UPR cells, albeit with many translational regulatory mechanisms featured in the Ire1p-Hac1p pathway. Thus, having an intron-encoded degron signal to specifically degrade undesired Hac1up production would protect cells against accidental activation of UPR. Expectedly, we found that the intronic degron sequence upstream of STOP1 is highly conserved by Saccharomyces yeasts. However, conservation at putative degron sequences was poor in all non-Saccharomyces yeast species surveyed. Nevertheless, the first four degron-encoded amino acids are highly conserved, and the putative degron was the most conserved intronic region among yeast species.
2. Materials and Methods
2.1. Retrieving the HAC1 Genes from 32 Yeast Species and Determining Their Introns
We retrieved the complete HAC1 gene and 18S rRNA sequences of 32 yeast species belonging to the Saccharomycetales order. These species were selected because all have available HAC1 and 18S rRNA gene records in the National Center for Biotechnology Information (NCBI) Gene Database. Of these species, 14 belong to the Saccharomycetaceae family and 18 belong to eight other families (Trichomonascaceae, Phaffomycetaceae, Ascoideaceae, Debaryomycetaceae, Metschnikowiaceae, Pichiaceae, Phaffomycetaceae, and Dipodascaceae). Furthermore, to show that the IRE1 gene is present in each yeast species surveyed, the IRE1 Gene ID was retrieved from NCBI Gene Database and listed in Supplemental File S1.
The HAC1 5′ and 3′ splice site information of Saccharomyces cerevisiae, Candida orthopsilosis, Candida albicans, Lodderomyces elongisporus, Clavispora lusitaniae, Scheffersomyces stipitis, Meyerozyma guilliermondii, and Debaryomyces hansenii were obtained from Iracane et al. [47] and those of Yarrowia lipolytica, Candida glabrata, Lachancea thermotolerans, Eremothecium gossypii, Kluyveromyces lactis, Zygosaccharomyces rouxii, Saccharomyces paradoxus, Naumovozyma castellii, and Candida dubliniensis were obtained from Hooks et al. [25]. Exon-intron junctions from these HAC1 sequences, along with those of Ogataea polymorpha which were annotated in the National Center for Biotechnology Information (NCBI) database, were extracted (15nt from each side) and scored by position weight matrix (PWM) [48] in Data Analysis in Molecular Biology and Evolution (DAMBE7) [49] with default settings. The resulting PWM scores (Supplemental File S1) were used to characterize the HAC1 5′ and 3′ splice sites of the remaining 14 species which do not have fully annotated HAC1 splice junctions in the NCBI GenBank.
2.2. Determining the Putative 3′ Degron Sequence and Its Degree of Conservation in 32 Yeast Species
The putative degron sequence was previously determined in the budding yeast S. cerevisiae by Di Santo et al. [38], which consists of 29nt from the 5′ end of the intron sequence followed by the first in-frame stop codon (the stop codon UGA designated as STOP1 in Figure 1). The truncated HAC1u in S. cerevisiae, which is 687nt from the start codon in the first exon until STOP1, translates into a 228 amino acid-long truncated Hac1up. To determine a putative degron sequence in the other 31 yeast species surveyed in this study, we manually identified the first in-frame stop codon (STOP1) in the intron of their HAC1 genes such that the nucleotide sequence of the truncated HAC1u, from start codon until STOP1, is translatable (divisible by 3), and the putative degron was determined as the sequence from the beginning of the intron until STOP1.
Most HAC1 introns contained an in-frame stop codon (STOP1). However, Candida dubliniensis, Candida albicans, Sugiyamaella lignohabitans, Ascoidea rubescens, and Yarrowia lipolytica did not contain an in-frame STOP1 (Supplemental File S1). Additionally, Cyberlindnerajadinii and Candida orthopsilosis contain in-frame stop codons TGA and TAA, respectively, after just one in-frame codon in intron, and one Candida haemulonii has an in-frame stop codon TGA after just two in-frame codons in intron; the sequence between the 5′ end of intron to STOP1 in these species are too short to constitute degron sequences. In brief, a total of 24 out of 32 species contain an in-frame STOP1 and the lengths of the degron sequences (including STOP1) range between 17nt to 101nt, coding between five to 33 sense codons.
To determine the degree of conservation between degrons in 32 species, the degron segments were retrieved as nucleotide sequences and as translated amino acids. In S. cerevisiae, the degron-encoded codons consist of 29nt upstream of STOP1 plus 1nt at the 3′ end of first exon; similarly, up to 2nt from the 3′ end of exon 1 was considered by the first degron-encoded codon for select yeast species (See Supplemental File S1) to ensure the degron is translatable as an amino acid sequence. The nucleotide and amino acid degron sequences were then aligned by Multiple Alignment using Fast Fourier Transform (MAFFT) with the slow but accurate G-INS-i option (global alignment approach using the Needleman-Wunsch algorithm) [50] implemented in DAMBE. Next, a heatmap was generated for the alignments with the degron in S. cerevisiae as reference, and a total similarity score (indicating the total number of matching nucleotides and amino acids at the degron) was calculated for each species.
2.3. Reconstructing the Phylogenetic Relationship of 32 Yeast Species and Determining Relative Conservation at HAC1 Intronic and Exonic Regions
Two phylogenetic trees were constructed for the 32 yeast species, one with complete HAC1 genes and the other with 18S rRNAs. Multiple sequence alignments (MSA) of HAC1 and 18S rRNA nucleotide sequences were performed using Multiple Sequence Comparison by Log-Expectation (MUSCLE) [51] implemented in DAMBE. Then, phylogenetic relationships were inferred from aligned sequences using PHYML (a phylogeny software based on the maximum-likelihood principle) approach [52] with bootstrap = 500, tree improvement = Nearest Neighbour Interchange (NNI), and best model selected by Smart Model Selection (SMS) [53] = Generalised time reversible model (GTR + G + I) based on Akaike Information Criterion (AIC). Then, the phylogenetic trees were illustrated using the Interactive Tree of Life (iTOL) v4 [54].
Relative degrees of site-specific conservation among aligned HAC1 nucleotide sequences were measured using the Phylogenetic Analysis with Space/Time models (PHAST) package [55]. The phylogenetic tree was fitted to the MSA by maximum likelihood using phyloFit, and the MSA and resulting neutral model file were used to score conservation with PhastCons. For both PHAST programs (phyloFit and PhastCons), parameters were left as default. PhastCons works by fitting a phylogenetic hidden Markov model (phylo-HMM) to the data by maximum likelihood, subject to constraints designed to calibrate the model across species groups, and then predicting and assigning log-odds scores to conserved elements based on this model [56]. Conservation scores are posterior probabilities generated at each site by their conserved state, and the score enables comparison of conservation across sites in a sequence alignment [56]. Relative conservation of the exonic and intronic (degron and downstream) regions was calculated as the average conservation score considering all nucleotide sites in the gene segment. To ensure that mean conservation results were not skewed (e.g., some exonic regions partially mapping to intronic regions of other sequences in the alignment), the process was repeated with coding DNA-sequence (CDS) and intronic HAC1 sequences separately.
2.4. Retrieving and Processing S. cerevisiae Transcriptomic Data
The experimental information (Supplemental File S1) and FASTQ (FASTA with quality score) transcriptomics data of S. cerevisiae from Van Dalfsen et al. [57] were retrieved from Gene Expression Omnibus (GEO) Datasets [58]. These include the datasets of untreated wild type cells (RNA-Seq: SRR7265163 and SRR7265164; Ribo-Seq: SRR7265151 and SRR7265152), cells treated with dithiothreitol (DTT) to induce UPR (RNA-Seq: SRR7265165 and SRR7265166; Ribo-Seq: SRR7265153 and SRR7265154), and cells treated with tunicamycin (TM) to induce UPR (RNA-Seq: SRR7265167 and SRR7265168; Ribo-Seq: SRR7265155 and SRR7265156).
Analyzing RNA-Seq Data (ARSDA) [59] was used to process the original FASTQ files in the following workflow: FASTQ → FASTA → FASTA+ → Basic Local Alignment Search Tool (BLAST) database (Figure 2). After initial conversion to FASTA format, the data was trimmed (‘FileǀRTrim Fasta Sequences’) to exclude the 3′-poly-A tails that were added to the Ribo-Seq sequences during library generation, with minimum length set to 50 and reads with ambiguous nucleotides excluded. When converting from FASTA to FASTA+, the minimum read length was specified as 50, so that all shorter sequences were excluded. The FASTA+ format groups identical sequences in the processed FASTA file under a new unique sequence ID (SeqID_#, where # is the number of identical copies), this reduces data size without loss of information [59].
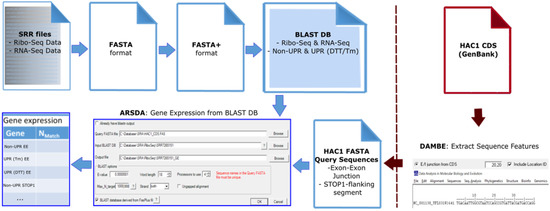
Figure 2.
Processing of RNA-Seq and Ribo-Seq data. ARSDA [59] was used to convert FASTQ datasets into the BLAST database files for profiling analyses. The query sequences were made from the HAC1 sequence file, retrieved from NCBI GenBank [60] using DAMBE7 [49].
2.5. Characterizing HAC1 Splicing Efficiency and Quantifying the Number of Translation Units Mapped to HAC1 Transcripts in S. cerevisiae
Empirical measurement of splicing efficiency (SE) is based on the quantification of spliced and unspliced forms of exonic and intronic sequences. An early attempt to characterize SE is by microarray, which quantifies the exon-exon junction (EE, featuring the spliced form) and exon-intron junction at the 5′ and 3′ sides of an intron (EI5 and EI3 respectively, featuring the unspliced forms) [61]. With the same rationale, a simple but accurate RNA-Seq-based approach to quantify SE has been developed and applied to yeast introns [62]. Here, we apply the RNA-Seq-based approach to quantify HAC1 SE in S. cerevisiae.
To make query sequence files, DAMBE7 [49] was used to extract the HAC1u sequence from S. cerevisiae HAC1 gene and three 40nt splice junction sequences were made: EE (20 nt from each exon, overall 40 nt long), EI5 (20 nt at the 3′ end of exon 1 and 20 nt at the 5′ end of intron), and EI3 (20nt at the 3′ end of intron and 20nt at the 5′ end of exon 2) in separate FASTA files. A third query sequence was made which contains the degron sequence: the HAC1u sequence was truncated to include only the intronic stop codon STOP1 and 20nt upstream of STOP 1 and 17nt downstream of STOP1 (overall 40nt long) and renamed as TargetHAC1u.
The three 40nt splice junction queries and TargetHAC1u were mapped to RNA-Seq and Ribo-Seq BLAST databases using BLAST in ARSDA. The BLAST critical E-value Ei where i is the BLAST database is calculated as follows: for i = SRR7265151 with 9699410 sequences of 50 nt each, the effective length for a match of 25 nt is m = 9699410×(50 − 25), and Ei of a read of 50 nt matching a query of 40 nt with an exact match of 25 consecutive bases is Ei = (40 − 25) × 9699410 × (50 − 25) × 2(−2 × 25) = 3.23 × 10−6. To prevent false matches, only mapped reads with match length ≥ 25 and match E-value < critical Ei were retained. Hence, the BLAST specifications are as follows: ungapped, minimum match length = 25, E value cutoff = 10−7, and Max number of target = 1,000,000. Furthermore, to ensure each reads map over the junction points in our queries, we manually checked each individual mapped region between q.start (beginning of map location on query site) and q.end (end of read map location on query site) to confirm that they overlap the junction points located at sites 20 and 21. All other mapped reads that do not fit these criteria were discarded.
To determine SE, we quantified the number of RNA-Seq reads mapped to the three splice junction queries. The number of mapped reads on EE junctions (NEE) quantifies the spliced mRNA, while the number of reads mapped to EI5 (NEI5) and EI3 (NEI3) junctions quantifies two independent measures of the unspliced mRNA. NEI5 is typically smaller than NEI3 for two reasons [62]. First, step 1 splicing reaction occurs before step 2 splicing reaction, and cleavage of EI5 occurs before EI3 to render less EI5 for sequencing. Second, EI3 is more prone to be included in a library generated with RNA-Seq data because a majority of the RNA-Seq data obtained from the library is enriched in poly(A) tail by oligo-dT. Thus, the total number of HAC1 mRNA is not measured as the average NTotal = NEE + (NEI5 + NEI3)/2, but is corrected as NTotal = NEE + pNEI5 + (1 − p)NEI3 with proportion p = NEI5/(NEI5 + NEI3), and SE = NEE/NTotal [41].
Translation efficiency of a given mRNA is correlated with the number of ribosomes involved with the mRNA [63]. Hence, to estimate the number of translation units on HAC1 transcripts, we quantified ribosome footprints mapped to the spliced HAC1i transcripts by the number of Ribo-Seq read matches at EE, and we quantified ribosome footprints mapped to the unspliced HAC1u transcript by the average number of Ribo-Seq reads mapped at EI5, EI3, and TARGETHAC1u.
3. Results
3.1. Efficient HAC1 Splicing Occurs in Unfolded Protein Response (UPR)-Induced S. cerevisiae Cells
Following an RNA-Seq-based approach to quantify SE, Table 1 shows the number of reads mapped to the three splice junctions in UPR-induced cells. HAC1u splicing is highly efficient in UPR-induced cells, with SE ranging from 0.955 to 1. NEE ranges from 42 to 140, but when mapped counts were adjusted for differences in Sequence Read Archive (SRA) data size (in number of kilobases in processed datasets), NEE/kb becomes more consistent among the four UPR-induced experiments, ranging from 2.7 × 10−4 to 4.2 × 10−4 (Supplemental File S1). However, in non-UPR cells, only one HAC1i transcript (NEE = 1, NEI5 = 0, and NEI3 = 0) was detected in both experiments (SRR7265163, 4) (Supplemental File S1). Consequently, non-UPR results were omitted from Table 1 because SE could not be calculated, and no statements can be made based on the results of a single (or zero) RNA-Seq reads for a gene product.

Table 1.
HAC1u is spliced efficiently in unfolded protein response (UPR)-induced cells. The number of RNA-Seq reads mapped to the three splice junctions were quantified by ungapped BLAST in ARSDA. BLAST databases were made for RNA-Seq datasets from WT yeast cells expressed under DTT-induced UPR (SRR7265165, 66) and TM-induced UPR (SRR7265167, 8) conditions.
3.2. Translation of HAC1u Is Constitutive in Both UPR-Induced and Non-UPR S. cerevisiae Cells
Ribosome profiling showed that HAC1i is translated only in UPR-induced cells, but that a constant and high rate of HAC1u translation occurs in both UPR-induced and non-UPR cells. Table 2 shows that the number of ribosome protected reads mapped to the EE junction (NEE, representing the spliced form) is substantially greater in UPR-induced cells (ranging from 85 to 178) than in non-UPR cells (2 and 5). When adjusted for difference in data size, NEE/kb ranges from 2.5 × 10−4 to 4.4 × 10−4 in UPR-induced experiments and it is 1.4 × 10−5 and 6.4 × 10−6 in the two non-UPR experiments. This is as expected because HAC1u splicing is efficient in UPR-induced cells (Table 1). In contrast, the number of translation units mapped to the unspliced HAC1u transcripts as estimated by TIHAC1u (the average number of ribosome-protected reads mapped to the three unspliced 40 nt queries EI5, EI3, and TargetHAC1u) are comparable between both cell types (UPR-induced and non-UPR). Surprisingly, TIHAC1u is not substantially lower than NEE in UPR-induced cells (Table 2). Considering all six experiments regardless of UPR, TIHAC1u ranges from 69 to 128. Additionally, when adjusted for differences in data size, TIHAC1u/kb ranges from 2.0 × 10−4 to 3.4 × 10−4 (Supplemental File S1) when all six Ribo-Seq experiments were considered. This range is comparable to NEE/kb ranging from 2.5 × 10−4 to 4.4 × 10−4 in the four UPR-induced experiments.

Table 2.
Ribosome profiling shows good HAC1u translation intensity in both cell types. The number of translation units mapped to the spliced HAC1i transcripts was estimated by NEE, whereas the number of translation units mapped to the unspliced HAC1u transcripts was estimated by TIHAC1u (the average of NEI5, NEI3, and N_TargetHAC1u). Number of ribosome-protected reads mapped to each region was determined by ungapped BLAST in ARSDA. BLAST databases were made from Ribo-Seq datasets of WT yeast cells expressed under non-UPR (SRR7265151, 2), DTT-induced UPR (SRR7265153, 4), and Tm-induced UPR (SRR7265155, 6) conditions.
3.3. The Putative Intron-Encoded Degron Is Conserved by Saccharomyces Yeasts but Not by Other Yeasts
Above results showed a constitutive degree of leaky HAC1u translation in both UPR-induced and non-UPR S. cerevisiae cells albeit the presence of a translation block to prevent undesirable HAC1u translation (Figure 1b). Hence, it would be beneficial if yeast gained an additional post-translational mechanism to rapidly degrade Hac1up. If the previously proposed degron sequence encoded in the last 10 amino acid of S. cerevisiae Hac1up [38] is important, then we expect this intronic sequence should be evolutionarily conserved among closely related yeast species.
Figure 3 shows the local amino acid alignments at putative degrons in 24 yeast species out of the 32 surveyed (See Materials and Methods for degron determination). As expected, we found that the intron-encoded degron is entirely conserved in the two Saccharomyces yeast species. Despite this, the degron in Saccharomyces yeast is poorly aligned to putative degrons determined in all 22 other yeast species. Two putative degrons that are most similar to that in S. cerevisiae are from Torulaspora delbrueckii of the Saccharomycetaceae family and Wickerhamomyces ciferii of the Phaffomycetaceae family, but both degrons only share 5 out of the 10 amino acid identities with degron of S. cerevisiae (Figure 3). Furthermore, three yeasts, Naumovozyma castellii and Naumovozyma dairenensis of the Saccharomycetaceae family and Clavispora lusitaniae of the Metschnikowiaceae family, encode putative degrons that share four out of the ten amino acid identities with degron of S. cerevisiae. It is worth noting that the degree of amino acid conservation in all five aforementioned species are mainly attributed to the first four amino acids (A, V, I, and T) which are the most conserved by yeast degrons. Lastly, 17 other yeasts encode putative degrons that share only three or fewer amino acid identities with degron of S. cerevisiae. Additionally, we performed a local alignment at degron nucleotide sequences (Supplemental Figure S1), which showed slightly higher overall similarities among degrons and relatively high degrees of conservation at the first 12 nucleotide sites (consistent with the conservation of the first four amino acids A, V, I, and T). To summarize, although the degron sequence in S. cerevisiae is poorly conserved by other yeasts at the genus level, the first four degron amino acids (A, V, I, T) are variably highly conserved by yeast species surveyed.
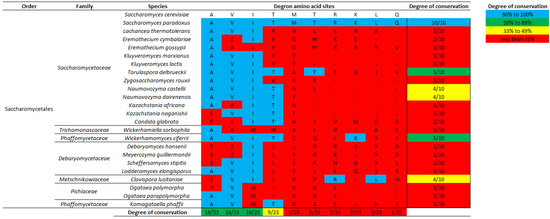
Figure 3.
Local amino acid alignments at 24 putative yeast degrons show that the degron sequence is conserved between two Saccharomyces species but not by any other yeast (See Materials and Methods for determination of degron-encoded amino acid sequences). Highlighted blue and red are matching and mis-matching amino acid sites, respectively, in yeast degrons against the reference S. cerevisiae degron. Degree of conservation designates the total number of matching amino acid residues, at the whole degron (last column) and at each amino acid site (last row), with scores in blue highlights high similarity (90–100%), in green highlights medium similarity (50–89%), in yellow highlights medium-low similarity (33–49%), and in red highlights low similarity (>33%).
We next reconstructed the phylogenetic relationships (see Materials and Methods) among 32 yeast species to further showcase that the degron sequences are not better conserved by lineages closely related to Saccharomyces species. Two phylogenetic trees were reconstructed, one with whole HAC1 nucleotide sequence alignment (Figure 4) and one with 18S rRNA alignment (Supplemental Figure S2), and both show two major clades representing the Saccharomycetacea family (red) and the Debaryomycetaceae family (blue). While the two trees are not completely identical in topology, they both illustrate that most, but not all, yeast species can be grouped appropriately by their hierarchical taxonomic ranks. For example, Ascoidea rubescens of the Ascoideaceae family and Clavispora lusitaniae and Candida haemulonii of the Pichiaceae family cannot be separated from the clade representing the Debaryomycetaceae family with HAC1 alignments. Figure 4 additionally shows that species closely related to the two Saccharomyces yeasts do not retain better conserved degron sequences (with degree of degron conservation indicated by the same color scheme as shown in Figure 3) than those more distantly related. A putative degron can be determined in all 14 Saccharomycetales, whereas among species of other yeast families, eight out of the 18 do not have an identifiable degron sequence. Nonetheless, most Saccharomycetales yeasts have poor similarities when compared to the S. cerevisiae degron (Figure 4). Even at the sister group of Saccharomyces species, the degron in two species (Zygosaccharomyces rouxii and Kazachstania africana) are poorly conserved (three shared amino acids) and the degron in the remaining three species (Naumovozyma castellii, Naumovozyma dairenensis, and Tourulaspora delbrueckii) are weakly conserved (four to five shared amino acids).
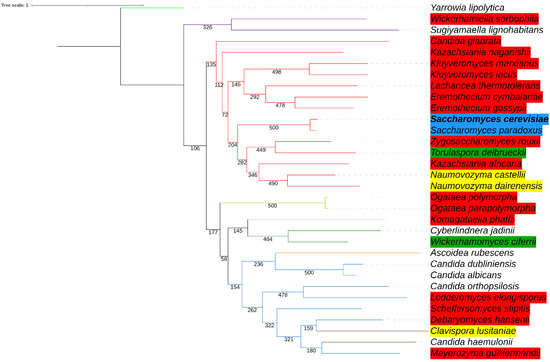
Figure 4.
Phylogenetic relationships of 32 yeast species with support values built with whole HAC1 alignments. The phylogenetic tree is constructed using the maximum-likelihood-based PHYML approach, with best model = GTR + G + I, Bootstrap = 500, and topology re-rooted at Yarrowia lipolytica. Yeast species belong to nine families with tree branches highlighted in color: Saccharomycetaceae (red), Trichomonascaceae (purple), Phaffomycetaceae (green), Ascoideaceae (orange), Debaryomycetaceae (blue), Metschnikowiaceae (brown), Pichiaceae (yellow), Phaffomycetaceae (grey), and Dipodascaceae (light green). Color highlighted at species name indicates the degree of amino acid conservation at the whole degron, with reference to the S. cerevisiae degron, as shown in Figure 3. Species in which a putative degron cannot be identified are not highlighted by color.
To further demonstrate that the degron is indeed highly conserved among Saccharomyces yeasts beyond the two examined above, we identified and retrieved the degron from the HAC1 gene of three additional Saccharomyces species (S. pastorianus, S. jueri, and S. kudriavzevii). Together, a local nucleotide alignment between five Saccharomyces species (Figure 5) at the degrons (encoded amino acids in bold, STOP1 in blue), flanked by nine sites at the 3′ terminus of exon 1 (grey) and nine sites downstream of STOP1, showed that the degron is almost entirely conserved among the five Saccharomyces yeasts. The nine sites on the 3′ end of exon 1 are fully conserved among the five Saccharomyces species. At the putative degron sequence (bold), only two nucleotide positions are not fully conserved, but all nucleotide differences are synonymous mutations. In comparison, intron sites downstream of STOP1 are poorly conserved, with only three conserved sites out of the nine examined.
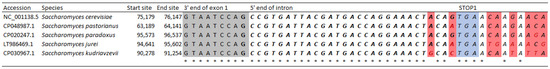
Figure 5.
Local sequence alignments shows that the putative degron sequence is well conserved between Saccharomyces cerevisiae and four other closely related Saccharomyces yeasts. The start and end site respectively denote the start and end locations of the entire HAC1 gene in the yeast genomes. Shaded grey are the nine sites on the 3′ terminus of exon 1. Sequence in bold is the putative degron sequence in S. cerevisiae. Shaded in blue is the intronic STOP1 used by the truncated Hac1up. Shaded in red are non-conserved sites and the * at the bottom denotes fully conserved sites. Nucleotide sequences were aligned using MAFFT G-INS-i.
3.4. The Degron Sequence Is the Most Conserved Segment at the HAC1 Intron
Above we showed that although the degron in Saccharomyces yeast is poorly conserved by other yeast species, the first 4 out of 10 amino acids are relatively highly conserved among putative degrons. To assess the degree of conservation at the degron sequences relative to other HAC1 gene segments, we next performed comparative sequence analyses using MUSCLE aligned HAC1 genes of the 32 species listed in Figure 3, with S. cerevisiae HAC1 gene as reference. Table 3 shows that, by average PhastCons score (See Materials and Methods), the putative intron-encoded degron is well conserved in comparison to the intronic region downstream of STOP1. Furthermore, exon 1 is better conserved than exon 2 and both are better conserved than the intron, but both exons are less conserved than the degron sequence.

Table 3.
The averaged nucleotide site conservation at HAC1 exons and introns (at Degrons and downstream sequences) as scored by PhastCons. The conservation scores at “Complete Alignment” were computed using entire HAC1 gene alignments, whereas scores at “Separate alignment” were computed using HAC1 CDS and intron sequence alignments separately.
Nonetheless, the above results are not surprising since the length of nucleotide sequence considered at the degron region (32nt including STOP1) was much shorter than that of the other three regions (exon 1, exon 2, downstream intronic region). To better assess the degree of conservation at the degron in comparison to the rest of the S. cerevisiae HAC1 gene, we computed the average PhastCons scores in short segments, with lengths equaling the length of the S. cerevisiae degron (window size = 32), that spanned the entire S. cerevisiae HAC1 gene (with step size = 1). Figure 6 shows that the degron sequence (purple) is indeed much more conserved in comparison to segments mapping to downstream intronic regions (red). In fact, the degron is comparably conserved against segments mapping to exon 2 (blue). At exon 1 however, segments spanning from sites 42 to 243 (step size # 42 to 212) and 622 to 661 (step size # 622 to 630) are highly conserved with average PhastCons scores higher than that of the degron.
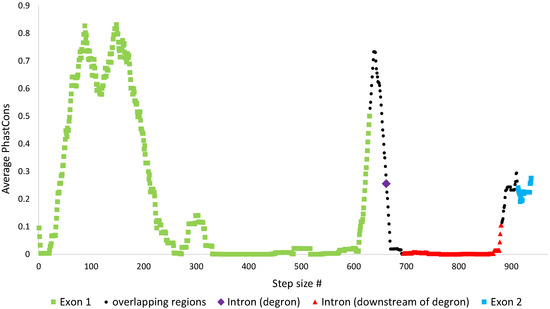
Figure 6.
The averaged PhastCons scores for 32 nt segments spanning the entire S. cerevisiae HAC1 gene, with window size = 32 (length of the degron) and step size = 1. The scores were computed using 32 MUSCLE aligned complete HAC1 genes and with S. cerevisiae HAC1 as reference. Scores are color-coded by region, for segments that mapped entirely within exon 1 (green), degron (purple), rest of intron (red), and exon 2 (blue). In addition, average PhastCons scores of 32 nt segments whose sequences overlapped exon-intron junctions or degron-downstream intron sequence are colored in black.
4. Discussion
HAC1 is constitutively transcribed in the S. cerevisiae yeast [4,9] and multiple studies have detected spliced HAC1i mRNAs in both non-UPR [19,44] and UPR-induced cells [19,44,64]. Yet splicing and translation of HAC1 is tightly controlled and evolution has placed multiple fail-safe mechanism against leaky HAC1 splicing and translation as these events would allow for the accumulation of Hac1up and Hac1ip to induce undesirable UPR [1]. First, Kar2p chaperone proteins hinder Ire1p oligomerization and prevent undesired HAC1u splicing during non-ER stress [8,42,43]. Second, HAC1 5′ UTR interacts with the intron to form a translation block against HAC1u translation. The Ire1p+Hac1p-mediated UPR signal in S. cerevisiae represents a beautiful translation control created by nature. Nonetheless, HAC1u mRNAs are detected in both non-UPR [38,44] and UPR-induced cells [44,64].
In spite of the fail-safe mechanisms mentioned above, a low level of leaky splicing and leaky translation of HAC1 transcripts may still occur [38]. Thus, a post-translation control would benefit the cell against undesired production of Hac1ip and Hac1up. Indeed, Hac1p protein expressions are tightly regulated. For example, while HAC1i and HAC1u mRNAs are actively translated during UPR, only Hac1ip was readily detected and Hac1up was quickly degraded [4,37,38]. Previous studies proposed that a degradation signal is encoded in the first exon [45], which helps explain why both Hac1up and Hac1ip are unstable and rapidly degraded [26,38,46,65,66] and why previous studies [4,26,67,68] were only able to detect Hac1ip under UPR conditions. In addition, the budding yeast has been proposed to have evolved a secondary degradation “degron” signal to specifically target and further facilitate the degradation of Hac1up [38].
Our investigation employed ribosome profiling and comparative sequence analyses to demonstrate that having an additional intron-encoded degron may indeed be a much-required element to specifically silence undesired Hac1up production. By ribosome profiling, we demonstrated the inability of translation fail-safe to completely inhibit Hac1up production. Both the spliced HAC1i and unspliced HAC1u isoforms are constitutively translated in both UPR-induced and non-UPR cells. In fact, Table 2 shows that the translation intensity of HAC1u is comparable to the translation intensity of HAC1i in UPR-induced cells regardless of UPR induction.
Furthermore, through comparative sequence analyses, we determined the intronic sequence encoding the degron to be fully conserved by closely related Saccharomyces yeasts (Figure 3 and Figure 5). However, degron conservation is limited to long introns in Saccharomyces yeasts and the degron is poorly conserved by other yeast lineages (Figure 3 and Figure 4). Nonetheless, a comparative sequence analysis showed that the first four degron amino acids, A, V, I, and T are highly conserved among putative degrons in 22 non-Saccharomyces yeasts (Figure 3), and the degron region is notably more conserved than the downstream intronic regions in yeast species (Figure 6). Similarly, a comparative gene study by Hooks and Griffiths-Jones [25] suggests that the 5′ UTR translation block may also be conserved only in HAC1 with long introns in Saccharomyces yeasts. Future transcriptomics studies on non-Saccharomyces yeasts in both UPR-induced and non-UPR conditions may pave the way for determining whether yeasts such as Candida species having short HAC1 introns have acquired alternative mechanisms to regulate UPR and HAC1 splicing.
It is noteworthy that sequence conservation at the degron in Saccharomyces yeasts is not a direct evidence of its degradation function and require further verification by application of mutagenesis and proteomics methods. Such experiments have been performed on the S. cerevisiae yeast [38], as summarized in the Introduction, but the need for a degron as a “fail-safe” against Hac1up expression remains to be tested in other yeast species. Our findings suggest a functional importance for the degron sequence in Saccharomyces yeast to the exclusion of other yeasts, and we expect our findings to motivate future studies to experimentally test the degron function in other Saccharomyces yeasts. The evolutionary pressure underlying degron conservation in Saccharomyces yeasts suggests a potentially important degron-mediated post-translational regulation to specifically prevent the accumulation of Hac1up, which when unchecked could trigger Ire1p-independent activation of the UPR, since Hac1up is constitutively translated (Table 2) and may be capable of functioning as an active transcription factor at UPRE promoters [38]. Lastly, it is interesting to note that if a degron is present in the last 10-aa of Hac1up, it is also possible that the degradation signal in Hac1ip and Hac1up are independently encoded. As visualized in Figure 7, an additional degron could potentially be encoded in the last 18 amino acids of exon 2 for Hac1ip.
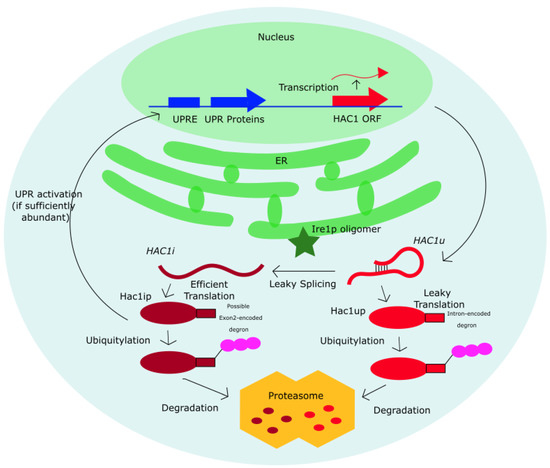
Figure 7.
A summary of the proposed independent post-translational control for Hac1ip and Hac1up. Both Hac1up and Hac1ip may independently encode a degron sequence that signals for their ubiquitylation and degradation, with the former located in the first 10 amino acid of the intronic sequence upstream of the intronic stop codon (STOP1) and the latter located within the 18 amino acid of exon 2.
The homologous XBP1 gene in humans and other metazoans also contains a conserved intron which undergoes similar Ire1p-mediated unconventional splicing for UPR activation [25]. Similar to HAC1, the protein translated from unspliced XBP1 mRNA, pXBP1(U), is truncated and unable to trigger UPR, while mRNA splicing causes a frame shift to allow pXBP1(S) to encode an effective transcription factor [69]. It has been shown that the C-terminus of pXBP1(U) is sufficient to direct rapid degradation of the protein [70,71], indicating a potential preservation of a degron-like sequence in higher eukaryotes [71].
The causes compelling Saccharomyces yeasts to rely on a pair of translational and post-translational regulation mechanisms remain unclear. It is possible that the leniency of the 5′ UTR-intron base-pairing interactions leading to leaky translation imparts an advantage on the cell, by allowing for the proper function of other processes regulating HAC1 mRNA, such as Ire1p-dependent splicing. On the other hand, it may be that this constitutive production and degradation of Hac1up itself serves a regulatory function, with the intriguing possibility that Hac1up can heterodimerize with residual levels of Hac1ip produced by leaky splicing to facilitate its degradation in the absence of ER stress. If Hac1up can function both as a translation factor and regulator of Hac1ip, it is possible that the metazoan ortholog of Hac1up, pXBP1(U) [72], can also act as a transcription factor, as pXBP1(U) is known to heterodimerize with its spliced form (pXBP1(S)) to accelerate pXBP1(S) degradation [71]. The Ire1p-Xbp1 pathway in humans may possess unexplored similarities with the Ire1p-Hac1p pathway in yeast, and future comparative research may provide novel insights into the regulation of the UPR in human cells.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/3/620/s1, Supplemental File S1 contain RNA-Seq and Ribo-Seq profiling analyses, sequence annotations and alignments, and computed PhastCons scores. Supplemental File S2 contains Supplemental Figures S1 and S2. The processed RNA-Seq datasets are available online at http://dambe.bio.uottawa.ca/Transcriptome.zip (accessed on 1 February 2021) and the processed Ribo-Seq datasets are available online at http://dambe.bio.uottawa.ca/RiboProf.zip (accessed on 1 February 2021).
Author Contributions
X.X. conceptualized the study. Y.W., A.T., and T.R. wrote the manuscript. A.T. and T.R. collected the data. A.T., T.R., and Y.W. analyzed the data and prepared all Figures. X.X. developed the software. All authors reviewed the manuscript. X.X. supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant to X.X. [RGPIN/2018-03878], and NSERC Doctoral Scholarship to Y.W. [CGSD/2019-535291].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://dambe.bio.uottawa.ca/Transcriptome.zip (accessed on 1 February 2021) and http://dambe.bio.uottawa.ca/RiboProf.zip (accessed on 1 February 2021). The data presented in this study are available in Supplemental Files S1 and S2.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gardner, B.M.; Pincus, D.; Gotthardt, K.; Gallagher, C.M.; Walter, P. Endoplasmic Reticulum Stress Sensing in the Unfolded Protein Response. Cold Spring Harb. Perspect. Biol. 2013, 5, a013169. [Google Scholar] [CrossRef]
- Mori, K. The unfolded protein response: The dawn of a new field. Proc. Jpn. Acad. Ser. B 2015, 91, 469–480. [Google Scholar] [CrossRef]
- Walter, P. Walking Along the Serendipitous Path of Discovery. Mol. Biol. Cell 2010, 21, 15–17. [Google Scholar] [CrossRef]
- Cox, J.S.; Walter, P. A Novel Mechanism for Regulating Activity of a Transcription Factor That Controls the Unfolded Protein Response. Cell 1996, 87, 391–404. [Google Scholar] [CrossRef]
- Korennykh, A.V.; Egea, P.F.; Korostelev, A.A.; Finer-Moore, J.; Zhang, C.; Shokat, K.M.; Stroud, R.M.; Walter, P. The unfolded protein response signals through high-order assembly of Ire1. Nat. Cell Biol. 2008, 457, 687–693. [Google Scholar] [CrossRef]
- Gardner, B.M.; Walter, P. Unfolded Proteins Are Ire1-Activating Ligands That Directly Induce the Unfolded Protein Response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Van Anken, E.; Pincus, D.; Coyle, S.; Aragón, T.; Osman, C.; Lari, F.; Puerta, S.G.; Korennykh, A.V.; Walter, P. Specificity in endoplasmic reticulum-stress signaling in yeast entails a step-wise engagement of HAC1 mRNA to clusters of the stress sensor Ire1. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Kimata, Y.; Oikawa, D.; Shimizu, Y.; Ishiwata-Kimata, Y.; Kohno, K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J. Cell Biol. 2004, 167, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Kawahara, T.; Yoshida, H.; Yanagi, H.; Yura, T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1996, 1, 803–817. [Google Scholar] [CrossRef]
- Shamu, C.E.; Walter, P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996, 15, 3028–3039. [Google Scholar] [CrossRef] [PubMed]
- Aragon, T.J.; Van Anken, E.; Pincus, D.H.; Serafimova, I.M.; Korennykh, A.V.; Rubio, C.A.; Walter, P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nat. Cell Biol. 2008, 457, 736–740. [Google Scholar] [CrossRef]
- Welihinda, A.A.; Tirasophon, W.; Green, S.R.; Kaufman, R.J. Protein Serine/Threonine Phosphatase Ptc2p Negatively Regulates the Unfolded-Protein Response by Dephosphorylating Ire1p Kinase. Mol. Cell. Biol. 1998, 18, 1967–1977. [Google Scholar] [CrossRef]
- Korennykh, A.V.; Korostelev, A.A.; Egea, P.F.; Finer-Moore, J.; Stroud, R.M.; Zhang, C.; Shokat, K.M.; Walter, P. Structural and functional basis for RNA cleavage by Ire1. BMC Biol. 2011, 9, 47. [Google Scholar] [CrossRef]
- Sidrauski, C.; Cox, J.S.; Walter, P. tRNA Ligase Is Required for Regulated mRNA Splicing in the Unfolded Protein Response. Cell 1996, 87, 405–413. [Google Scholar] [CrossRef]
- Sidrauski, C.; Walter, P. The Transmembrane Kinase Ire1p Is a Site-Specific Endonuclease That Initiates mRNA Splicing in the Unfolded Protein Response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Gonzalez, T.N.; Sidrauski, C.; Dörfler, S.; Walter, P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999, 18, 3119–3132. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, R.J. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999, 13, 1211–1233. [Google Scholar] [CrossRef]
- Welihinda, A.A.; Tirasophon, W.; Kaufman, R.J. The Cellular Response to Protein Misfolding in the Endoplasmic Reticulum. Gene Expr. 2018, 7, 293–300. [Google Scholar]
- Welihinda, A.A.; Tirasophon, W.; Kaufman, R.J. The Transcriptional Co-activator ADA5 Is Required for HAC1 mRNA Processing in Vivo. J. Biol. Chem. 2000, 275, 3377–3381. [Google Scholar] [CrossRef]
- Wimalasena, T.T.; Enjalbert, B.; Guillemette, T.; Plumridge, A.; Budge, S.; Yin, Z.; Brown, A.J.; Archer, D.B. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet. Biol. 2008, 45, 1235–1247. [Google Scholar] [CrossRef]
- Hernández-Elvira, M.; Torres-Quiroz, F.; Escamilla-Ayala, A.; Domínguez-Martin, E.; Escalante, R.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. The Unfolded Protein Response Pathway in the Yeast Kluyveromyces lactis. A Comparative View among Yeast Species. Cells 2018, 7, 106. [Google Scholar] [CrossRef]
- Guerfal, M.; Ryckaert, S.; Jacobs, P.P.; Ameloot, P.; Van Craenenbroeck, K.; DeRycke, R.; Callewaert, N. The HAC1 gene from Pichia pastoris: Characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb. Cell Factories 2010, 9, 49. [Google Scholar] [CrossRef]
- Moon, H.-Y.; Cheon, S.A.; Kim, H.; Agaphonov, M.O.; Kwon, O.; Oh, D.-B.; Kim, J.-Y.; Kang, H.A. Hansenula polymorpha Hac1p Is Critical to Protein N-Glycosylation Activity Modulation, as Revealed by Functional and Transcriptomic Analyses. Appl. Environ. Microbiol. 2015, 81, 6982–6993. [Google Scholar] [CrossRef]
- Oh, M.H.; Cheon, S.A.; Kang, H.A.; Kim, J.-Y. Functional characterization of the unconventional splicing of Yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast 2010, 27, 443–452. [Google Scholar] [CrossRef]
- Hooks, K.B.; Griffiths-Jones, S. Conserved RNA structures in the non-canonical Hac1/Xbp1 intron. RNA Biol. 2011, 8, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.E.; Walter, P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 1997, 7, 850–859. [Google Scholar] [CrossRef]
- Nikawa, J. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996, 24, 4222–4226. [Google Scholar] [CrossRef] [PubMed]
- Goffin, L.; Vodala, S.; Fraser, C.; Ryan, J.; Timms, M.; Meusburger, S.; Catimel, B.; Nice, E.C.; Silver, P.A.; Xiao, C.-Y.; et al. The Unfolded Protein Response Transducer Ire1p Contains a Nuclear Localization Sequence Recognized by Multiple β Importins. Mol. Biol. Cell 2006, 17, 5309–5323. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Sant, A.; Kohno, K.; Normington, K.; Gething, M.; Sambrook, J. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992, 11, 2583–2593. [Google Scholar] [CrossRef]
- Kohno, K.; Normington, K.; Sambrook, J.; Gething, M.J.; Mori, K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993, 13, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Fordyce, P.M.; Pincus, D.; Kimmig, P.; Nelson, C.S.; El-Samad, H.; Walter, P.; DeRisi, J.L. Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc. Natl. Acad. Sci. USA 2012, 109, E3084–E3093. [Google Scholar] [CrossRef] [PubMed]
- Patil, C.K.; Li, H.; Walter, P. Gcn4p and Novel Upstream Activating Sequences Regulate Targets of the Unfolded Protein Response. PLoS Biol. 2004, 2, e246. [Google Scholar] [CrossRef] [PubMed]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and Genomic Analyses Reveal an Essential Coordination between the Unfolded Protein Response and ER-Associated Degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef]
- Badis, G.; Chan, E.T.; van Bakel, H.; Pena-Castillo, L.; Tillo, D.; Tsui, K.; Carlson, C.D.; Gossett, A.J.; Hasinoff, M.J.; Warren, C.L.; et al. A Library of Yeast Transcription Factor Motifs Reveals a Widespread Function for Rsc3 in Targeting Nucleosome Exclusion at Promoters. Mol. Cell 2008, 32, 878–887. [Google Scholar] [CrossRef]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 2000, 97, 4660–4665. [Google Scholar] [CrossRef]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. Palindrome with Spacer of One Nucleotide Is Characteristic of the cis-Acting Unfolded Protein Response Element inSaccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 9912–9920. [Google Scholar] [CrossRef]
- Rüegsegger, U.; Leber, J.H.; Walter, P. Block of HAC1 mRNA Translation by Long-Range Base Pairing Is Released by Cytoplasmic Splicing upon Induction of the Unfolded Protein Response. Cell 2001, 107, 103–114. [Google Scholar] [CrossRef]
- Di Santo, R.; Aboulhouda, S.; Weinberg, D.E. The Fail-Safe Mechanism of Post-Transcriptional Silencing of Unspliced HAC1 MRNA. eLife 2016, 5. [Google Scholar] [CrossRef]
- Payne, T.; Hanfrey, C.; Bishop, A.L.; Michael, A.J.; Avery, S.V.; Archer, D.B. Transcript-specific translational regulation in the unfolded protein response ofSaccharomyces cerevisiae. FEBS Lett. 2008, 582, 503–509. [Google Scholar] [CrossRef]
- Kuhn, K.M.; DeRisi, J.L.; Brown, P.O.; Sarnow, P. Global and Specific Translational Regulation in the Genomic Response of Saccharomyces cerevisiae to a Rapid Transfer from a Fermentable to a Nonfermentable Carbon Source. Mol. Cell. Biol. 2001, 21, 916–927. [Google Scholar] [CrossRef]
- Xia, X. Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2019, 20, 2860. [Google Scholar] [CrossRef]
- Pincus, D.; Chevalier, M.W.; Aragón, T.; Van Anken, E.; Vidal, S.E.; El-Samad, H.; Walter, P. BiP Binding to the ER-Stress Sensor Ire1 Tunes the Homeostatic Behavior of the Unfolded Protein Response. PLoS Biol. 2010, 8, e1000415. [Google Scholar] [CrossRef]
- Oikawa, D.; Kimata, Y.; Kohno, K. Self-association and BiP dissociation are not sufficient for activation of the ER stress sensor Ire1. J. Cell Sci. 2007, 120, 1681–1688. [Google Scholar] [CrossRef] [PubMed]
- Bicknell, A.A.; Babour, A.; Federovitch, C.M.; Niwa, M. A novel role in cytokinesis reveals a housekeeping function for the unfolded protein response. J. Cell Biol. 2007, 177, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Pal, B.; Chan, N.C.; Helfenbaum, L.; Tan, K.; Tansey, W.P.; Gething, M.-J. SCFCdc4-mediated Degradation of the Hac1p Transcription Factor Regulates the Unfolded Protein Response inSaccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 426–440. [Google Scholar] [CrossRef][Green Version]
- Kawahara, T.; Yanagi, H.; Yura, T.; Mori, K. Endoplasmic Reticulum Stress-induced mRNA Splicing Permits Synthesis of Transcription Factor Hac1p/Ern4p That Activates the Unfolded Protein Response. Mol. Biol. Cell 1997, 8, 1845–1862. [Google Scholar] [CrossRef]
- Iracane, E.; Donovan, P.D.; Ola, M.; Butler, G.; Holland, L.M. Identification of an Exceptionally Long Intron in theHAC1Gene ofCandida parapsilosis. mSphere 2018, 3, e00532-18. [Google Scholar] [CrossRef] [PubMed]
- Stormo, G.D.; Schneider, T.D.; Gold, L.; Ehrenfeucht, A. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982, 10, 2997–3011. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 2009, 537, 39–64. [Google Scholar]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Pollard, K.S.; Siepel, A.C. PHAST and RPHAST: Phylogenetic analysis with space/time models. Brief. Bioinform. 2010, 12, 41–51. [Google Scholar] [CrossRef]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef] [PubMed]
- Van Dalfsen, K.M.; Hodapp, S.; Keskin, A.; Otto, G.M.; Berdan, C.A.; Higdon, A.; Cheunkarndee, T.; Nomura, D.K.; Jovanovic, M.; Brar, G.A. Global Proteome Remodeling during ER Stress Involves Hac1-Driven Expression of Long Undecoded Transcript Isoforms. Dev. Cell 2018, 46, 219–235.e8. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef]
- Xia, X. ARSDA: A New Approach for Storing, Transmitting and Analyzing Transcriptomic Data. G3 Genes Genomes Genet. 2017, 7, 3839–3848. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef] [PubMed]
- Pleiss, J.A.; Whitworth, G.B.; Bergkessel, M.; Guthrie, C. Rapid, Transcript-Specific Changes in Splicing in Response to Environmental Stress. Mol. Cell 2007, 27, 928–937. [Google Scholar] [CrossRef]
- Xia, X. RNA-Seq approach for accurate characterization of splicing efficiency of yeast introns. Methods 2020, 176, 25–33. [Google Scholar] [CrossRef]
- Rodnina, M.V. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci. 2016, 25, 1390–1406. [Google Scholar] [CrossRef]
- Leber, J.H.; Bernales, S.; Walter, P. IRE1-Independent Gain Control of the Unfolded Protein Response. PLoS Biol. 2004, 2, e235. [Google Scholar] [CrossRef]
- Okamura, K.; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/BiP from an ER Sensory Molecule, Ire1p, Triggers the Unfolded Protein Response in Yeast. Biochem. Biophys. Res. Commun. 2000, 279, 445–450. [Google Scholar] [CrossRef]
- Sarkar, D.; Paira, S.; Das, B. Nuclear mRNA degradation tunes the gain of the unfolded protein response in Saccharomyces cerevisiae. Nucleic Acids Res. 2017, 46, 1139–1156. [Google Scholar] [CrossRef]
- Arava, Y.; Wang, Y.; Storey, J.D.; Liu, C.L.; Brown, P.O.; Herschlag, D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2003, 100, 3889–3894. [Google Scholar] [CrossRef] [PubMed]
- Sathe, L.; Bolinger, C.; Mannan, M.A.-U.; Dever, T.E.; Dey, M. Evidence That Base-pairing Interaction between Intron and mRNA Leader Sequences Inhibits Initiation of HAC1 mRNA Translation in Yeast. J. Biol. Chem. 2015, 290, 21821–21832. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA Is Induced by ATF6 and Spliced by IRE1 in Response to ER Stress to Produce a Highly Active Transcription Factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Tirosh, B.; Iwakoshi, N.N.; Glimcher, L.H.; Ploegh, H.L. Rapid Turnover of Unspliced Xbp-1 as a Factor That Modulates the Unfolded Protein Response. J. Biol. Chem. 2006, 281, 5852–5860. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Oku, M.; Suzuki, M.; Mori, K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 2006, 172, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Iwakoshi, N.N.; Lee, A.-H.; Glimcher, L.H. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 2003, 194, 29–38. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).