Abstract
The genus Gemmobacter grows phototrophically, aerobically, or anaerobically, and utilizes methylated amine. Here, we present two high-quality complete genomes of the strains con4 and con5T isolated from a culture of Anabaena. The strains possess sMMO (soluble methane monooxygenase)-oxidizing alkanes to carbon dioxide. Functional genes for methane-oxidation (prmAC, mimBD, adh, gfa, fdh) were identified. The genome of strain con5T contains nirB, nirK, nirQ, norB, norC, and norG genes involved in dissimilatory nitrate reduction. The presence of nitrite reductase gene (nirK) and the nitric-oxide reductase gene (norB) indicates that it could potentially use nitrite as an electron acceptor in anoxic environments. Taxonomic investigations were also performed on two strains through polyphasic methods, proposing two isolates as a novel species of the genus Gemmobacter. The findings obtained through the whole genome analyses provide genome-based evidence of complete oxidation of methane to carbon dioxide. This study provides a genetic blueprint of Gemmobacter fulva con5T and its biochemical characteristics, which help us to understand the evolutionary biology of the genus Gemmobacter.
1. Introduction
Gemmobacter is of interest because of its metabolic pathways and its habitats. Members of the Gemmobacter species are able to grow phototrophically, aerobically, and anaerobically [1,2,3,4,5,6]. The first species, Gemmobacter changlensis, isolated from a snow sample collected in the Indian Himalayas, was proposed as psychrotolerant and phototrophic bacteria [1]. Another phototrophically growing species, Gemmobacter aestuarii, recovered from estuarine surface water, was described as containing a complete gene cluster for photosynthesis [3]. The members of the genus Gemmobacter have been discovered in a wide range of natural environments, such as freshwater, sulphuric cave waters, estuaries, forest ponds, artificial fountains, tidal flats, activated sludge, a white stork nestling, coastal planktonic seaweed, and conserved forages, indicating that members of this genus are widely distributed in natural and artificial environments [2,4,5,6,7,8,9,10,11,12,13,14,15].
Comparative genomics analyses revealed that members of the genus Gemmobacter, including Gemmobacter aquatilis, Gemmobacter lutimaris, Gemmobacter sp. HYN0069, Gemmobacter caeni, and Gemmobacter sp. LW-1, utilize methylated amine. These species have related genes encoding the enzymes trimethylamine (TMA) dehydrogenase, TMA monooxygenase, and TMA demethylase in their genome, indicating metabolic potential of using the TMA oxidation pathway to convert trimethylamine to dimethylamine [3,8]. In a recent study, we isolated two alphaproteobacterial strains, con4 and con5T, from an Anabaena culture that contained all genes related to the oxidization of methane to carbon dioxide.
The species in the genus Gemmobacter were characterized as Gram-negative, oxidase and catalase-positive, non-spore-forming, and rod-shaped containing ubiquinone-10 as the major respiratory quinone, with G + C content in the range of 61.4–69.1 mol% [1,3,4,5]. Here, we report a comparative analysis of the genomes of two strains, con4 and con5T, together with a taxonomic proposal based on their phylogenetic, genomic, physiological, and chemotaxonomic characteristics.
2. Material and Methods
2.1. Isolation and Culture Conditions of the Strains
Strains con5T and con4 were recovered from a culture of Anabaena using a serial dilution method [16]. For the Anabaena culture, Anabaena variabilis FBCC010004 strain was obtained from the Freshwater Bioresources Culture Collection (FBCC, https://fbcc.nibr.re.kr/fbcc, accessed on 25 August 2021) of the Nakdonggang National Institute of Biological Resources (NNIBR, South Korea). Anabaena cells were cultured in 500 mL standard cell culture flasks with Blue–Green (BG11) broth (Merck, St. Louis, MO, USA) under the conditions: 20 °C, 35% humidity, 12 h:12 h light-dark photoperiod, 20 μmol m−2 s−1 irradiance, and 200 rpm agitation [17]. From the Anabaena culture, a 100 μL sub-sample of the suspended material was aseptically spread onto modified R2A agar (Difco, NJ, USA) [18] and incubated at 25 °C under heterotrophic conditions. Two yellow-pigmented strains, con5T and con4, were isolated after three days and routinely sub-cultivated on R2A agar at 30 °C for 48 h and kept in a glycerol solution (20%, v/v) at −70 °C for long-term preservation. For most experiments, all strains were cultivated on R2A agar at 30 °C.
2.2. Morphological, Physiological, and Chemotaxonomic Characteristics
Cell motility and morphology were examined via a phase-contrast microscope (Nikon Optiphot, 1000 × magnification) and transmission electron microscopy (Philips CM-20) using cells grown for 48 h in R2A media at 30 °C. The growth temperature was tested at 4, 8, 15, 20, 30, 37, and 45 °C. The pH range for growth was determined using different buffering systems: citrate–phosphate for pH 4–7, phosphate for pH 8, and glycine–NaOH for pH 9–11 [19]. Salt tolerance was checked on NaCl-free R2A agar with different NaCl concentrations 1.0–5.0% (w/v; at intervals of 1.0%) NaCl. Anaerobic growth was examined after incubating strains con4 and con5T on R2A agar in an anaerobic glove box (Coy Laboratory Products, Grass Lake, MI, USA) containing an atmosphere of 99% N2 and 1% H2 and equipped with palladium catalyst packs to remove oxygen and a gas analyzer to monitor oxygen concentrations. Cell growth at different media was investigated in different media of R2A agar, tryptic soy agar (TSA; Difco, NJ, USA), nutrient agar (NA; Difco, NJ, USA), and Luria–Bertani (LB; Difco, NJ, USA) medium. The antimicrobial susceptibility of the strains was checked on R2A agar medium using the filter-paper disk method [20], where the disks contained the following: amikacin (30 µg mL−1), ampicillin/sulbactam (20 µg mL−1, 1:1), chloramphenicol (30 µg mL−1), erythromycin (30 µg mL−1), gentamicin (30 µg mL−1), kanamycin (30 µg mL−1), lincomycin (15 µg mL−1), nalidixic acid (30 µg mL−1), rifampicin (30 µg mL−1), spectinomycin (25 µg mL−1), streptomycin (25 µg mL−1), teicoplanin (30 µg mL−1), tetracycline (30 µg mL−1), and vancomycin (30 µg mL−1). The haloes for susceptibility were recorded when the diameters were more than 10 mm after incubation at 30 °C for 2 days. Other biochemical activities were examined using API 20NE, API ID 32GN, and API ZYM kits (bioMérieux, l’Etoile, France) according to the manufacturer’s instructions.
The whole-cell fatty acid profile was analyzed by gas chromatography (Hewlett Packard 6890, Kyoto, Japan) with the TSBA 6 database using Sherlock software v6.1 at KCTC (Korean Collection for Type Cultures) service center. The cells of the strains con5T and con4 and four Gemmobacter-type strains were collected after being grown at 30 °C for 48 h on the same sectors of R2A agar plates. Cell-harvesting standardization was done following the method described by Jin et al. [18]. The extraction of respiratory quinones and polar lipids was completed following the method described previously [21,22]. The respiratory quinones were identified using HPLC (Shimadzu, Kyoto, Japan) with an YMC-Pack ODS-A column, and the polar lipids were identified with two-dimensional TLC plates (60 F254, Merck).
2.3. Phylogenetic and Genomic Analyses
For a 16S rRNA gene analysis, genomic DNA of strains con5T and con4 was extracted using a FastDNATM SPIN kit. Amplification of the 16S rRNA genes was done by PCR with 27F/1492R primer sets [23]. The 16S rRNA gene sequences of strains con5T and con4 were compared with the available sequences in the EzBioCloud server [24]. Evolutionary analyses were performed with mega7 software [25]. Sequence edition and multiple alignment were done using the programs bioedit [26] and clustal x [27], respectively. Phylogenetic trees were reconstructed using three algorithms: Neighbor–Joining (NJ), Maximum–Parsimony (MP), and Maximum–Likelihood (ML) [28,29,30]. The ranch robustness of the phylogenetic trees was estimated by bootstrap analyses based on 1000 re-samplings of the sequences [31]. A phylogenomic tree was re-constructed from the available type strains of species of the genus Gemmobacter with whole genome sequences on the TYGS (Type Strain Genome Server) [32]. The phylogenomic tree was inferred using FastME 2.1.4 [33] from GBDP (Genome Blast Distance Phylogeny) distances calculated from genome sequences. Branch lengths were calculated using the GBDP distance formula d5 [34]. The whole genomes of strains con5T and con4 were sequenced using both the PacBio RSII platform and Illumina next-generation sequencing technology at Macrogen Inc. (Seoul, South Korea). The chromosomes and plasmids were assembled using the software package of SMRT portal (v.3.2.0), Pacific Biosciences, (CA, USA) [35]. The de novo genome annotation of strains con5T and con4 was performed with the Prokka (v.1.4.6) pipeline [36], and a sequence-based comparison was made using the SEED Viewer [37]. The predicted coding sequences (CDSs) were submitted to the COG database to create the functional categories [38,39]. The values of average nucleotide identity (ANI) were calculated by using the OrthoANI tool in EZBioCloud server [40], and the digital DNA–DNA hybridization (dDDH) values were calculated using the genome-to-genome distance calculator (GGDC 2.1) based on draft genome sequences [34].
3. Results and Discussion
3.1. Physiological Tests
The two strains con5T and con4 were Gram-negative, non-motile, aerobic, and rod-shaped (Supplementary Figure S1). The colonies appeared yellow, convex, circular, and smooth, with entire edges, after being grown for two days at 30 °C on R2A agar. Cell growth was observed at temperatures ranging from 4 to 37 °C and at pH 5–10 (weak at pH 5). Oxidase and catalase activities were present. The cells were found to assimilate N-acetyl-glucosamine, d-glucose, d-mannitol, d-mannose, malate, and maltose but not adipate, l-arabinose, caprate, citrate, gluconate, and phenyl acetate (API 20NE). The cells were found to be positive for the following enzyme activities (API ZYM test strip): esterase (C4), esterase lipase (C8), α-glucosidase, leucine arylamidase, and naphthol-AS-BI-phosphohydrolase. However, the cells were found to be negative for N-acetyl-β-glucosaminidase, acid phosphatase, alkaline phosphatase, α-chymotrypsin, cystine arylamidase, α-fucosidase, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, lipase (C14), α-mannosidase, trypsin, and valine arylamidase (Table 1). The cells were found to be susceptible to amikacin (30 µg mL−1), ampicillin/sulbactam (1:1; µg mL−1), chloramphenicol (30 µg mL−1), erythromycin (30 µg mL−1), gentamicin (30 µg mL−1), kanamycin (30 µg mL−1), nalidixic acid (30 µg mL−1), rifampicin (30 µg mL−1), spectinomycin (25 µg mL−1), streptomycin (25 µg mL−1), teicoplanin (30 µg mL−1), tetracycline (30 µg mL−1), and vancomycin (30 µg mL−1) but resistant to lincomycin (15 µg mL−1).

Table 1.
Features that differentiate strains con5T and con4 from type strains of recognized Gemmobacter species: Strains: 1, con5T; 2, con4; 3, G. changlensis KCTC 5728T; 4, G. aquatilis DSM 3857T; 5, G. lutimaris KCTC 62715T; 6, G. tilapiae KCTC 23310T. All data are from this study unless indicated. +, positive; −, negative; nd, not determined.
The major fatty acids were summed feature 8 (comprising C18:1 ω7c and/or C18:1 ω6c) for both strains (data not shown). The major fatty acids in strains con5T and con4 were consistent with the major fatty acid components in species from the genus Gemmobacter. Notably, con5T and con4 differed from the four closest relatives in the proportions of some minor fatty acids. The major predominant respiratory ubiquinone was Q-10. The polar lipids consisted of phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylcholine (PC), two unidentified glycolipids (GL1, GL2), three phospholipids (PL1, PL2, and PL3), and three unidentified aminophospholipids (APL1, APL2, and APL3) for the type strain con5T; and phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylcholine (PC), two unidentified glycolipids (GL1, GL2), three unidentified phospholipids (PL1, PL2, and PL3), and two unidentified aminophospholipids (APL1, APL2) for the strain con4 (Supplementary Figure S2). This profile is similar to that of closely related species G. changlensis and G. aquaticus, with major components of PE, PG, and PC, and strains con5T and con4 contain two unidentified glycolipids that differentiate these two novel strains from close members in the genus Gemmobacter.
3.2. Phylogenetic and Genomic Analysis: The Taxonomic Status
The 16S rRNA sequences of strains con5T and con4 share 100% identity between them and 94.9–97.9% identity with those of the closest species within the genus Gemmobacter (Table 2). The 16S rRNA gene sequences of strains con5T and con4 were compared with the 16S rRNA gene sequences of representative species within the genus Gemmobacter and related genera in the EzTaxon-e server. The two strains share over 97.0% similarity with G. aquaticus A1-9T (97.9%), G. caeruleus N8T (97.7%), G. lutimaris YJ-T1-11T (97.4%), and G. tilapiae KCTC 23310T (97.3%) and less than 97% with the remaining species within the genus Gemmobacter. Strains con5T and con4 also share high similarities with other species than members of Gemmobacter: 96.7% with Cypionkella collinsensis 4-T-34T, 96.7% with Cypionkella psychrotolerans PAMC 27389T, and 96.4% with Cypionkella aquatica DC2N1-10T. However, it was clear from the topology of the phylogenetic tree (Figure 1) that strains con5T and con4 clustered clearly with the species of Gemmobacter. In addition, the phylogenomic tree reconstructed on the TYGS provided clearer evidence for the taxonomic position of the two strains within the genus Gemmobacter (Supplementary Figure S3). The genomic DNA G + C content of the two strains was 64.1 mol%, which is in the range reported previously for the genus Gemmobacter (61.4–69.4 mol%) [3,4]. The ANI and dDDH values of strains con5T and con4 with other available type strains of Gemmobacter were 73.31–80.61 % and 19.03–23.15 % (Table 2, Supplementary Figure S4), respectively, which were much lower than the species boundaries of ANI or dDDH of 95–96% and 70%, respectively, and fall in the intergeneric range [41,42,43].

Table 2.
General features and relationship of the genomes of strains con5T and con4 with the closely related species of the genus Gemmobacter.
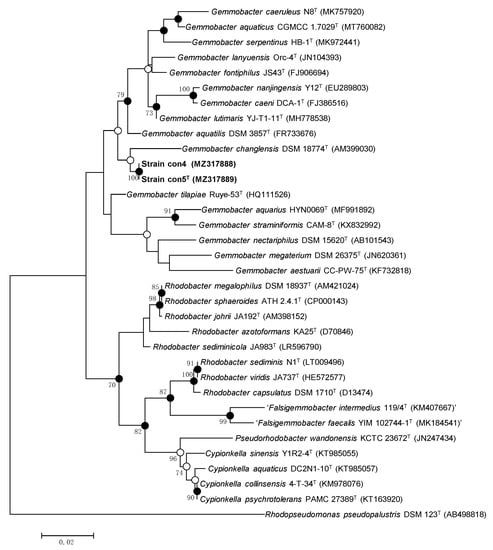
Figure 1.
Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences showing the position of strains con5T and con4 within Gemmobacter. Sequences of related species in the Rhodobacteraceae family are shown. The numbers at the branch-nodes represent the percentage of 1000 bootstrap replicates; only values above 70% are depicted in the tree. Filled circles indicate the corresponding nodes recovered by ML, MP, and NJ phylogenetic trees. Open circles indicate the corresponding nodes were also recovered with either the ML or MP algorithm. The analysis involved a total of 1308 aligned nucleotide positions in the final dataset.
3.3. Genome Properties
The genome of strain con5T was 4.7 Mb and contained a circular chromosome of 3.4 Mb and six plasmids sized 34.0–425.5 kb (CP076361–CP076367) (Supplementary Tables S1 and S2). Of 4534 genes, 4472 were protein-coding genes and 62 were RNA genes (nine rRNA genes, 52 tRNA genes, and one tmRNA gene). For strain con4 (JAHHWR000000000), of 4417 genes, 4317 were protein-coding genes and 53 were RNA genes (four rRNA genes, 46 tRNA genes, one tmRNA gene, and two ncRNA genes) (Supplementary Figure S5). The DNA G + C content of both strains was 64.1 mol% (Supplementary Tables S1 and S2).
3.4. Genome Analyses for Denitrification and Methane Oxidation
It is generally understood that methanotrophic bacteria are mostly active at the oxic-anoxic transition zone in stratified lakes, using oxygen to oxidize methane. The methanotrophs produce methane monooxygenase to utilize methane as a carbon source [44,45]. The methanotrophs express two kinds of methane monooxygenase, soluble methane monooxygenase (sMMO) and particulate methane monooxygenase (pMMO), and these enzymes can also oxidize various alkanes [46,47,48].
Strain con5T possesses sMMO (prmAC, mimBD) but does not have a pMMO gene in its genome sequence. The presence of one PQQ-dependent alcohol dehydrogenase (ADH) (PQQ-MDH: adh, adh1, and adhB), two NAD+-dependent ADH (adh, adh1), and another alcohol dehydrogenase (adhB) in strain con5T makes it a good candidate for the conversion of methane to aldehyde (Figure 2). Genes involved in the glutathione (GSH)-dependent pathway to metabolize formaldehyde are identified. Formaldehyde (HCHO)-activating enzyme (gfa), a GSH-dependent formaldehyde dehydrogenase (fdh), and S-formyl-GSH hydrolase (fgh) convert formaldehyde to formate, and then the formate is oxidized by formate dehydrogenase (fdh) to the final product, carbon dioxide (CO2) (Figure 2 and Figure 3). Those genes were also found in the strain con4 (Figure 4).
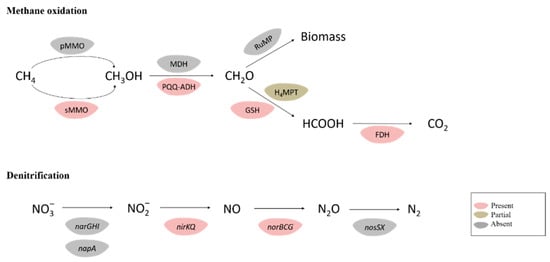
Figure 2.
Metabolic pathways of methane oxidation and denitrification in the strain con5T. Enzymes and pathways indicated in light-pink were encoded by the genome; light-green indicates the presence of an incomplete pathway. Grey pathways and genes were not detected. FDH, formate dehydrogenase; GSH, glutathione (GSH)-dependent pathway; H4F, methylene tetrahydrofolate pathway; H4MPT, tetrahydromethanopterin pathway; MDH, methanol dehydrogenase; PQQ-ADH, PQQ-dependent alcohol dehydrogenase; RuMP, ribulose monophosphate pathway.
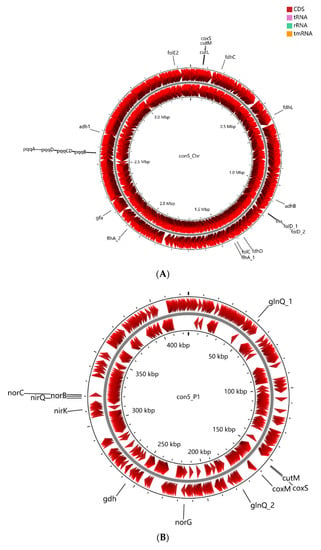
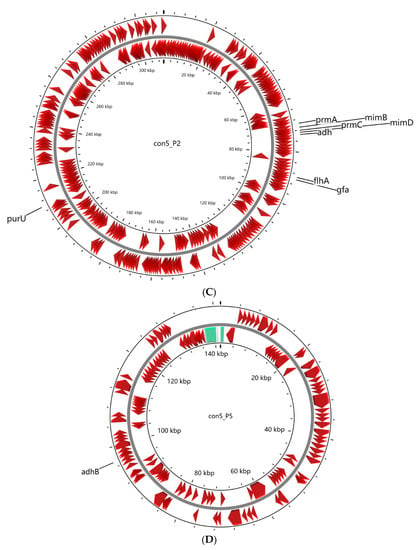
Figure 3.
Graphic representation of circular chromosome (A) and plasmids (B–D) of strain con5T. The locations of genes involved in methylotrophy are indicated at the outside of the map. The locations of genes involved in methylotrophy are indicated at the outside of the map. The CDS (dark red), tRNA (purple), rRNA (green), and tmRNA (golden) were shown on the reverse and forward strand, respectively.
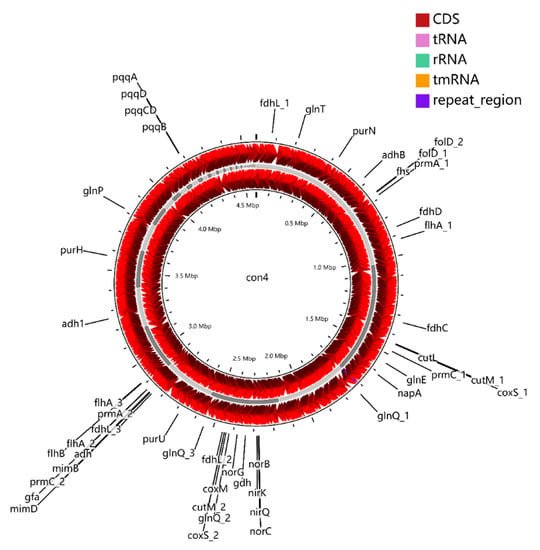
Figure 4.
Graphic representation of circular genome of strain con4. The locations of genes involved in methylotrophy are indicated at the outside of the map. The locations of genes involved in methylotrophy are indicated at the outside of the map. The CDS (dark red), tRNA (purple), rRNA (green), and tmRNA (golden) were shown on the reverse and forward strand, respectively.
Some methanotrophs comprise genes encoding enzymes for the nitrate reduction pathway, which was confirmed to be related to methane oxidation under anoxic conditions [49], and these methanotrophic species encode a complete nitrate reduction pathway to use nitrate as a terminal electron acceptor when oxygen is depleted [50]. The genome of strain con5T contains nirB, nirK, nirQ, norB, norC, and norG genes involved in dissimilatory nitrate reduction, but no dissimilatory nitrate reductase (narG) or nitrous oxide reductase (nosZ) genes are detected, which is incomplete for the denitrification pathway (Figure 2 and Figure 3). This suggests that strain con5T cannot perform nitrate reduction, itself, unless it utilizes unknown or incorrectly classified reduction pathways up to date. The genome of strain con5T contains the nitrite reductase gene, nirK, and the nitric-oxide reductase gene norB, and it could potentially use nitrite as an electron acceptor in anoxic environments.
4. Conclusions
In this study, the strains con5T and con4, representing methane oxidizing species from an Anabaena culture belonging to the genus Gemmobacter, were investigated using genomic and polyphasic methods. The findings obtained through the whole genome analyses provide genome-based evidence of complete oxidation of methane to carbon dioxide. This study provides a genetic blueprint of Gemmobacter fulva con5T and its biochemical characteristics, which help us to understand the evolutionary biology of the genus Gemmobacter. Based on the phylogenetic position and the genotypic, chemotaxonomic, and physiological differences, we propose that strains con5T and con4, Gemmobacter fulva sp. nov., should be assigned as a novel species of the genus Gemmobacter in the family Rhodobacteraceae (Table 3).

Table 3.
Descriptions of Gemmobacter fulva sp. nov.
5. Nucleotide Sequence Accession Numbers
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of strains con5T and con4 are MZ317888 and MZ317889, respectively. Accession numbers for the genome sequences of strains con5T and con4 are CP076361–CP076367 and JAHHWR000000000, respectively.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9122423/s1, Figure S1. Transmission electron micrograph of strains con5T (A) and con4 (B) grown on R2A for 48 h at 30 °C. Bar,1 μm. Figure S2. Polar lipid profile for strains con5T (A) and con4 (B). All the polar lipids were stained with 5% molybdatophosphoric acid (for total lipids), molybdenum blue (for phospholipids), ninhydrin (for amino lipids), and anisaldehyde/sulfuric acid (glycolipids). AL, unidentified aminolipid; APL, unidentified aminophospholipid; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PC, phosphatidylcholine; GL, unidentified glycolipid; PL, unidentified phospholipid. 1, first dimension of TLC; 2, second dimension of TLC. Figure S3. Phylogenomic tree based on genome sequences of the strains con5T and con4 in the TYGS (https://tygs.dsmz.de/, accessed on 25 August 2021). The branch lengths are calculated in terms of GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 95.1%. The tree was rooted at the midpoint [34]. Figure S4. Cluster analysis of the profiles obtained from ANI (A) and DDH values (B). Dendrograms were calculated on the basis of the average nucleotide identity scores based on the whole genomes using the unweighted pair group method with the arithmetic averages clustering algorithm (UPGMA). Figure S5. Graphic representation of circular genome plot of strain con5T. The circles from inside to outside represent GC skew ([G − C]/[G + C]) plot of the genome, GC content, CDS-reverse, CDS-forward, contigs, and position label. Table S1. General genome features of strains con5T and con4. Table S2. Genomic feature of type strain con5T.
Author Contributions
Conceptualization, L.J.; Data curation, C.-Z.J. and H.-G.L.; Formal analysis, H.-G.L.; Funding acquisition, C.S.L.; Investigation, L.J., C.-Z.J. and C.S.L.; Writing—original draft, L.J. and C.S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the research program of the Nakdonggang National Institute of Biological Resources funded by the Ministry of Environment of Korea (NNIBR202102102), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Korea Environment Industry & Technology Institute (KEITI) through project to develop eco-friendly new materials and processing technology derived from wildlife, funded by Korea Ministry of Environment (MOE) (2021003240004). The authors dedicate this article to Kwang Kyu Kim, who was a warm and creative scientist but unfortunately passed away on 27 July 2021.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anil Kumar, P.; Srinivas, T.-N.-R.; Sasikala, C.; Ramana, C.-V. Rhodobacter changlensis sp. nov., a psychrotolerant, phototrophic alphaproteobacterium from the Himalayas of India. Int. J. Syst. Evol. Microbiol. 2007, 57, 2568–2571. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, W.-M.; Cho, N.-T.; Huang, W.-C.; Young, C.-C.; Sheu, S.-Y. Description of Gemmobacter fontiphilus sp. nov., isolated from a freshwater spring, reclassification of Catellibacterium nectariphilum as Gemmobacter nectariphilus comb. nov., Catellibacterium changlense as Gemmobacter changlensis comb. nov., Catellibacterium aquatile as Gemmobacter aquaticus nom. nov., Catellibacterium caeni as Gemmobacter caeni comb. nov., Catellibacterium nanjingense as Gemmobacter nanjingensis comb. nov., and emended description of the genus Gemmobacter and of Gemmobacter aquatilis. Int. J. Syst. Evol. Microbiol. 2013, 63, 470–478. [Google Scholar] [CrossRef]
- Hameed, A.; Shahina, M.; Lin, S.-Y.; Chen, W.-M.; Hsu, Y.-H.; Lai, W.-A.; Young, C.-C. Description of Gemmobacter aestuarii sp. nov., isolated from estuarine surface water and reclassification of Cereibacter changlensis as Gemmobacter changlensis Chen et al. 2013. Arch. Microbiol. 2020, 202, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.; Kannan, A.D.; Shobnam, N.; Mahmood, M.; Lee, J.; Im, J. Gemmobacter serpentinus sp. nov., isolated from conserved forages. Int. J. Syst. Evol. Microbiol. 2020, 70, 4224–4232. [Google Scholar] [CrossRef] [PubMed]
- Rothe, B.; Fisher, A.; Hirsch, P.; Sittig, M.; Stackebrandt, E. The phylogenetic position of the budding bacteria Blustobacter aggregatus and Gemmobacter aquatilis gen. nov., sp. nov. Arch. Microbiol. 1987, 1147, 92–99. [Google Scholar] [CrossRef]
- Sheu, S.-Y.; Shiau, Y.-W.; Wei, Y.-T.; Chen, W.-M. Gemmobacter lanyuensis sp. nov., isolated from a freshwater spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 4039–4045. [Google Scholar] [CrossRef]
- Kang, J.Y.; Kim, M.-J.; Chun, J.; Son, K.P.; Jahng, K.Y. Gemmobacter straminiformis sp. nov., isolated from an artificial fountain. Int. J. Syst. Evol. Microbiol. 2017, 67, 5019–5025. [Google Scholar] [CrossRef]
- Kröber, E.; Cunningham, M.R.; Peixoto, J.; Spurgin, L.; Wischer, D.; Kruger, R.; Kumaresan, D. Comparative genomics analyses indicate differential methylated amine utilization trait within members of the genus Gemmobacter. Environ. Microbiol. Rep. 2021, 13, 195–208. [Google Scholar] [CrossRef]
- Liu, J.-J.; Zhang, X.-Q.; Chi, F.-T.; Pan, J.; Sun, C.; Wu, M. Gemmobacter megaterium sp. nov., isolated from coastal planktonic seaweeds. Int. J. Syst. Evol. Microbiol. 2014, 64, 66–71. [Google Scholar] [CrossRef]
- Qu, J.-H.; Ma, W.-W.; Zhou, J.; Wang, X.-F.; Lu, W.-L.; Qu, L.-B.; Wang, L.-F. Gemmobacter caeruleus sp. nov., a novel species originating from lake sediment. Int. J. Syst. Evol. Microbiol. 2020, 70, 1987–1992. [Google Scholar] [CrossRef]
- Sheu, S.-Y.; Sheu, D.-S.; Sheu, F.-S.; Chen, W.-M. Gemmobacter tilapiae sp. nov., a poly-β-hydroxybutyrate-accumulating bacterium isolated from a freshwater pond. Int. J. Syst. Evol. Microbiol. 2013, 63, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Hanada, S.; Manome, A.; Tsuchida, T.; Kurane, R.; Nakamura, K.; Kamagata, Y. Catellibacterium nectariphilum gen. nov., sp. nov., which requires a diffusible compound from a strain related to the genus Sphingomonas for vigorous growth. Int. J. Syst. Evol. Microbiol. 2004, 54, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.; Lee, D.W.; Lee, H.; Kwon, B.-O.; Khim, J.S.; Yim, U.H.; Park, H.; Park, B.; Choi, I.-G.; Kim, B.S.; et al. Gemmobacter lutimaris sp. nov., a marine bacterium isolated from a tidal flat. Int. J. Syst. Evol. Microbiol. 2019, 69, 1676–1681. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.-A.; Zheng, J.-W.; Cai, S.; Hang, B.-J.; He, J.; Li, S.-P. Catellibacterium nanjingense sp. nov., a propanil-degrading bacterium isolated from activated sludge, and emended description of the genus Catellibacterium. Int. J. Syst. Evol. Microbiol. 2012, 62, 495–499. [Google Scholar] [CrossRef]
- Zheng, J.-W.; Chen, Y.-G.; Zhang, J.; Ni, Y.-Y.; He, J.; Li, S.-P. Description of Catellibacterium caeni sp. nov., reclassification of Thodobacter changlensis Anil Kumar; et al. 2007 as Catellibacterium changlense comb. nov. and emended description of the genus Catellibacterium. Int. J. Syst. Evol. Microbiol. 2011, 61, 1921–1926. [Google Scholar] [CrossRef]
- Jin, L.; Wu, X.; Ko, S.-R.; Jin, F.-J.; Li, T.; Ahn, C.-Y.; Oh, H.-M.; Lee, H.-G. Description of Hymenobacter daejeonensis sp. nov., isolated from grass soil, based on multilocus sequence analysis of the 16S rRNA gene, gyrB and tuf genes. Antonie van Leeuwenhoek 2018, 111, 2283–2292. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Kim, M.; Park, T.; Lee, C.-S. Effect of cryopreservation on the bacterial community structure of filamentous cyanobacteria, Trichormus variabilis (Nostocales, Cyanobacteria). Cryobiology 2021, 98, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Ko, S.-R.; Lee, C.-S.; Ahn, C.-Y.; Oh, H.-M.; Lee, H.-G. Asprobacter aquaticus gen. nov., sp. nov., a prosthecate alphaproteobacterium isolated from fresh water. Int. J. Syst. Evol. Microbiol. 2017, 67, 4443–4448. [Google Scholar] [CrossRef] [PubMed]
- Gomori, G. [16] Preparation of buffers for use in enzyme studies. Science 1955, 1, 138–146. [Google Scholar] [CrossRef]
- Chaudhary, D.-K.; Kim, D.-U.; Kim, D.; Kim, J. Flavobacterium petrolei sp. nov., a novel psychrophilic, diesel-degrading bacterium isolated from oil-contaminated Arctic soil. Sci. Rep. 2019, 9, 1–9. [Google Scholar]
- Komagata, K.; Suzuki, K.-I. 4 Lipid and Cell-Wall Analysis in Bacterial Systematics. Methods Microbiol. 1988, 19, 161–207. [Google Scholar] [CrossRef]
- Tindall, B.-J. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 1990, 66, 199–202. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. mega7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.-A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.-D.; Gibson, T.-J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.-G. The Clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 24, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Fitch, W.-M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsentein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinf. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Devoid, S.; Disz, T.; Edwards, R.A.; Henry, C.S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. SEED Servers: High-Performance Access to the SEED Genomes, Annotations, and Metabolic Models. PLoS ONE 2012, 7, e48053. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Hanson, R.-S.; Hanson, T.-E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef]
- Kang, T.J.; Lee, E.Y. Metabolic versatility of microbial methane oxidation for biocatalytic methane conversion. J. Ind. Eng. Chem. 2016, 35, 8–13. [Google Scholar] [CrossRef]
- Banerjee, R.; Jones, J.-C.; Lipscomb, J.-D. Soluble Methane Monooxygenase. Annu. Rev. Biochem. 2019, 88, 409–431. [Google Scholar] [CrossRef]
- Burrows, K.J.; Cornish, A.; Scott, D.; Higgins, I.J. Substrate Specificities of the Soluble and Particulate Methane Mono-oxygenases of Methylosinus trichosporium OB3b. Microbiology 1984, 130, 3327–3333. [Google Scholar] [CrossRef]
- Elliott, S.J.; Zhu, M.; Tso, L.; Nguyen, H.-H.T.; Yip, J.H.-K.; Chan, S.I. Regio- and Stereoselectivity of Particulate Methane Monooxygenase from Methylococcus capsulatus (Bath). J. Am. Chem. Soc. 1997, 119, 9949–9955. [Google Scholar] [CrossRef]
- Smith, G.J.; Angle, J.C.; Solden, L.M.; Borton, M.A.; Morin, T.H.; Daly, R.A.; Johnston, M.D.; Stefanik, K.C.; Wolfe, R.; Gil, B.; et al. Members of the Genus Methylobacter Are Inferred to Account for the Majority of Aerobic Methane Oxidation in Oxic Soils from a Freshwater Wetland. mBio 2018, 9, e00815-18. [Google Scholar] [CrossRef]
- Kits, K.-D.; Klotz, M.-G.; Stein, L.-Y. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ. Microbiol. 2015, 17, 3219–3232. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).