Loss and Gain of Gut Bacterial Phylotype Symbionts in Afrotropical Stingless Bee Species (Apidae: Meliponinae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. DNA Extraction
2.3. 16S rRNA Gene Amplification, Sequencing, and Gut Community Analysis
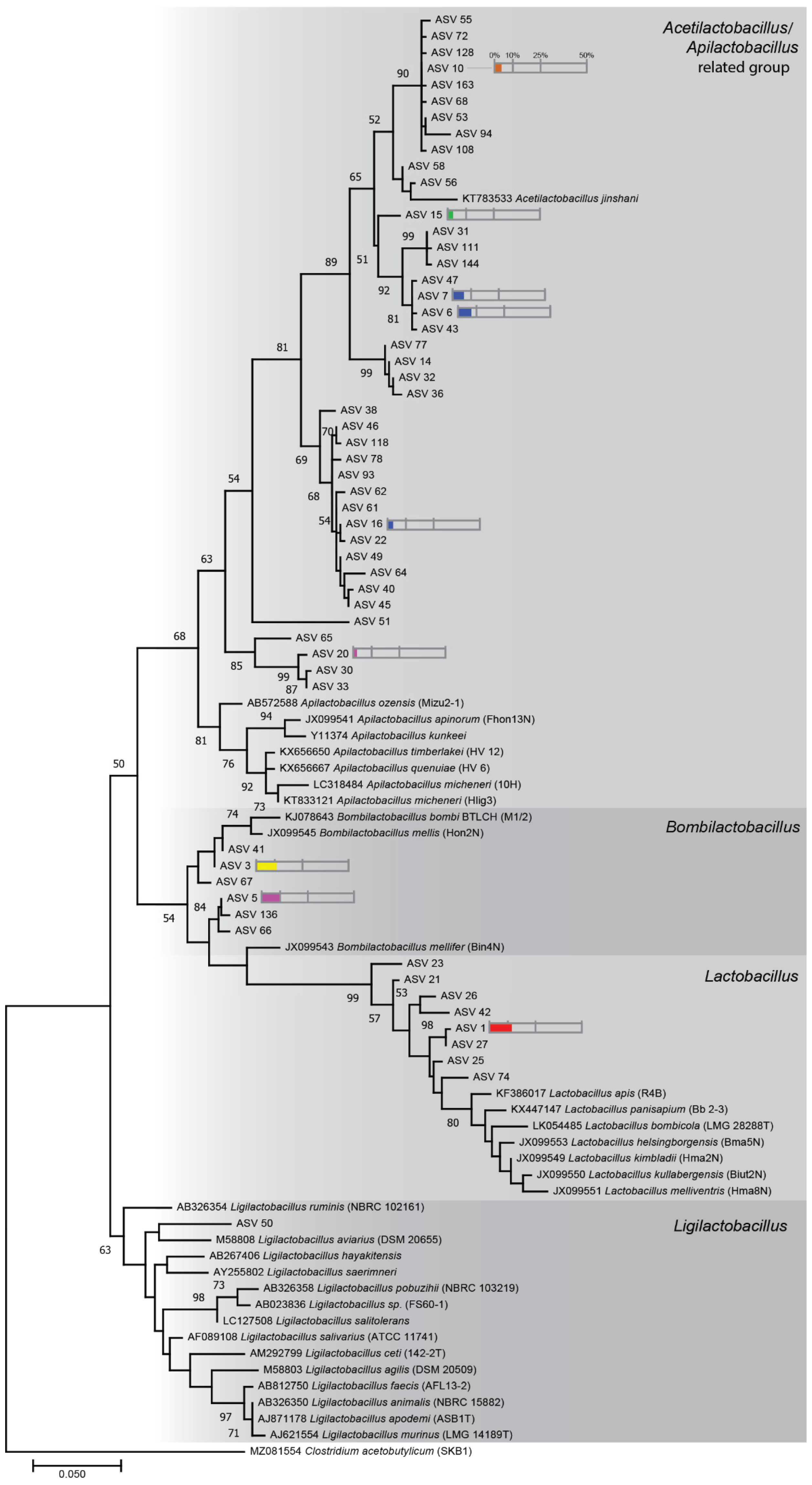
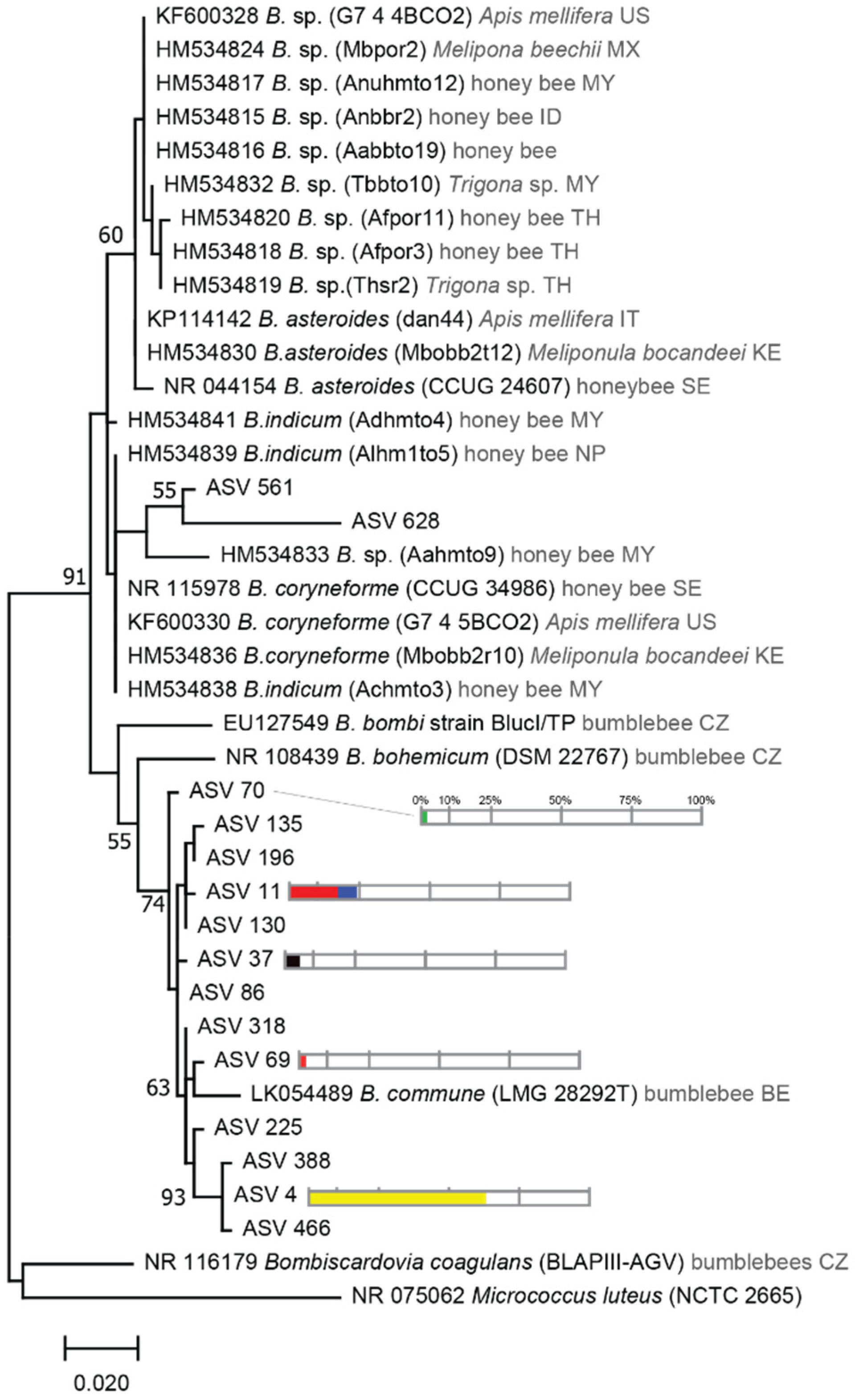
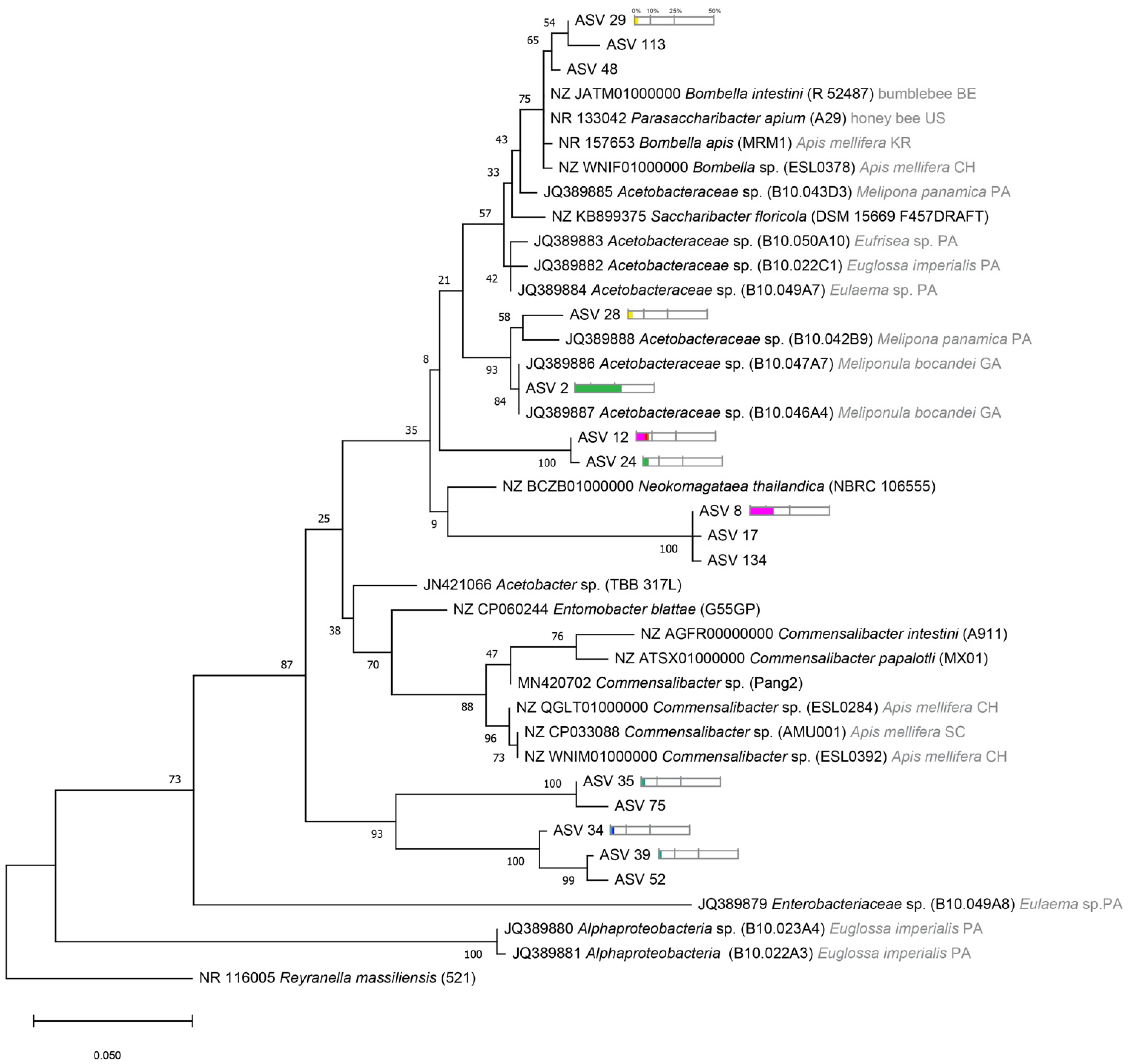
2.4. Phylogenetic Tree Analysis
3. Results
3.1. Bacterial Communities Associated with Stingless Bee Guts
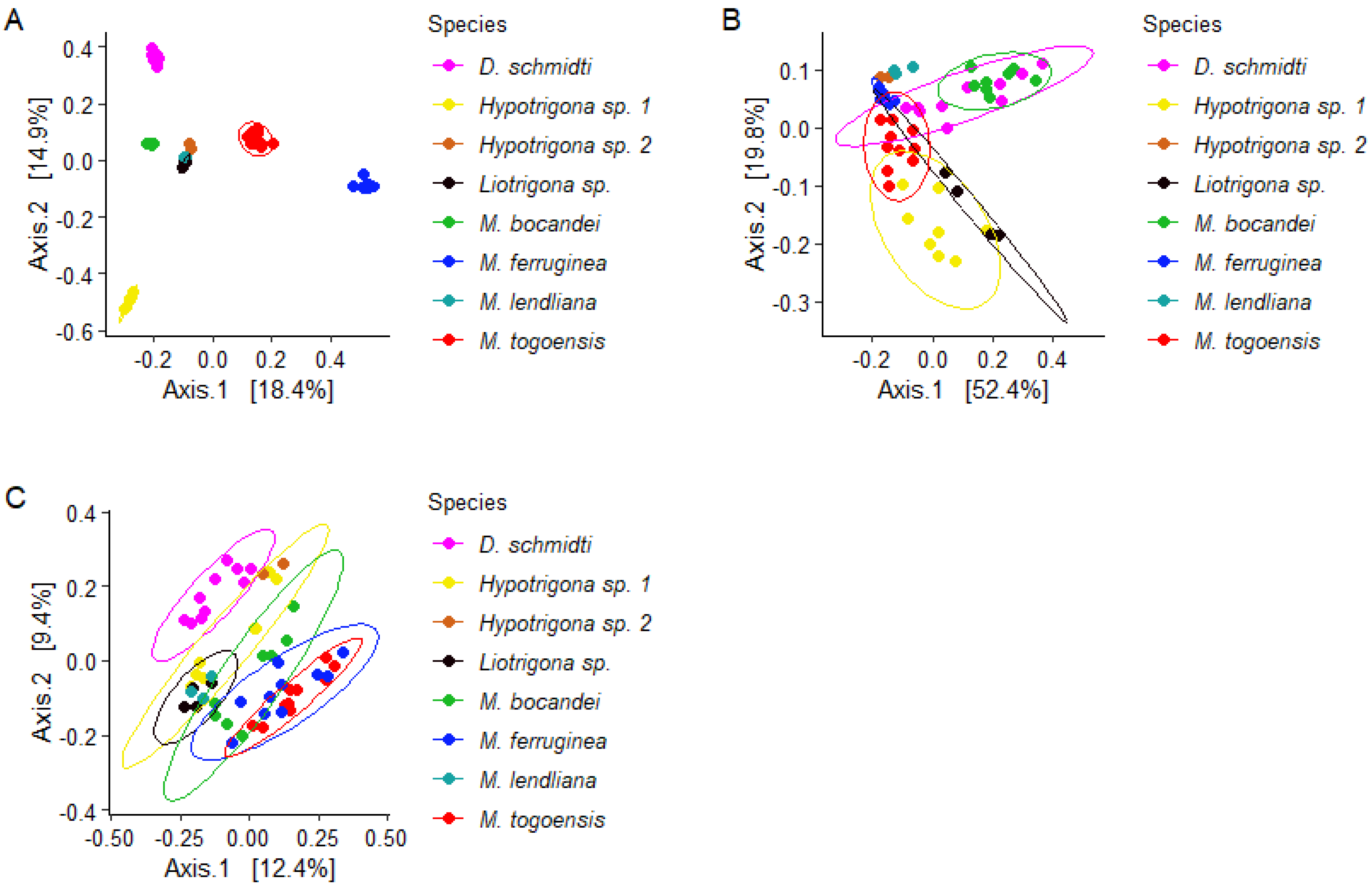
3.2. Bacterial Communities Varied across Stingless Bee Species
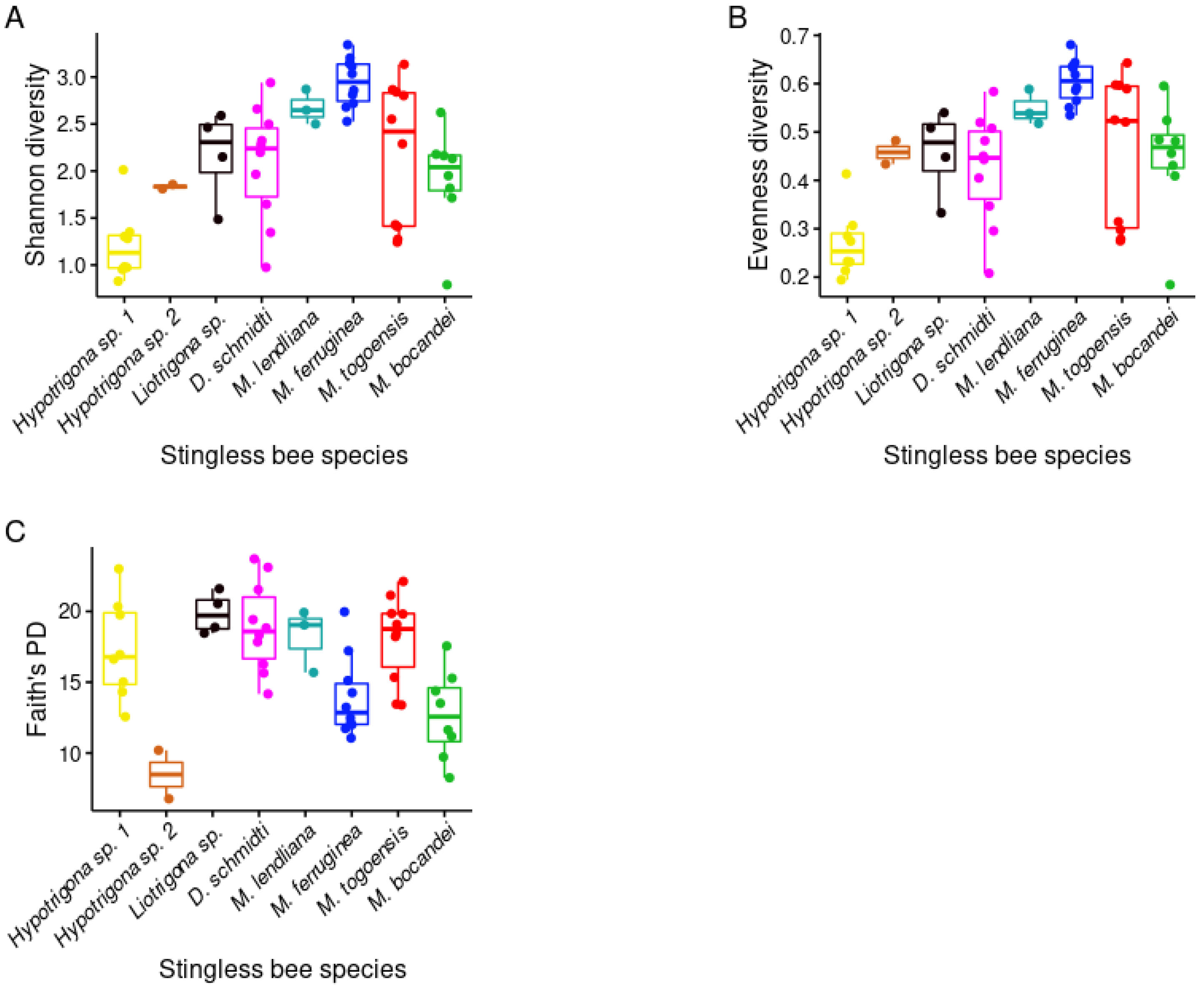
3.3. Phylogenetic Diversity of Stingless Bee Gut Bacterial Microbiota
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bawa, K.S. Plant-Pollinator Interactions in Tropical Rain Forests. Annu. Rev. Ecol. Syst. 1990, 21, 399–422. [Google Scholar] [CrossRef]
- Van Dulmen, A. Pollination and phenology of flowers in the canopy of two contrasting rain forest types in Amazonia, Colombia. Plant Ecol. 2001, 153, 73–85. [Google Scholar] [CrossRef]
- Garibaldi, L.A.; Carvalheiro, L.G.; Vaissière, B.E.; Gemmill-Herren, B.; Hipólito, J.; Freitas, B.M.; Ngo, H.T.; Azzu, N.; Sáez, A.; Åström, J.; et al. Mutually beneficial pollinator diversity and crop yield outcomes in small and large farms. Science 2016, 351, 388–391. [Google Scholar] [CrossRef]
- Grüter, C. Stingless Bees, Their Behaviour, Ecology and Evolution; Springer Nature Switzerland AG: Cham, Switzerland, 2020; ISBN 9783030600891. [Google Scholar]
- Jalil, A.H. Beescape for Meliponines: Conservation of Indo-Malayan Stingless Bees; Partridge Publishing Singapore: Singapore, 2014; ISBN 9781482823622. [Google Scholar]
- Michener, C.D. The Bees of the World; The Johns Hopkins University Press: Baltimore, MD, USA, 2000; Volume 58, ISBN 0-8018-6133-0. [Google Scholar]
- Quezada-Euán, J.J.G. Stingless Bees of Mexico; Springer: Berlin/Heidelberg, Germany, 2018; ISBN 9783319777849. [Google Scholar]
- Rasmussen, C.; Cameron, S.A. Global stingless bee phylogeny supports ancient divergence, vicariance, and long distance dispersal. Biol. J. Linn. Soc. 2010, 99, 206–232. [Google Scholar] [CrossRef]
- Roig-Alsina, A.; Alvarez, L.J. Southern distributional limits of Meliponini bees (Hymenoptera, Apidae) in the Neotropics: Taxonomic notes and distribution of Plebeia droryana and P. emerinoides in Argentina. Zootaxa 2017, 4244, 261–268. [Google Scholar] [CrossRef]
- Eardley, C.D. Taxonomic revision of the African stingless bees (Apoidea: Apidae: Apinae: Meliponini). Afr. Plant Prot. 2004, 10, 63–96. [Google Scholar]
- Fabre Anguilet, E.; Nguyen, B.K.; Bengone Ndong, T.; Haubruge, É.; Francis, F. Meliponini and Apini in Africa (Apidae: Apinae): A review on the challenges and stakes bound to their diversity and their distribution. Biotechnol. Agron. Soc. Environ. 2015, 19, 382–391. [Google Scholar]
- Ndungu, N.N.; Nkoba, K.; Sole, C.; Pirk, C.; Abdullahi, A.Y.; Raina, S.K.; Masiga, D.K. Resolving taxonomic ambiguity and cryptic speciation of Hypotrigona species through morphometrics and DNA barcoding. J. Apic. Res. 2018, 57, 354–363. [Google Scholar] [CrossRef]
- Ndungu, N.N.; Kiatoko, N.; Ciosi, M.; Salifu, D.; Nyansera, D.; Masiga, D.; Raina, S.K. Identification of stingless bees (Hymenoptera: Apidae) in Kenya using morphometrics and DNA barcoding. J. Apic. Res. 2017, 56, 341–353. [Google Scholar] [CrossRef]
- Nkoba, K.; Raina, S.K.; Muli, E.; Mithöfer, K.; Mueke, J. Species richness and nest dispersion of some tropical meliponine bees (Apidae: Meliponinae) in six habitat types in the Kakamega forest, western Kenya. Int. J. Trop. Insect Sci. 2012, 32, 194–202. [Google Scholar] [CrossRef]
- Azmi, W.A.; Sembok, W.Z.W.; Yusuf, N.; Hatta, M.F.M.; Salleh, A.F.; Hamzah, M.A.H.; Ramli, S.N. Effects of Pollination by the Indo-Malaya Stingless Bee (Hymenoptera: Apidae) on the Quality of Greenhouse-Produced Rockmelon. J. Econ. Entomol. 2019, 112, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.; Biesmeijer, J.C.; Breeze, T.; Dicks, L.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Kiatoko, N.; Raina, S.K.; Muli, E.; Mueke, J. Enhancement of fruit quality in Capsicum annum through pollination by Hypotrigona gribodoi in Kakamega, Western Kenya. Entomol. Sci. 2014, 17, 106–110. [Google Scholar] [CrossRef]
- Slaa, E.J.; Chaves, L.A.S.; Malagodi-Braga, K.S.; Hofstede, F.E. Stingless bees in applied pollination: Practice and perspectives. Apidologie 2006, 37, 293–315. [Google Scholar] [CrossRef]
- Nkoba, K.; Raina, S.K.; Langevelde, F. A vertical compartmented hive design for reducing post-harvest colony losses in three afrotropical stingless bee species (Apidae: Meliponinae). Int. J. Dev. Res. 2016, 6, 9026–9034. [Google Scholar]
- Kishan Tej, M.; Srinivasan, M.R.; Rajashree, V.; Thakur, R.K. Stingless bee Tetragonula iridipennis Smith for pollination of greenhouse cucumber. J. Entomol. Zool. Stud. 2017, 5, 1729–1733. [Google Scholar]
- Nishio, E.K.; Ribeiro, J.; de Oliveira, A.G.; Andrade, C.G.T.J.; Proni, E.A.; Kobayashi, R.K.T.; Nakazato, G. Antibacterial synergic effect of honey from two stingless bees: Scaptotrigona bipunctata Lepeletier, 1836, and S. postica Latreille, 1807. Sci. Rep. 2016, 6, 21641. [Google Scholar] [CrossRef]
- Dos Santos, S.B.; Roselino, A.; Hrncir, M.; Bego, L. Pollination of tomatoes by the stingless bee Melipona quadrifasciata and the honey bee Apis mellifera (Hymenoptera, Apidae). Genet. Mol. Res. 2009, 8, 751–757. [Google Scholar] [CrossRef]
- Mokaya, H.O.; Nkoba, K.; Ndunda, R.M.; Vereecken, N.J. Characterization of honeys produced by sympatric species of Afrotropical stingless bees (Hymenoptera, Meliponini). Food Chem. 2021, 366, 130597. [Google Scholar] [CrossRef]
- Moran, N.A.; Tran, P.; Gerardo, N.M.; Tannock, G.W.; Ghazally, S.; Walter, J.; Loach, D.; Brooks, H.; Cook, G.; Surette, M.; et al. Symbiosis and Insect Diversification: An Ancient Symbiont of Sap-Feeding Insects from the Bacterial Phylum Bacteroidetes. Appl. Environ. Microbiol. 2005, 71, 8419–8425. [Google Scholar] [CrossRef]
- Gerardo, N.; Hurst, G. Q&A: Friends (but sometimes foes) within: The complex evolutionary ecology of symbioses between host and microbes. BMC Biol. 2017, 15, 126. [Google Scholar] [CrossRef]
- Schmidt, K.; Engel, P. Mechanisms underlying gut microbiota–host interactions in insects. J. Exp. Biol. 2021, 224, jeb207696. [Google Scholar] [CrossRef]
- Salem, H.; Kaltenpoth, M. Beetle–Bacterial Symbioses: Endless Forms Most Functional. Annu. Rev. Entomol. 2022, 67, 201–219. [Google Scholar] [CrossRef]
- Weisskopf, L.; Schulz, S.; Garbeva, P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat. Rev. Microbiol. 2021, 19, 391–404. [Google Scholar] [CrossRef]
- Duplais, C.; Sarou-Kanian, V.; Massiot, D.; Hassan, A.; Perrone, B.; Estevez, Y.; Wertz, J.T.; Martineau, E.; Farjon, J.; Giraudeau, P.; et al. Gut bacteria are essential for normal cuticle development in herbivorous turtle ants. Nat. Commun. 2021, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Cui, Y.; Chu, X.; Li, G.; Yang, M.; Wang, R.; Liang, G.; Wu, S.; Tigabu, M.; Zhang, F.; et al. Gut Bacterial Communities of Lymantria xylina and Their Associations with Host Development and Diet. Microorganisms 2021, 9, 1860. [Google Scholar] [CrossRef]
- Chakraborty, A.; Roy, A. Microbial influence on plant–insect interaction. In Plant-Pest Interactions: From Molecular Mechanisms to Chemical Ecology; Springer: Singapore, 2021. [Google Scholar]
- Emery, O.; Schmidt, K.; Engel, P. Immune system stimulation by the gut symbiont Frischella perrarain the honey bee (Apis mellifera). Mol. Ecol. 2017, 26, 2576–2590. [Google Scholar] [CrossRef]
- Engel, P.; Bartlett, K.D.; Moran, N.A. The Bacterium Frischella perrara Causes Scab Formation in the Gut of its Honeybee Host. MBio 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. USA 2011, 108, 19288–19292. [Google Scholar] [CrossRef] [PubMed]
- Vernier, C.L.; Chin, I.M.; Adu-Oppong, B.; Krupp, J.J.; Levine, J.; Dantas, G.; Ben-Shahar, Y. The gut microbiome defines social group membership in honey bee colonies. Sci. Adv. 2020, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef]
- Zheng, H.; Nishida, A.; Kwong, W.K.; Koch, H.; Engel, P.; Steele, M.I.; Moran, N.A. Metabolism of Toxic Sugars by Strains of the Bee Gut Symbiont Gilliamella apicola. MBio 2016, 7, e01326-16. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Perreau, J.; Powell, J.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.A.; Brettell, L.E.; Liu, H.; Nacko, S.; Spooner-Hart, R.; Riegler, M.; Cook, J.M. Temporal changes in the microbiome of stingless bee foragers following colony relocation. FEMS Microbiol. Ecol. 2020, 97, fiaa236. [Google Scholar] [CrossRef]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef]
- Leonhardt, S.D.; Kaltenpoth, M. Microbial Communities of Three Sympatric Australian Stingless Bee Species. PLoS ONE 2014, 9, e105718. [Google Scholar] [CrossRef]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as Major Modulators of Insect Health: Lactic Acid Bacteria and Honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Tang, Q.-H.; Miao, C.-H.; Chen, Y.-F.; Dong, Z.-X.; Cao, Z.; Liao, S.-Q.; Wang, J.-X.; Wang, Z.-W.; Guo, J. The composition of bacteria in gut and beebread of stingless bees (Apidae: Meliponini) from tropics Yunnan, China. Antonie Leeuwenhoek 2021, 114, 1293–1305. [Google Scholar] [CrossRef]
- Cerqueira, A.E.S.; Hammer, T.J.; Moran, N.A.; Santana, W.C.; Kasuya, M.C.M.; da Silva, C.C. Extinction of anciently associated gut bacterial symbionts in a clade of stingless bees. ISME J. 2021, 15, 2813–2816. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Urbano, S.D.S.; Caesar, L.; Blochtein, B.; Sattler, A.; Zuge, V.; Haag, K.L. Report on the microbiota of Melipona quadrifasciata affected by a recurrent disease. J. Invertebr. Pathol. 2017, 143, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Vit, P.; Roubik, D.W.; Pedro, S.R.M. Taxonomy as a Tool for Conservation of African Stingless Bees and Their Honey. In Eardley, Connal Kwapong, Peter; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–654. ISBN 9781461449607. [Google Scholar]
- Engel, P.; James, R.R.; Koga, R.; Kwong, W.K.; Mcfrederick, Q.S.; Moran, N.A. Standard methods for research on Apis mellifera gut symbionts. J. Apic. Res. 2013, 524, 1–24. [Google Scholar] [CrossRef]
- Tola, Y.; Waweru, J.; Hurst, G.; Slippers, B.; Paredes, J. Characterization of the Kenyan Honey Bee (Apis mellifera) Gut Microbiota: A First Look at Tropical and Sub-Saharan African Bee Associated Microbiomes. Microorganisms 2020, 8, 1721. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Rao, V.D.; Yeoman, C.J.; Wilson, B.A.; White, B.A.; Goldenfeld, N.; Robinson, G.E. Caste-Specific Differences in Hindgut Microbial Communities of Honey Bees (Apis mellifera). PLoS ONE 2015, 10, e0123911. [Google Scholar] [CrossRef]
- Killer, J.; Dubná, S.; Sedláček, I.; Švec, P. Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int. J. Syst. Evol. Microbiol. 2014, 64, 152–157. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Vásquez, A. Detection and Identification of a Novel Lactic Acid Bacterial Flora within the Honey Stomach of the Honeybee Apis mellifera. Curr. Microbiol. 2008, 57, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Praet, J.; Meeus, I.; Cnockaert, M.; Houf, K.; Smagghe, G.; Vandamme, P. Novel lactic acid bacteria isolated from the bumble bee gut: Convivina intestini gen. nov., sp. nov., Lactobacillus bombicola sp. nov., and Weissella bombi sp. nov. Antonie Leeuwenhoek 2015, 107, 1337–1349. [Google Scholar] [CrossRef]
- Anderson, K.E.; Johansson, A.; Sheehan, T.H.; Mott, B.M.; Corby-Harris, V.; Johnstone, L.; Sprissler, R.; Fitz, W. Draft genome sequences of two Bifidobacterium sp. from the honey bee (Apis mellifera). Gut Pathog. 2013, 5, 42. [Google Scholar] [CrossRef]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Johnson, M.M.; Jolley, M. Composition and acquisition of the microbiome in solitary, ground-nesting alkali bees. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Killer, J.; Kopečný, J.; Mrazek, J.; Rada, V.; Benada, O.; Koppová, I.; Havlik, J.; Straka, J. Bifidobacterium bombi sp. nov., from the bumblebee digestive tract. Int. J. Syst. Evol. Microbiol. 2009, 59, 2020–2024. [Google Scholar] [CrossRef][Green Version]
- Yun, J.-H.; Lee, J.-Y.; Hyun, D.-W.; Jung, M.-J.; Bae, J.-W. Bombella apis sp. nov., an acetic acid bacterium isolated from the midgut of a honey bee. Int. J. Syst. Evol. Microbiol. 2017, 67, 2184–2188. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Illeghems, K.; Van Kerrebroeck, S.; Borremans, W.; Cleenwerck, I.; Smagghe, G.; De Vuyst, L.; Vandamme, P. Whole-Genome Sequence Analysis of Bombella intestini LMG 28161T, a Novel Acetic Acid Bacterium Isolated from the Crop of a Red-Tailed Bumble Bee, Bombus lapidarius. PLoS ONE 2016, 11, e0165611. [Google Scholar] [CrossRef]
- Yukphan, P.; Malimas, T.; Muramatsu, Y.; Potacharoen, W.; Tanasupawat, S.; Nakagawa, Y.; Tanticharoen, M.; Yamada, Y. Neokomagataea gen. Nov., with descriptions of Neokomagataea thailandica sp. nov. and Neokomagataea tanensis sp. Nov., osmotolerant acetic acid bacteria of the α-proteobacteria. Biosci. Biotechnol. Biochem. 2011, 75, 419–426. [Google Scholar] [CrossRef]
- Ellegaard, K.; Engel, P. New Reference Genome Sequences for 17 Bacterial Strains of the Honey Bee Gut Microbiota. Microbiol. Resour. Announc. 2018, 7, 4–6. [Google Scholar] [CrossRef]
- Siozios, S.; Moran, J.; Chege, M.; Hurst, G.D.D.; Paredes, J.C. Complete Reference Genome Assembly for Commensalibacter sp. Strain AMU001, an Acetic Acid Bacterium Isolated from the Gut of Honey Bees. Microbiol. Resour. Announc. 2019, 8, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Corby-Harris, V.; Maes, P.; Anderson, K.E. The Bacterial Communities Associated with Honey Bee (Apis mellifera) Foragers. PLoS ONE 2014, 9, e95056. [Google Scholar] [CrossRef] [PubMed]
- Rosso, G.B.; Engel, P. Functional roles and metabolic niches in the honey bee gut microbiota. Curr. Opin. Microbiol. 2018, 43, 69–76. [Google Scholar] [CrossRef]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and Virus Protection in Insects. Science 2008, 322, 702. [Google Scholar] [CrossRef]
- Teixeira, L.; Ferreira, Á.; Ashburner, M. The Bacterial Symbiont Wolbachia Induces Resistance to RNA Viral Infections in Drosophila melanogaster. PLoS Biol. 2008, 6, 2753–2763. [Google Scholar] [CrossRef]
- Galbraith, D.A.; Fuller, Z.L.; Ray, A.M.; Brockmann, A.; Frazier, M.; Gikungu, M.W.; Martinez, J.F.I.; Kapheim, K.M.; Kerby, J.T.; Kocher, S.D.; et al. Investigating the viral ecology of global bee communities with high-throughput metagenomics. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Genet. 2008, 6, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Luna-Lucena, D.; Rabico, F.; Simoes, Z.L. Reproductive capacity and castes in eusocial stingless bees (Hymenoptera: Apidae). Curr. Opin. Insect Sci. 2019, 31, 20–28. [Google Scholar] [CrossRef]
- Vollet-Neto, A.; Koffler, S.; Dos Santos, C.F.; Menezes, C.; Nunes, F.; Hartfelder, K.; Imperatriz-Fonseca, V.L.; Alves, D.A. Recent advances in reproductive biology of stingless bees. Insectes Sociaux 2018, 65, 201–212. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tola, Y.H.; Waweru, J.W.; Ndungu, N.N.; Nkoba, K.; Slippers, B.; Paredes, J.C. Loss and Gain of Gut Bacterial Phylotype Symbionts in Afrotropical Stingless Bee Species (Apidae: Meliponinae). Microorganisms 2021, 9, 2420. https://doi.org/10.3390/microorganisms9122420
Tola YH, Waweru JW, Ndungu NN, Nkoba K, Slippers B, Paredes JC. Loss and Gain of Gut Bacterial Phylotype Symbionts in Afrotropical Stingless Bee Species (Apidae: Meliponinae). Microorganisms. 2021; 9(12):2420. https://doi.org/10.3390/microorganisms9122420
Chicago/Turabian StyleTola, Yosef Hamba, Jacqueline Wahura Waweru, Nelly N. Ndungu, Kiatoko Nkoba, Bernard Slippers, and Juan C. Paredes. 2021. "Loss and Gain of Gut Bacterial Phylotype Symbionts in Afrotropical Stingless Bee Species (Apidae: Meliponinae)" Microorganisms 9, no. 12: 2420. https://doi.org/10.3390/microorganisms9122420
APA StyleTola, Y. H., Waweru, J. W., Ndungu, N. N., Nkoba, K., Slippers, B., & Paredes, J. C. (2021). Loss and Gain of Gut Bacterial Phylotype Symbionts in Afrotropical Stingless Bee Species (Apidae: Meliponinae). Microorganisms, 9(12), 2420. https://doi.org/10.3390/microorganisms9122420






