Transcriptome Signatures in Pseudomonas simiae WCS417 Shed Light on Role of Root-Secreted Coumarins in Arabidopsis-Mutualist Communication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Root Exudates Collection
2.3. Bacteria Cultivation and Inoculation
2.4. Bacterial Sample Collection
2.5. cDNA Library Preparation
2.6. RNA Sequencing
2.7. Bacterial Motility Assay
3. Results
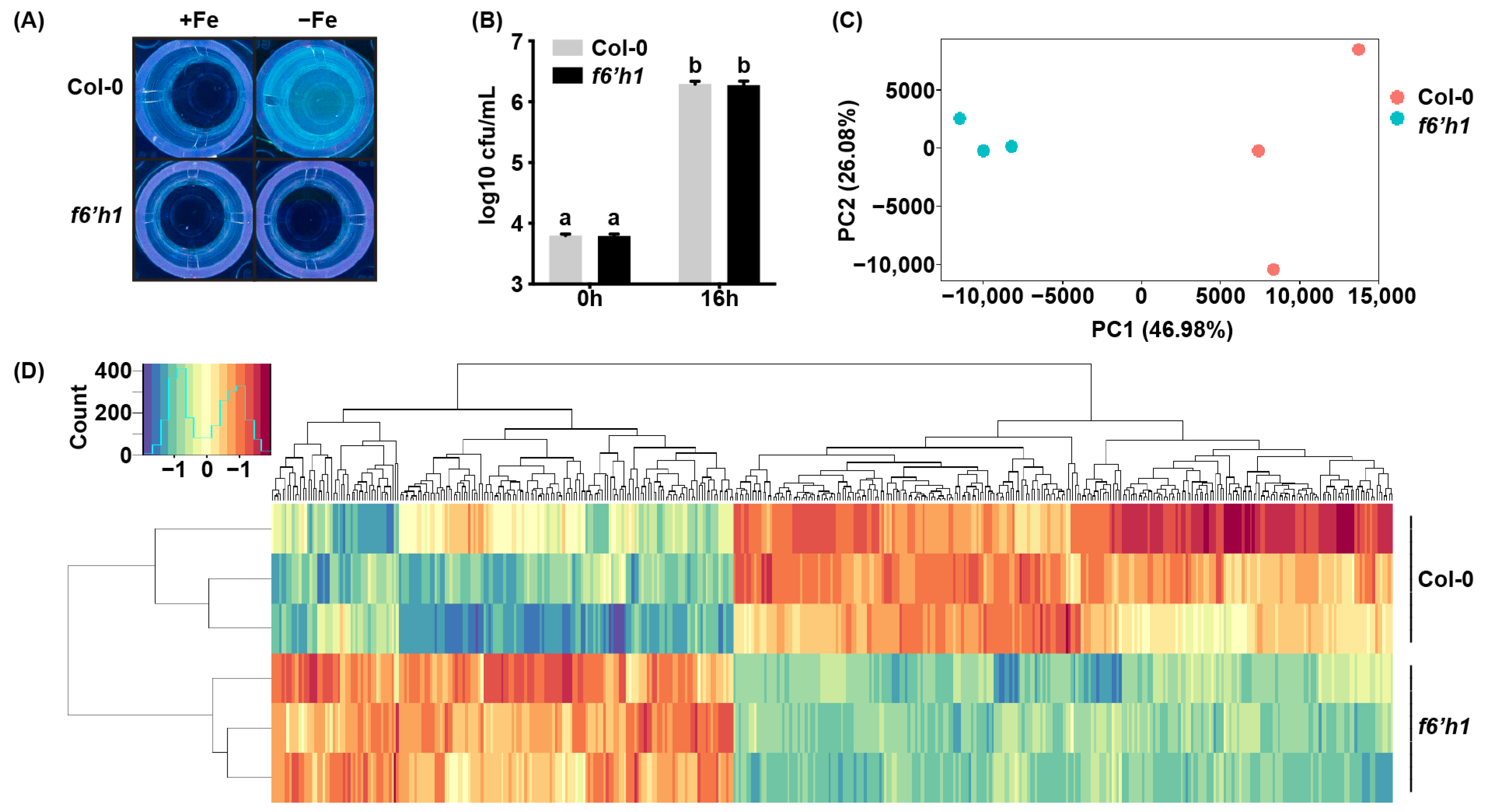
3.1. F6′H1-Dependent Coumarins Induce Transcriptional Changes in WCS417
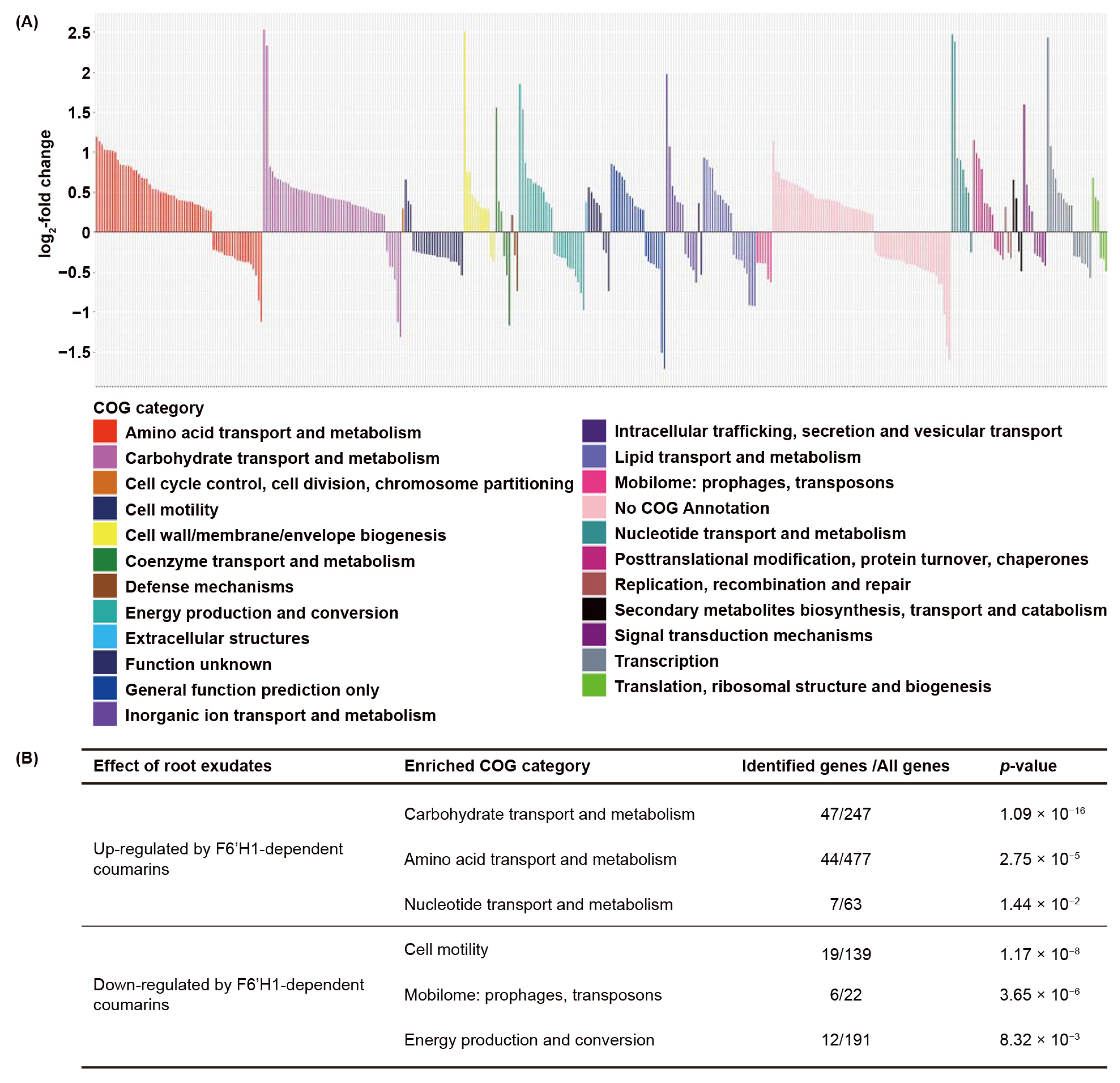
3.2. Biological Functions Affected by F6′H1-Dependent Root Exudates
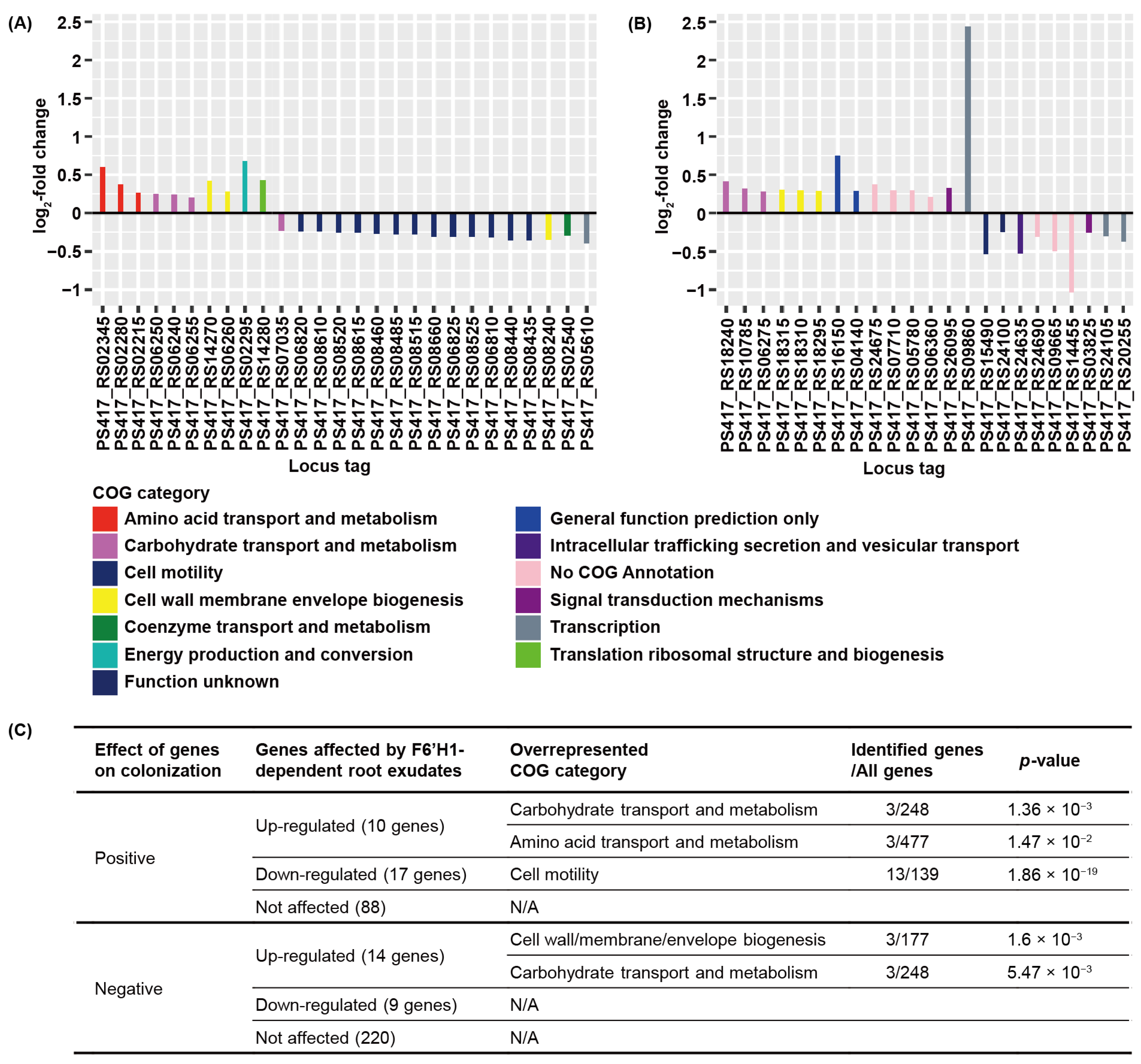
3.3. F6′H1-Dependent Root Exudates Affect Bacterial Motility Required for Root Colonization
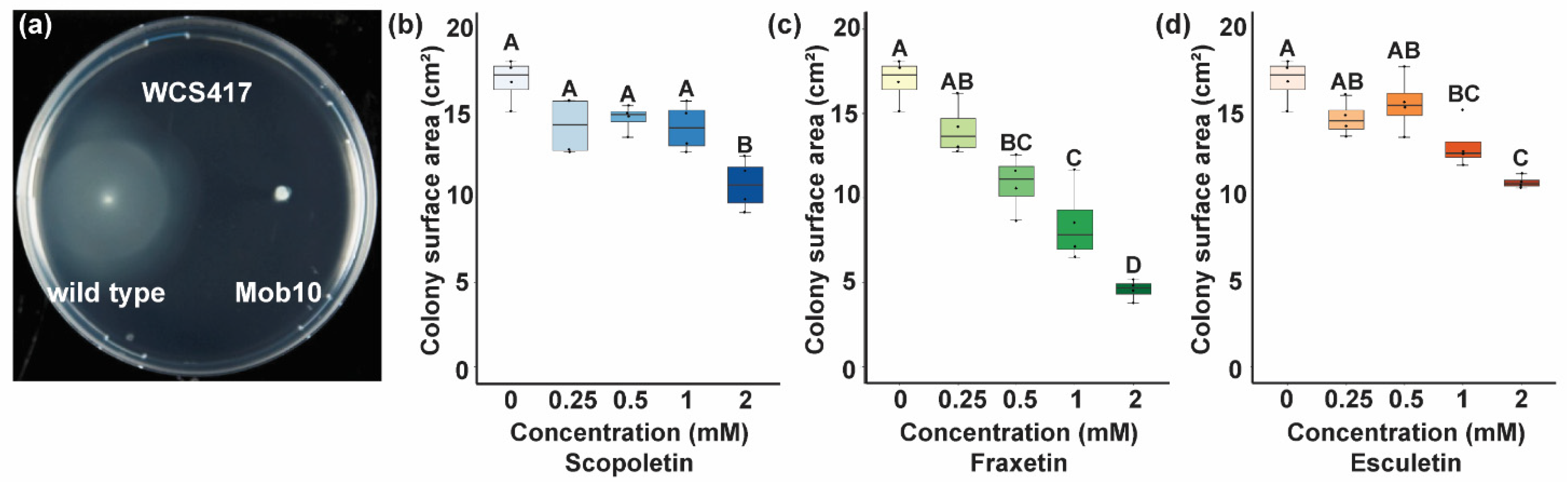
3.4. Coumarins Affect Bacterial Motility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oldroyd, G.E.; Murray, J.D.; Poole, P.S.; Downie, J.A. The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [Green Version]
- Sasse, J.; Martinoia, E.; Northen, T. Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Stringlis, I.A.; Yu, K.; Feussner, K.; De Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Zhao, J.; Wen, T.; Zhao, M.; Li, R.; Goossens, P.; Huang, Q.; Bai, Y.; Vivanco, J.M.; Kowalchuk, G.A.; et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 2018, 6, 156. [Google Scholar] [CrossRef] [Green Version]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; Da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019, 49, 73–82. [Google Scholar] [CrossRef]
- Yu, K.; Pieterse, C.M.J.; Bakker, P.A.H.M.; Berendsen, R.L. Beneficial microbes going underground of root immunity. Plant Cell Environ. 2019, 42, 2860–2870. [Google Scholar] [CrossRef] [Green Version]
- Stringlis, I.A.; Proietti, S.; Hickman, R.; Van Verk, M.C.; Zamioudis, C.; Pieterse, C.M.J. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018, 93, 166–180. [Google Scholar] [CrossRef] [Green Version]
- Yu, K.; Liu, Y.; Tichelaar, R.; Savant, N.; Lagendijk, E.; Van Kuijk, S.J.; Stringlis, I.A.; Van Dijken, A.J.; Pieterse, C.M.; Bakker, P.A.; et al. Rhizosphere-Associated Pseudomonas Suppress Local Root Immune Responses by Gluconic Acid-Mediated Lowering of Environmental pH. Curr. Biol. 2019, 29, 3913–3920.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berendsen, R.L.; Van Verk, M.C.; Stringlis, I.A.; Zamioudis, C.; Tommassen, J.; Pieterse, C.M.J.; Bakker, P.A.H.M. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genom. 2015, 16, 539. [Google Scholar] [CrossRef] [Green Version]
- Van Der Ent, S.; Verhagen, B.W.M.; Van Doorn, R.; Bakker, D.; Verlaan, M.G.; Pel, M.J.C.; Joosten, R.G.; Proveniers, M.C.G.; Van Loon, L.C.; Ton, J.; et al. MYB72 Is Required in Early Signaling Steps of Rhizobacteria-Induced Systemic Resistance in Arabidopsis. Plant Physiol. 2008, 146, 1293–1304. [Google Scholar] [CrossRef] [Green Version]
- Segarra, G.; Van Der Ent, S.; Trillas, I.; Pieterse, C.M.J. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 2009, 11, 90–96. [Google Scholar] [CrossRef] [Green Version]
- Zamioudis, C.; Hanson, J.; Pieterse, C.M.J. β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 2014, 204, 368–379. [Google Scholar] [CrossRef]
- Zamioudis, C.; Korteland, J.; Van Pelt, J.A.; Van Hamersveld, M.; Dombrowski, N.; Bai, Y.; Hanson, J.; Van Verk, M.C.; Ling, H.; Schulze-Lefert, P.; et al. Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB 72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses. Plant J. 2015, 84, 309–322. [Google Scholar] [CrossRef] [Green Version]
- Verbon, E.H.; Trapet, P.L.; Stringlis, I.A.; Kruijs, S.; Bakker, P.A.; Pieterse, C.M. Iron and Immunity. Annu. Rev. Phytopathol. 2017, 55, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Verbon, E.H.; Trapet, P.L.; Kruijs, S.; Temple-Boyer-Dury, C.; Rouwenhorst, T.G.; Pieterse, C.M.J. Rhizobacteria-Mediated Activation of the Fe Deficiency Response in Arabidopsis Roots: Impact on Fe Status and Signaling. Front. Plant Sci. 2019, 10, 909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stringlis, I.A.; De Jonge, R. The Age of Coumarins in Plant–Microbe Interactions. Plant Cell Physiol. 2019, 60, 1405–1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, H.H.; Schmidt, W. Mobilization of Iron by Plant-Borne Coumarins. Trends Plant Sci. 2017, 22, 538–548. [Google Scholar] [CrossRef]
- Palmer, C.M.; Hindt, M.N.; Schmidt, H.; Clemens, S.; Guerinot, M.L. MYB10 and MYB72 Are Required for Growth under Iron-Limiting Conditions. PLoS Genet. 2013, 9, e1003953. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Celma, J.; Lin, W.-D.; Fu, G.-M.; Abadía, J.; López-Millán, A.-F.; Schmidt, W. Mutually Exclusive Alterations in Secondary Metabolism Are Critical for the Uptake of Insoluble Iron Compounds by Arabidopsis and Medicago truncatula. Plant Physiol. 2013, 162, 1473–1485. [Google Scholar] [CrossRef]
- Fourcroy, P.; Siso-Terraza, P.; Sudre, D.; Saviron, M.; Reyt, G.; Gaymard, F.; Abadia, A.; Abadia, J.; Alvarez-Fernandez, A.; Briat, J.F. Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 2014, 201, 155–167. [Google Scholar] [CrossRef]
- Schmid, N.B.; Giehl, R.F.; Doll, S.; Mock, H.P.; Strehmel, N.; Scheel, D.; Kong, X.; Hider, R.C.; Von Wiren, N. Feruloyl-CoA 6′-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 2014, 164, 160–172. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, H.; Günther, C.; Weber, M.; Spörlein, C.; Loscher, S.; Böttcher, C.; Schobert, R.; Clemens, S. Metabolome Analysis of Arabidopsis thaliana Roots Identifies a Key Metabolic Pathway for Iron Acquisition. PLoS ONE 2014, 9, e102444. [Google Scholar] [CrossRef] [Green Version]
- Rajniak, J.; Giehl, R.F.H.; Chang, E.; Murgia, I.; Von Wirén, N.; Sattely, E.S. Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat. Chem. Biol. 2018, 14, 442–450. [Google Scholar] [CrossRef]
- Tsai, H.-H.; Rodríguez-Celma, J.; Lan, P.; Wu, Y.-C.; Vélez-Bermúdez, I.C.; Schmidt, W. Scopoletin 8-Hydroxylase-Mediated Fraxetin Production Is Crucial for Iron Mobilization. Plant Physiol. 2018, 177, 194–207. [Google Scholar] [CrossRef] [Green Version]
- Gnonlonfin, G.J.B.; Sanni, A.; Brimer, L. Review Scopoletin—A Coumarin Phytoalexin with Medicinal Properties. Crit. Rev. Plant Sci. 2012, 31, 47–56. [Google Scholar] [CrossRef]
- Beyer, S.F.; Beesley, A.; Rohmann, P.F.; Schultheiss, H.; Conrath, U.; Langenbach, C.J. The Arabidopsis non-host defence-associated coumarin scopoletin protects soybean from Asian soybean rust. Plant J. 2019, 99, 397–413. [Google Scholar] [CrossRef]
- Kai, K.; Shimizu, B.-I.; Mizutani, M.; Watanabe, K.; Sakata, K. Accumulation of coumarins in Arabidopsis thaliana. Phytochemistry 2006, 67, 379–386. [Google Scholar] [CrossRef]
- Großkinsky, D.K.; Naseem, M.; Abdelmohsen, U.R.; Plickert, N.; Engelke, T.; Griebel, T.; Zeier, J.; Novák, O.; Strnad, M.; Pfeifhofer, H.; et al. Cytokinins Mediate Resistance against Pseudomonas syringae in Tobacco through Increased Antimicrobial Phytoalexin Synthesis Independent of Salicylic Acid Signaling. Plant Physiol. 2011, 157, 815–830. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Wang, L.; Zhang, B.; Ma, J.; Hettenhausen, C.; Cao, G.; Sun, G.; Wu, J.; Wu, J. Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J. Exp. Bot. 2014, 65, 4305–4315. [Google Scholar] [CrossRef]
- Chezem, W.R.; Memon, A.; Li, F.-S.; Weng, J.-K.; Clay, N.K. SG2-Type R2R3-MYB Transcription Factor MYB15 Controls Defense-Induced Lignification and Basal Immunity in Arabidopsis. Plant Cell 2017, 29, 1907–1926. [Google Scholar] [CrossRef] [Green Version]
- Voges, M.J.E.E.E.; Bai, Y.; Schulze-Lefert, P.; Sattely, E.S. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. USA 2019, 116, 12558–12565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Cole, B.J.; Feltcher, M.E.; Waters, R.J.; Wetmore, K.M.; Mucyn, T.S.; Ryan, E.M.; Wang, G.; Ul-Hasan, S.; McDonald, M.; Yoshikuni, Y.; et al. Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 2017, 15, e2002860. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Bakker, P.A.H.M.; Van Der Heijdt, W.H.W.; Wendehenne, D.; Pugin, A. Early Responses of Tobacco Suspension Cells to Rhizobacterial Elicitors of Induced Systemic Resistance. Mol. Plant Microbe Interact. 2008, 21, 1609–1621. [Google Scholar] [CrossRef] [Green Version]
- Tatusov, R.L.; Natale, D.A.; Garkavtsev, I.V.; Tatusova, T.A.; Shankavaram, U.T.; Rao, B.S.; Kiryutin, B.; Galperin, M.Y.; Fedorova, N.D.; Koonin, E.V. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef]
- Robe, K.; Izquierdo, E.; Vignols, F.; Rouached, H.; Dubos, C. The Coumarins: Secondary Metabolites Playing a Primary Role in Plant Nutrition and Health. Trends Plant Sci. 2021, 26, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Stassen, M.J.J.; Hsu, S.; Pieterse, C.M.J.; Stringlis, I.A. Coumarin Communication Along the Microbiome–Root–Shoot Axis. Trends Plant Sci. 2021, 26, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Weir, T.L.; Van Der Lelie, D.; Vivanco, J.M. Rhizosphere chemical dialogues: Plant–microbe interactions. Curr. Opin. Biotechnol. 2009, 20, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Chagas, F.O.; Pessotti, R.D.C.; Caraballo-Rodríguez, A.M.; Pupo, M.T. Chemical signaling involved in plant–microbe interactions. Chem. Soc. Rev. 2018, 47, 1652–1704. [Google Scholar] [CrossRef]
- Bakker, P.A.; Pieterse, C.M.; De Jonge, R.; Berendsen, R.L. The Soil-Borne Legacy. Cell 2018, 172, 1178–1180. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [Green Version]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2019, 10, 1741. [Google Scholar] [CrossRef] [PubMed]
- Rossez, Y.; Wolfson, E.B.; Holmes, A.; Gally, D.L.; Holden, N.J. Bacterial Flagella: Twist and Stick, or Dodge across the Kingdoms. PLOS Pathog. 2015, 11, e1004483. [Google Scholar] [CrossRef]
- De Weger, L.A.; Van Der Vlugt, C.; Wijfjes, A.H.; Bakker, P.; Schippers, B.; Lugtenberg, B. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 1987, 169, 2769–2773. [Google Scholar] [CrossRef] [Green Version]
- Kusumoto, A.; Kamisaka, K.; Yakushi, T.; Terashima, H.; Shinohara, A.; Hommay, M. Regulation of Polar Flagellar Number by the flhF and flhG Genes in Vibrio alginolyticus. J. Biochem. 2006, 139, 113–121. [Google Scholar] [CrossRef]
- Pandza, S.; Baetens, M.; Park, C.H.; Au, T.; Keyhan, M.; Matin, A. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 2000, 36, 414–423. [Google Scholar] [CrossRef]
- Schuhmacher, J.S.; Thormann, K.M.; Bange, G. How bacteria maintain location and number of flagella? FEMS Microbiol. Rev. 2015, 39, 812–822. [Google Scholar] [CrossRef] [Green Version]
- Salvetti, S.; Ghelardi, E.; Celandroni, F.; Ceragioli, M.; Giannessi, F.; Senesi, S. FlhF, a signal recognition particle-like GTPase, is involved in the regulation of flagellar arrangement, motility behaviour and protein secretion in Bacillus cereus. Microbiology 2007, 153, 2541–2552. [Google Scholar] [CrossRef] [Green Version]
- Millet, Y.A.; Danna, C.H.; Clay, N.K.; Songnuan, W.; Simon, M.D.; Werck-Reichhart, D.; Ausubel, F.M. Innate Immune Responses Activated in Arabidopsis Roots by Microbe-Associated Molecular Patterns. Plant Cell 2010, 22, 973–990. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Emonet, A.; Tendon, V.D.; Marhavy, P.; Wu, D.; Lahaye, T.; Geldner, N. Co-incidence of Damage and Microbial Patterns Controls Localized Immune Responses in Roots. Cell 2020, 180, 440–453.e18. [Google Scholar] [CrossRef] [Green Version]
- Danhorn, T.; Fuqua, C. Biofilm Formation by Plant-Associated Bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef]
- Rudrappa, T.; Biedrzycki, M.L.; Bais, H.P. Causes and consequences of plant-associated biofilms. FEMS Microbiol. Ecol. 2008, 64, 153–166. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, D.; Wang, D.; Miao, Y.; Shao, J.; Zhou, X.; Xu, Z.; Li, Q.; Feng, H.; Li, S.; et al. Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genom. 2015, 16, 685. [Google Scholar] [CrossRef] [Green Version]
- Rudrappa, T.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. Root-Secreted Malic Acid Recruits Beneficial Soil Bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belas, R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014, 22, 517–527. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation inPseudomonas fluorescensWCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Guttenplan, S.B.; Kearns, D.B. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef] [Green Version]
- Blair, K.M.; Turner, L.; Winkelman, J.T.; Berg, H.C.; Kearns, D.B. A Molecular Clutch Disables Flagella in the Bacillus subtilis Biofilm. Science 2008, 320, 1636–1638. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, I.; Nygren, E.; Khalid, F.; Myint, S.L.; Uhlin, B.E. A Cyclic-di-GMP signalling network regulates biofilm formation and surface associated motility of Acinetobacter baumannii 17978. Sci. Rep. 2020, 10, 1991. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Li, X.; Gunawardana, M.; Maguire, K.; Guerrero-Given, D.; Schaudinn, C.; Wang, C.; Baum, M.M.; Webster, P. Beta- Lactam Antibiotics Stimulate Biofilm Formation in Non-Typeable Haemophilus influenzae by Up-Regulating Carbohydrate Metabolism. PLoS ONE 2014, 9, e99204. [Google Scholar] [CrossRef] [Green Version]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef]
- Wen, Z.T.; Burne, R.A. Functional Genomics Approach to Identifying Genes Required for Biofilm Development by Streptococcus mutans. Appl. Environ. Microbiol. 2002, 68, 1196–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, G.A.; Gibbs, K.A.; Hager, P.W.; Phibbs, P.V., Jr.; Kolter, R. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 425–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanholme, R.; Sundin, L.; Seetso, K.C.; Kim, H.; Liu, X.; Li, J.; De Meester, B.; Hoengenaert, L.; Goeminne, G.; Morreel, K.; et al. COSY catalyses trans–cis isomerization and lactonization in the biosynthesis of coumarins. Nat. Plants 2019, 5, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
| Locus Tag | COG Description | Product Description | Colonization | Log2-Fold Change | |
|---|---|---|---|---|---|
| Cellulose biosynthesis | PS417_RS08660 | Cellulose biosynthesis protein BcsQ | Cobyric acid synthase | + | −0.31 |
| Flagellar biosynthesis | PS417_RS06810 | Flagellar basal body rod protein FlgB | Flagellar basal body rod protein FlgB | + | −0.32 |
| PS417_RS06820 | Flagellar hook assembly protein FlgD | Flagellar basal body rod modification protein FlgD | + | −0.24 | |
| PS417_RS06825 | Flagellar hook protein FlgE | Flagellar hook protein FlgE | + | −0.31 | |
| PS417_RS08430 | Flagellar basal body rod protein FlgF | Flagellar basal body rod protein FlgF | −0.31 | ||
| PS417_RS08435 | Flagellar basal body rod protein FlgG | Flagellar basal body rod protein FlgG | + | −0.36 | |
| PS417_RS08440 | Flagellar basal body L-ring protein FlgH | Flagellar basal body L-ring protein | + | −0.36 | |
| PS417_RS08460 | Flagellin and related hook-associated protein FlgL | Flagellar hook-associated protein FlgL | + | −0.27 | |
| PS417_RS08485 | Flagellin-specific chaperone FliS | Flagellar biosynthesis protein FliS | + | −0.27 | |
| PS417_RS08515 | Flagellar biosynthesis/type III secretory pathway M-ring protein FliF/YscJ | Flagellar M-ring protein FliF | + | −0.28 | |
| PS417_RS08520 | Flagellar motor switch protein FliG | Flagellar motor switch protein FliG | + | −0.26 | |
| PS417_RS08525 | Flagellar biosynthesis/type III secretory pathway protein FliH | Flagellar assembly protein FliH | + | −0.31 | |
| PS417_RS08610 | Flagellar biosynthesis pathway, component FlhA | Flagellar biosynthesis protein FlhA | + | −0.24 | |
| PS417_RS08615 | Flagellar biosynthesis GTPase FlhF | Flagellar biosynthesis regulator FlhF | + | −0.26 | |
| Chemotaxis | PS417_RS00895 | Methyl-accepting chemotaxis protein | Chemotaxis protein | 0.66 | |
| PS417_RS05665 | Methyl-accepting chemotaxis protein | Methyl-accepting chemotaxis protein | −0.41 | ||
| PS417_RS07665 | Methyl-accepting chemotaxis protein | Chemotaxis protein | −0.23 | ||
| PS417_RS13440 | Methyl-accepting chemotaxis protein | Chemotaxis protein | −0.36 | ||
| PS417_RS15490 | Methyl-accepting chemotaxis protein | Methyl-accepting chemotaxis protein | −0.53 | ||
| PS417_RS18840 | Methyl-accepting chemotaxis protein | Chemotaxis protein | 0.35 | ||
| PS417_RS18940 | Methyl-accepting chemotaxis protein | Methyl-accepting chemotaxis protein | 0.39 | ||
| PS417_RS23840 | Methyl-accepting chemotaxis protein | Methyl-accepting chemotaxis protein | −0.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, K.; Stringlis, I.A.; van Bentum, S.; de Jonge, R.; Snoek, B.L.; Pieterse, C.M.J.; Bakker, P.A.H.M.; Berendsen, R.L. Transcriptome Signatures in Pseudomonas simiae WCS417 Shed Light on Role of Root-Secreted Coumarins in Arabidopsis-Mutualist Communication. Microorganisms 2021, 9, 575. https://doi.org/10.3390/microorganisms9030575
Yu K, Stringlis IA, van Bentum S, de Jonge R, Snoek BL, Pieterse CMJ, Bakker PAHM, Berendsen RL. Transcriptome Signatures in Pseudomonas simiae WCS417 Shed Light on Role of Root-Secreted Coumarins in Arabidopsis-Mutualist Communication. Microorganisms. 2021; 9(3):575. https://doi.org/10.3390/microorganisms9030575
Chicago/Turabian StyleYu, Ke, Ioannis A. Stringlis, Sietske van Bentum, Ronnie de Jonge, Basten L. Snoek, Corné M. J. Pieterse, Peter A. H. M. Bakker, and Roeland L. Berendsen. 2021. "Transcriptome Signatures in Pseudomonas simiae WCS417 Shed Light on Role of Root-Secreted Coumarins in Arabidopsis-Mutualist Communication" Microorganisms 9, no. 3: 575. https://doi.org/10.3390/microorganisms9030575
APA StyleYu, K., Stringlis, I. A., van Bentum, S., de Jonge, R., Snoek, B. L., Pieterse, C. M. J., Bakker, P. A. H. M., & Berendsen, R. L. (2021). Transcriptome Signatures in Pseudomonas simiae WCS417 Shed Light on Role of Root-Secreted Coumarins in Arabidopsis-Mutualist Communication. Microorganisms, 9(3), 575. https://doi.org/10.3390/microorganisms9030575






