Structural Diversity, Fitness Cost, and Stability of a BlaNDM-1-Bearing Cointegrate Plasmid in Klebsiella pneumoniae and Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Filter Mating Assay and S1-PFGE
2.3. Growth Curve Measurements
2.4. Pairwise Competition Assay
2.5. Galleria Mellonella Larval Infection Assay
2.6. Biofilm Formation
2.7. Plasmid Stability Experiments
2.8. DNA Sequencing and Bioinformatics Analysis
2.9. Statistics
3. Results and Discussion
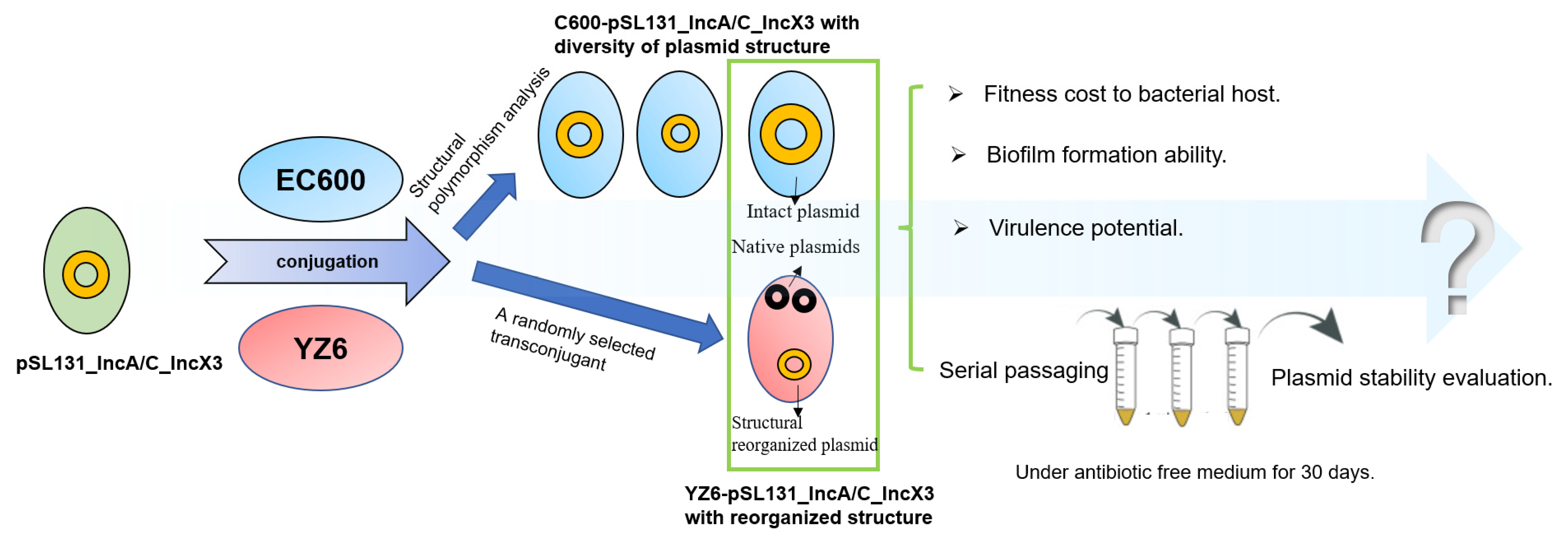
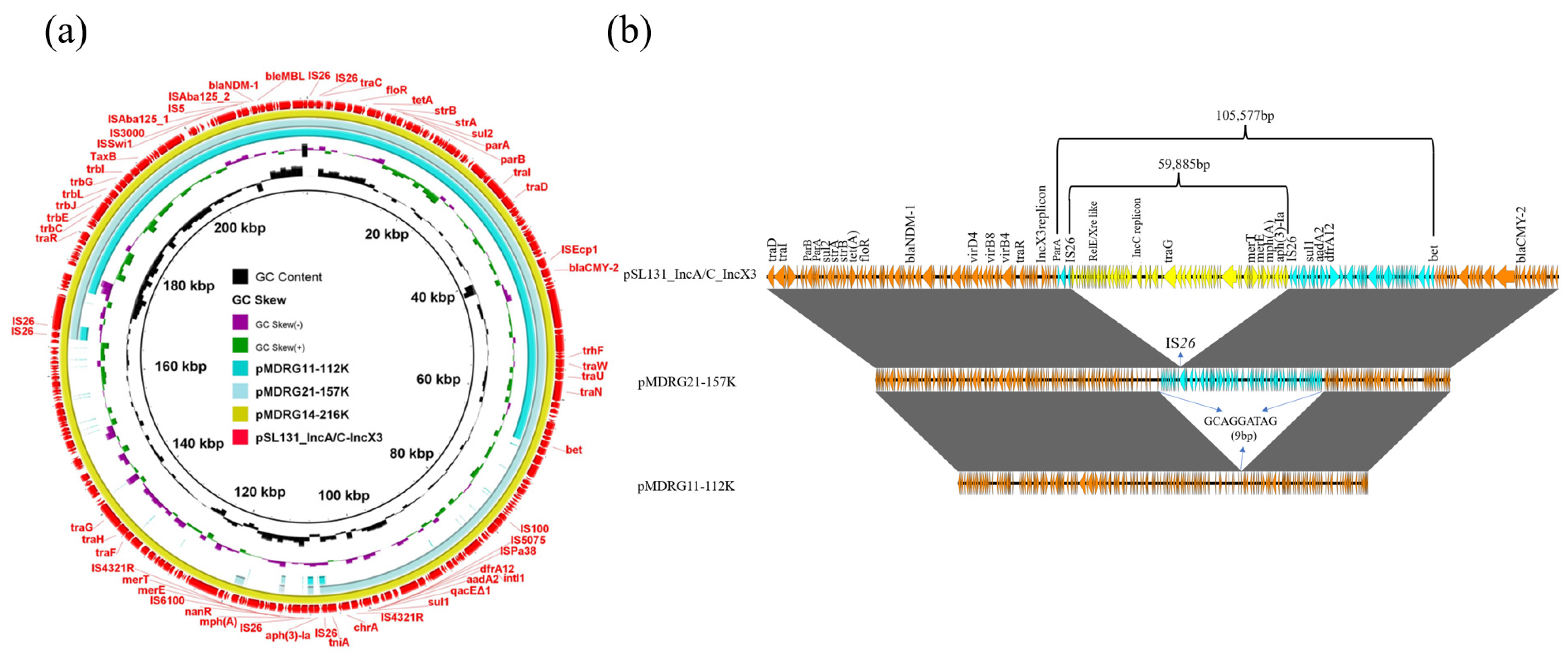
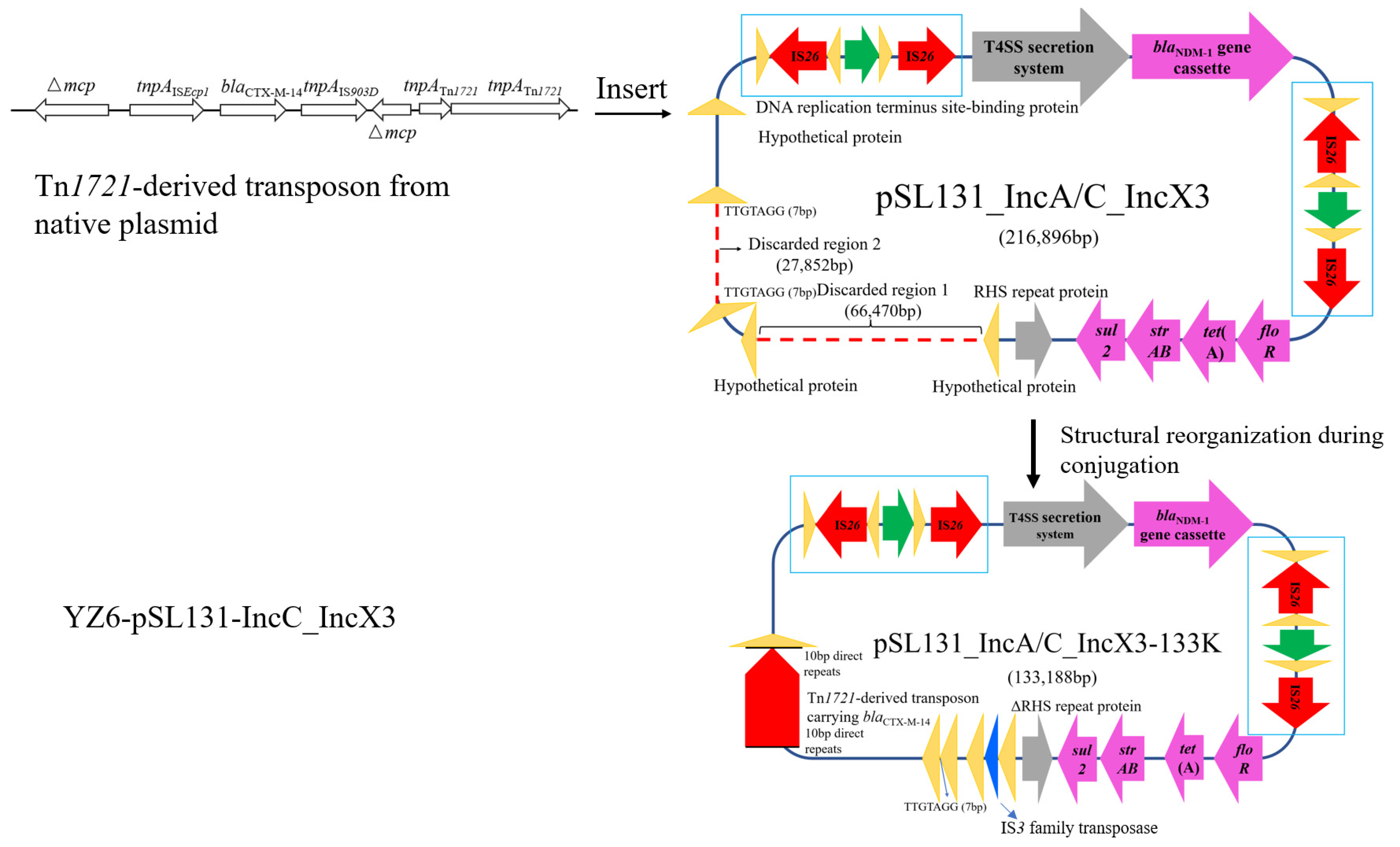
3.1. Structural Diversity of Cointegrate Plasmid pSL131_IncA/C_IncX3 in EC600 after Conjugation
3.2. Cointegrate Plasmid pSL131_IncA/C_IncX3 Experienced Structural Reorganization in K. pneumoniae YZ6 after Conjugation
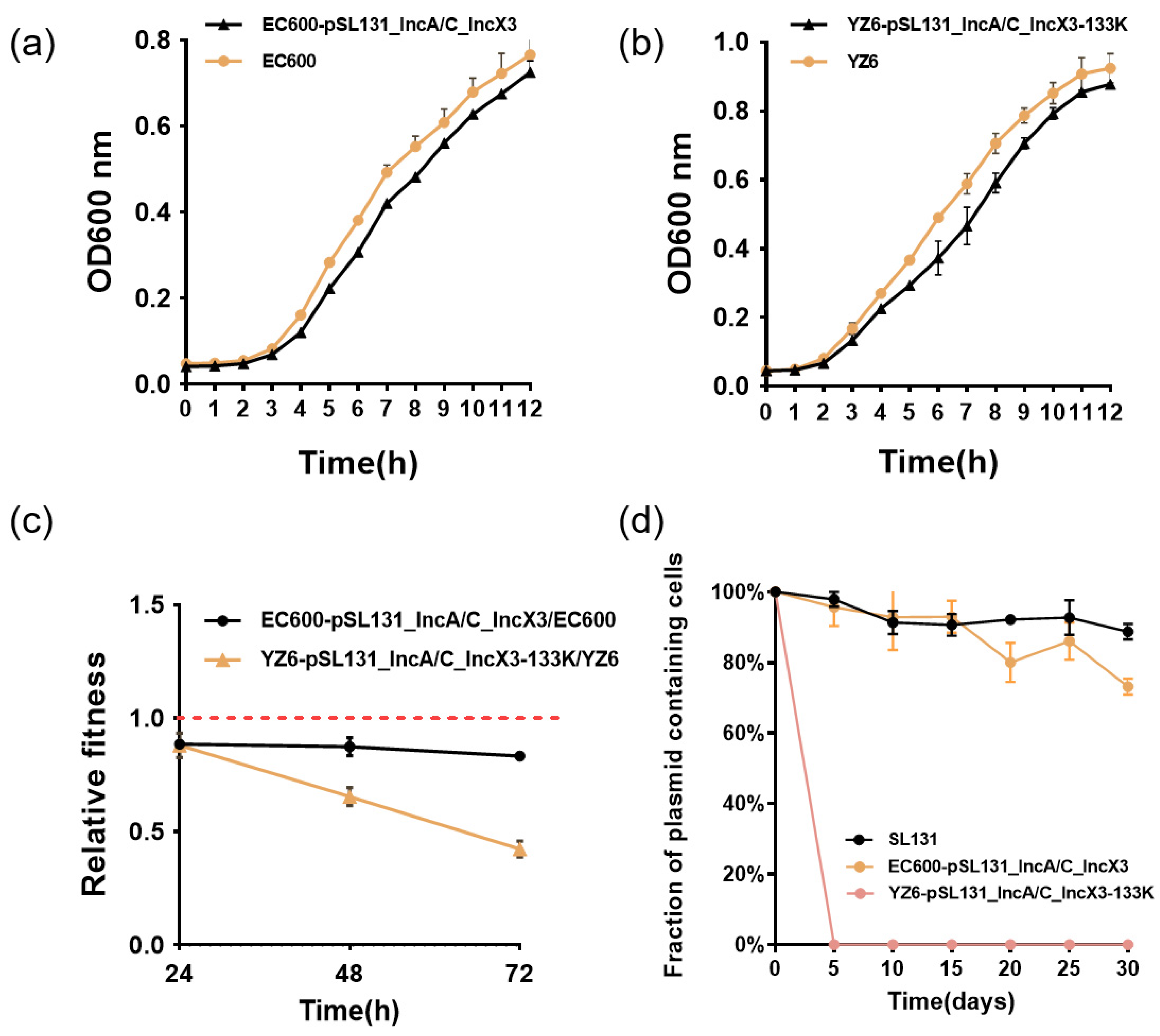
3.3. Cointegrate Plasmid pSL131_IncA/C_IncX3 Could Impose Different Levels of Fitness Cost in Klebsiella pneumoniae and E. coli
3.4. Stability of Plasmids in YZ6, EC600, and Natural Host in Antibiotic-Free Medium
3.5. Stability of Structural Deficiency Plasmid Was Improved in YZ6 under Positive Selection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- San Millan, A.; Pena-Miller, R.; Toll-Riera, M.; Halbert, Z.V.; McLean, A.R.; Cooper, B.S.; MacLean, R.C. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 2014, 5, 5208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Baltrus, D.A. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 2013, 28, 489–495. [Google Scholar] [CrossRef]
- Harrison, P.W.; Lower, R.P.; Kim, N.K.; Young, J.P. Introducing the bacterial ’chromid’: Not a chromosome, not a plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef]
- Carroll, A.C.; Wong, A. Plasmid persistence: Costs, benefits, and the plasmid paradox. Can. J. Microbiol. 2018, 64, 293–304. [Google Scholar] [CrossRef]
- Cottell, J.L.; Webber, M.A.; Piddock, L.J. Persistence of transferable extended-spectrum-beta-lactamase resistance in the absence of antibiotic pressure. Antimicrob. Agents Chemother. 2012, 56, 4703–4706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porse, A.; Schonning, K.; Munck, C.; Sommer, M.O. Survival and Evolution of a Large Multidrug Resistance Plasmid in New Clinical Bacterial Hosts. Mol. Biol. Evol. 2016, 33, 2860–2873. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Wu, A.Y.; Iredell, J.R. Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria. Microorganisms 2021, 9, 1276. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.Y.; Kamruzzaman, M.; Iredell, J.R. Specialised functions of two common plasmid mediated toxin-antitoxin systems, ccdAB and pemIK, in Enterobacteriaceae. PLoS ONE 2020, 15, e0230652. [Google Scholar] [CrossRef]
- Xie, M.; Li, R.; Liu, Z.; Chan, E.W.C.; Chen, S. Recombination of plasmids in a carbapenem-resistant NDM-5-producing clinical Escherichia coli isolate. J. Antimicrob. Chemother. 2018, 73, 1230–1234. [Google Scholar] [CrossRef]
- Liu, Z.; Xiao, X.; Liu, Y.; Li, R.; Wang, Z. Recombination of NDM-5-producing plasmids mediated by IS26 among Escherichia coli. Int. J. Antimicrob. Agents 2020, 55, 105815. [Google Scholar] [CrossRef]
- Li, R.; Lu, X.; Peng, K.; Liu, Z.; Li, Y.; Liu, Y.; Xiao, X.; Wang, Z. Deciphering the Structural Diversity and Classification of the Mobile Tigecycline Resistance Gene tet(X)-Bearing Plasmidome among Bacteria. Msystems 2020, 5, e00134-20. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Lu, X.; Peng, K.; Liu, Y.; Xiao, X.; Wang, Z. Reorganization of mcr-1-bearing large MDR plasmids resolved by nanopore sequencing. J. Antimicrob. Chemother. 2020, 75, 1645–1647. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Zhu, Y.; Li, R.; Pan, Y.; Liu, J.; Yuan, L.; Hu, G. Emergence of a hybrid plasmid derived from IncN1-F33:A-:B- and mcr-1-bearing plasmids mediated by IS26. J. Antimicrob. Chemother. 2019, 74, 3184–3189. [Google Scholar] [CrossRef] [PubMed]
- Chavda, K.D.; Chen, L.; Jacobs, M.R.; Rojtman, A.D.; Bonomo, R.A.; Kreiswirth, B.N. Complete sequence of a bla(KPC)-harboring cointegrate plasmid isolated from Escherichia coli. Antimicrob. Agents Chemother. 2015, 59, 2956–2959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Li, P.; Jiang, X.; Bi, D.; Xie, Y.; Tai, C.; Deng, Z.; Rajakumar, K.; Ou, H.Y. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 2012, 194, 1841–1842. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Xie, M.; Liu, L.; Huang, Y.; Wu, X.; Wang, Z.; Chan, E.W.C.; Chen, S. Characterisation of a cointegrate plasmid harbouring blaNDM-1 in a clinical Salmonella Lomita strain. Int. J. Antimicrob. Agents 2020, 55, 105817. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, Y.; Fan, Y.; Han, J.; Xiong, Z.; Liu, X.; Li, B.; Lu, B.; Cao, B. Characterization of an NDM-5-producing hypervirulent Klebsiella pneumoniae sequence type 65 clone from a lung transplant recipient. Emerg. Microbes Infect. 2021, 10, 396–399. [Google Scholar] [CrossRef]
- Li, X.; Mu, X.; Zhang, P.; Zhao, D.; Ji, J.; Quan, J.; Zhu, Y.; Yu, Y. Detection and characterization of a clinical Escherichia coli ST3204 strain coproducing NDM-16 and MCR-1. Infect. Drug Resist. 2018, 11, 1189–1195. [Google Scholar] [CrossRef]
- Hunter, S.B.; Vauterin, P.; Lambert-Fair, M.A.; Van Duyne, M.S.; Kubota, K.; Graves, L.; Wrigley, D.; Barrett, T.; Ribot, E. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: Converting the national databases to the new size standard. J. Clin. Microbiol. 2005, 43, 1045–1050. [Google Scholar] [CrossRef] [Green Version]
- Bertani, G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 2004, 186, 595–600. [Google Scholar] [CrossRef] [Green Version]
- Ma, T.; Fu, J.; Xie, N.; Ma, S.; Lei, L.; Zhai, W.; Shen, Y.; Sun, C.; Wang, S.; Shen, Z.; et al. Fitness Cost of blaNDM-5-Carrying p3R-IncX3 Plasmids in Wild-Type NDM-Free Enterobacteriaceae. Microorganisms 2020, 8, 377. [Google Scholar] [CrossRef] [Green Version]
- De Gelder, L.; Ponciano, J.M.; Joyce, P.; Top, E.M. Stability of a promiscuous plasmid in different hosts: No guarantee for a long-term relationship. Microbiology 2007, 153, 452–463. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. snippy: Fast Bacterial Variant Calling from NGS Reads. Available online: https://github.com/tseemann/snippy (accessed on 4 August 2020).
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Li, B.; Tang, K.; Yao, J.; Wood, T.K.; Wang, P.; Wang, X. Conjugative plasmid-encoded toxin-antitoxin system PrpT/PrpA directly controls plasmid copy number. Proc. Natl. Acad. Sci. USA 2021, 118, e2011577118. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Ye, L.; Chan, E.W.; Zhang, R.; Chen, S. Tracking Recombination Events That Occur in Conjugative Virulence Plasmid p15WZ-82_Vir during the Transmission Process. Msystems 2020, 5, e00140-20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.Y.; Li, W.; Cai, R.M.; Lu, Y.W.; Zhang, Y.; Cai, P.; Webber, M.A.; Jiang, H.X. Mobilization of Tn1721-like structure harboring blaCTX-M-27 between P1-like bacteriophage in Salmonella and plasmids in Escherichia coli in China. Vet. Microbiol. 2021, 253, 108944. [Google Scholar] [CrossRef]
- Tang, Y.; Li, G.; Liang, W.; Shen, P.; Zhang, Y.; Jiang, X. Translocation of Carbapenemase Gene blaKPC-2 both Internal and External to Transposons Occurs via Novel Structures of Tn1721 and Exhibits Distinct Movement Patterns. Antimicrob. Agents Chemother. 2017, 61, e01151-17. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhang, Y.; Bi, D.; Shen, P.; Ai, F.; Liu, H.; Tian, Y.; Ma, Y.; Wang, B.; Rajakumar, K.; et al. First report of a clinical, multidrug-resistant Enterobacteriaceae isolate coharboring fosfomycin resistance gene fosA3 and carbapenemase gene blaKPC-2 on the same transposon, Tn1721. Antimicrob. Agents Chemother. 2015, 59, 338–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frech, G.; Schwarz, S. Plasmid-encoded tetracycline resistance in Salmonella enterica subsp. enterica serovars choleraesuis and typhimurium: Identification of complete and truncated Tn1721 elements. FEMS Microbiol. Lett. 1999, 176, 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, X.; Zhang, L.; Moran, R.A.; Xu, Q.; Sun, L.; van Schaik, W.; Yu, Y. Cointegration as a mechanism for the evolution of a KPC-producing multidrug resistance plasmid in Proteus mirabilis. Emerg. Microbes Infect. 2020, 9, 1206–1218. [Google Scholar] [CrossRef]
- Lee, H.; Shin, J.; Chung, Y.J.; Park, M.; Kang, K.J.; Baek, J.Y.; Shin, D.; Chung, D.R.; Peck, K.R.; Song, J.H.; et al. Co-introduction of plasmids harbouring the carbapenemase genes, blaNDM-1 and blaOXA-232, increases fitness and virulence of bacterial host. J. Biomed. Sci. 2020, 27, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Gallego, M.; Torrens, G.; Castillo-Vera, J.; Moya, B.; Zamorano, L.; Cabot, G.; Hultenby, K.; Alberti, S.; Mellroth, P.; Henriques-Normark, B.; et al. Impact of AmpC Derepression on Fitness and Virulence: The Mechanism or the Pathway? mBio 2016, 7, e01783-16. [Google Scholar] [CrossRef] [Green Version]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [Green Version]
- Weingarten, R.A.; Johnson, R.C.; Conlan, S.; Ramsburg, A.M.; Dekker, J.P.; Lau, A.F.; Khil, P.; Odom, R.T.; Deming, C.; Park, M.; et al. Genomic Analysis of Hospital Plumbing Reveals Diverse Reservoir of Bacterial Plasmids Conferring Carbapenem Resistance. MBio 2018, 9, e02011-17. [Google Scholar] [CrossRef] [Green Version]
- Savage, V.J.; Chopra, I.; O’Neill, A.J. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 2013, 57, 1968–1970. [Google Scholar] [CrossRef] [Green Version]
- Wein, T.; Hulter, N.F.; Mizrahi, I.; Dagan, T. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat. Commun. 2019, 10, 2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouma, J.E.; Lenski, R.E. Evolution of a bacteria/plasmid association. Nature 1988, 335, 351–352. [Google Scholar] [CrossRef]
- Harrison, E.; Guymer, D.; Spiers, A.J.; Paterson, S.; Brockhurst, M.A. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 2015, 25, 2034–2039. [Google Scholar] [CrossRef] [Green Version]
- Heuer, H.; Fox, R.E.; Top, E.M. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol. Ecol. 2007, 59, 738–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knopp, M.; Andersson, D.I. Amelioration of the Fitness Costs of Antibiotic Resistance Due To Reduced Outer Membrane Permeability by Upregulation of Alternative Porins. Mol. Biol. Evol. 2015, 32, 3252–3263. [Google Scholar] [CrossRef] [Green Version]
- Bredin, J.; Saint, N.; Mallea, M.; De, E.; Molle, G.; Pages, J.M.; Simonet, V. Alteration of pore properties of Escherichia coli OmpF induced by mutation of key residues in anti-loop 3 region. Biochem. J. 2002, 363, 521–528. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Wang, Z.; Lu, X.; Peng, K.; Chen, S.; He, S.; Li, R. Structural Diversity, Fitness Cost, and Stability of a BlaNDM-1-Bearing Cointegrate Plasmid in Klebsiella pneumoniae and Escherichia coli. Microorganisms 2021, 9, 2435. https://doi.org/10.3390/microorganisms9122435
Liu Z, Wang Z, Lu X, Peng K, Chen S, He S, Li R. Structural Diversity, Fitness Cost, and Stability of a BlaNDM-1-Bearing Cointegrate Plasmid in Klebsiella pneumoniae and Escherichia coli. Microorganisms. 2021; 9(12):2435. https://doi.org/10.3390/microorganisms9122435
Chicago/Turabian StyleLiu, Ziyi, Zhiqiang Wang, Xiaoyu Lu, Kai Peng, Sheng Chen, Susu He, and Ruichao Li. 2021. "Structural Diversity, Fitness Cost, and Stability of a BlaNDM-1-Bearing Cointegrate Plasmid in Klebsiella pneumoniae and Escherichia coli" Microorganisms 9, no. 12: 2435. https://doi.org/10.3390/microorganisms9122435
APA StyleLiu, Z., Wang, Z., Lu, X., Peng, K., Chen, S., He, S., & Li, R. (2021). Structural Diversity, Fitness Cost, and Stability of a BlaNDM-1-Bearing Cointegrate Plasmid in Klebsiella pneumoniae and Escherichia coli. Microorganisms, 9(12), 2435. https://doi.org/10.3390/microorganisms9122435







