Challenging the Conventional Interpretation of HCMV Seronegativity
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Immunological Responses to HCMV
2.3. vIL-10 ELISA
2.4. Detection of CMV DNA in Saliva
2.5. Detection of CMV-Encoded miRNA in Saliva
3. Results
3.1. Three Individuals Were Seronegative despite Detectable HCMV DNA
3.2. Four Seronegative Individuals Had Detectable HCMV vIL-10 in Plasma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuhair, M.; Smit, G.S.A.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef]
- Jacobson, M.A.; Mills, J. Serious cytomegalovirus disease in the acquired immunodeficiency syndrome (AIDS). Clinical findings, diagnosis, and treatment. Ann. Intern. Med. 1988, 108, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Rubin, R.H.; Tolkoff-Rubin, N.E.; Oliver, D.; Rota, T.R.; Hamilton, J.; Betts, R.F.; Pass, R.F.; Hillis, W.; Szmuness, W.; Farrell, M.L.; et al. Multicenter seroepidemiologic study of the impact of cytomegalovirus infection on renal transplantation. Transplantation 1985, 40, 243–249. [Google Scholar] [CrossRef]
- Cannon, M.J.; Hyde, T.B.; Schmid, D.S. Review of cytomegalovirus shedding in bodily fluids and relevance to congenital cytomegalovirus infection. Rev. Med. Virol. 2011, 21, 240–255. [Google Scholar] [CrossRef]
- Amin, M.M.; Stowell, J.D.; Hendley, W.; Garcia, P.; Schmid, D.S.; Cannon, M.J.; Dollard, S.C. CMV on surfaces in homes with young children: Results of PCR and viral culture testing. BMC Infect. Dis. 2018, 18, 391. [Google Scholar] [CrossRef] [PubMed]
- Thom, J.T.; Walton, S.M.; Torti, N.; Oxenius, A. Salivary gland resident APCs are Flt3L- and CCR2-independent macrophage-like cells incapable of cross-presentation. Eur. J. Immunol. 2014, 44, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Lee, S.; Lloyd, M.; Irish, A.; Price, P. The Detection of CMV in Saliva Can Mark a Systemic Infection with CMV in Renal Transplant Recipients. Int. J. Mol. Sci. 2019, 20, 5230. [Google Scholar] [CrossRef] [PubMed]
- Mayer, B.T.; Krantz, E.M.; Swan, D.; Ferrenberg, J.; Simmons, K.; Selke, S.; Huang, M.-L.; Casper, C.; Corey, L.; Wald, A.; et al. Transient Oral Human Cytomegalovirus Infections Indicate Inefficient Viral Spread from Very Few Initially Infected Cells. J. Virol. 2017, 91, e00380-17. [Google Scholar] [CrossRef] [PubMed]
- Farrell, H.; Lawler, C.; Tan, C.S.E.; MacDonald, K.; Bruce, K.; Mach, M.; Davis-Poynter, N.; Stevenson, P.G. Murine Cytomegalovirus Exploits Olfaction to Enter New Hosts. mBio 2016, 7, e00251-16. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J. Infectious diseases in schools and child care facilities. Pediatr. Rev. 2001, 22, 39–46. [Google Scholar] [CrossRef]
- Poole, E.; Sinclair, J. Sleepless latency of human cytomegalovirus. Med. Microbiol. Immunol. 2015, 204, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.; Abendroth, A.; Slobedman, B. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 2004, 78, 1440–1447. [Google Scholar] [CrossRef]
- Poole, E.; Neves, T.C.; Oliveira, M.T.; Sinclair, J.; Da Silva, M.C.C. Human Cytomegalovirus interleukin 10 Homologs: Facing the Immune System. Front. Cell. Infect. Microbiol. 2020, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Morooka, M.; Hashimoto, S.; Ihra, M.; Yoshikawa, T. Analysis of the shedding of three beta-herpesviruses in urine and saliva of children with renal disease. J. Med. Virol. 2014, 86, 505–511. [Google Scholar] [CrossRef]
- Larsson, S.; Soderberg-Naucler, C.; Wang, F.; Moller, E. Cytomegalovirus DNA can be detected in peripheral blood mononuclear cells from all seropositive and most seronegative healthy blood donors over time. Transfusion 1998, 38, 271–278. [Google Scholar] [CrossRef]
- Litjens, N.H.R.; Huang, L.; Dedeoglu, B.; Meijers, R.W.J.; Kwekkeboom, J.; Betjes, M.G.H. Protective Cytomegalovirus (CMV)-Specific T-Cell Immunity Is Frequent in Kidney Transplant Patients without Serum Anti-CMV Antibodies. Front. Immunol. 2017, 8, 1137. [Google Scholar] [CrossRef]
- Young, V.P.; Mariano, M.C.; Tu, C.C.; Allaire, K.M.; Avdic, S.; Slobedman, B.; Spencer, J.V. Modulation of the Host Environment by Human Cytomegalovirus with Viral Interleukin 10 in Peripheral Blood. J. Infect. Dis. 2017, 215, 874–882. [Google Scholar] [CrossRef]
- Lee, S.; Affandi, J.S.; Irish, A.B.; Price, P. Cytomegalovirus infection alters phenotypes of different gammadelta T-cell subsets in renal transplant recipients with long-term stable graft function. J. Med. Virol. 2017, 89, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Doualeh, M.; Affandi, J.S.; Makwana, N.; Irish, A.; Price, P. Functional and clinical consequences of changes to natural killer cell phenotypes driven by chronic cytomegalovirus infections. J. Med. Virol. 2019, 91, 1120–1127. [Google Scholar] [CrossRef]
- Young, V.P.; Mariano, M.C.; Faure, L.; Spencer, J.V. Detection of Cytomegalovirus Interleukin 10 (cmvIL-10) by Enzyme-Linked Immunosorbent Assay (ELISA). Methods Mol. Biol. 2021, 2244, 291–299. [Google Scholar]
- Watzinger, F.; Suda, M.; Preuner, S.; Baumgartinger, R.; Ebner, K.; Baskova, L.; Niesters, H.; Lawitschka, A.; Lion, T. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J. Clin. Microbiol. 2004, 42, 5189–5198. [Google Scholar] [CrossRef]
- Barbi, M.; Binda, S.; Caroppo, S.; Primache, V.; Didò, P.; Guidotti, P.; Corbetta, C.; Melotti, D. CMV gB genotypes and outcome of vertical transmission: Study on dried blood spots of congenitally infected babies. J. Clin. Virol. 2001, 21, 75–79. [Google Scholar] [CrossRef]
- Waters, S.; Lee, S.; Munyard, K.; Irish, A.; Price, P.; Wang, B.H. Human Cytomegalovirus-Encoded microRNAs Can Be Found in Saliva Samples from Renal Transplant Recipients. Noncoding RNA 2020, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Brook, E.; Affandi, J.; Howson, P.; Tanudjaja, S.A.; Dhaliwal, S.; Irish, A.; Price, P. A high burden of cytomegalovirus marks poor vascular health in transplant recipients more clearly than in the general population. Clin. Transl. Immunol. 2019, 8, e1043. [Google Scholar] [CrossRef] [PubMed]
- Price, P.; Lee, S.; Affandi, J.; Parsons, R.; Naylor, L.H.; Watts, G.F.; Irish, A. Cytomegalovirus antibody and vascular pathology in renal transplant recipients. J. Med. Virol. 2017, 89, 177–181. [Google Scholar] [CrossRef]
- Estiasari, R.; Aryanto, I.; Lee, S.; Pramana, S.; Djauzi, S.; Price, P. Determinants of cognitive health in Indonesian HIV patients beginning antiretroviral therapy. J. Neurovirol. 2020, 26, 32–40. [Google Scholar] [CrossRef]
- Costa-Garcia, M.; Vera, A.; Moraru, M.; Vilches, C.; López-Botet, M.; Muntasell, A. Antibody-mediated response of NKG2Cbright NK cells against human cytomegalovirus. J. Immunol. 2015, 194, 2715–2724. [Google Scholar] [CrossRef] [PubMed]
- Slifkin, M.; Tempesti, P.; Poutsiaka, D.D.; Snydman, D.R. Late and atypical cytomegalovirus disease in solid-organ transplant recipients. Clin. Infect. Dis. 2001, 33, E62–E68. [Google Scholar] [CrossRef][Green Version]
- Cha, T.A.; Tom, E.; Kemble, G.W.; Duke, G.M.; Mocarski, E.S.; Spaete, R.R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 1996, 70, 78–83. [Google Scholar] [CrossRef]
- Wang, D.; Shenk, T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA 2005, 102, 18153–18158. [Google Scholar] [CrossRef]
- Biolatti, M.; Dell’Oste, V.; Pautasso, S.; von Einem, J.; Marschall, M.; Plachter, B.; Gariglio, M.; De Andrea, M.; Landolfo, S. Regulatory Interaction between the Cellular Restriction Factor IFI16 and Viral pp65 (pUL83) Modulates Viral Gene Expression and IFI16 Protein Stability. J. Virol. 2016, 90, 8238–8250. [Google Scholar] [CrossRef] [PubMed]
- Alunno, A.; Caneparo, V.; Carubbi, F.; Bistoni, O.; Caterbi, S.; Gariglio, M.; Bartoloni, E.; Landolfo, S.; Gerli, R. Interferon gamma-inducible protein 16 (IFI16) and anti-IFI16 antibodies in primary Sjogren’s syndrome: Findings in serum and minor salivary glands. Reumatismo 2015, 67, 85–90. [Google Scholar] [CrossRef][Green Version]
- Kaminski, H.; Garrigue, I.; Couzi, L.; Taton, B.; Bachelet, T.; Moreau, J.F.; Dechanet-Merville, J.; Thiebaut, R.; Merville, P. Surveillance of gammadelta T Cells Predicts Cytomegalovirus Infection Resolution in Kidney Transplants. J. Am. Soc. Nephrol. 2016, 27, 637–645. [Google Scholar] [CrossRef]
- Tu, C.C.; Arnolds, K.L.; O’Connor, C.M.; Spencer, J.V. Human Cytomegalovirus UL111A and US27 Gene Products Enhance the CXCL12/CXCR4 Signaling Axis via Distinct Mechanisms. J. Virol. 2018, 92, e01981-17. [Google Scholar] [CrossRef] [PubMed]
- Avdic, S.; McSharry, B.P.; Slobedman, B. Modulation of dendritic cell functions by viral IL-10 encoded by human cytomegalovirus. Front. Microbiol. 2014, 5, 337. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Munoz, F.M.; Callahan, S.T.; Rupp, R.; Wootton, S.H.; Edwards, K.M.; Turley, C.B.; Stanberry, L.R.; Patel, S.M.; Mcneal, M.M.; et al. Safety and efficacy of a cytomegalovirus glycoprotein B (gB) vaccine in adolescent girls: A randomized clinical trial. Vaccine 2016, 34, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Rezzouk, B.; Bouattar, T.; Belkadi, B.; Razine, R.; Bayahia, R.; Ouzeddoun, N.; Benamar, L.; Rhou, H.; Bouihat, N.; Ibrahimi, A.; et al. Characteristics and Outcomes of Cytomegalovirus Infection in Seropositive Kidney Transplant Recipients in the Era of Antiviral Prophylaxis with Valacyclovir: A Single-Center Study in Morocco. Transpl. Res. Risk Manag. 2021, 13, 1. [Google Scholar] [CrossRef]
| Target | Primer | Sequence (5′-3′) | Product (Base Pairs) |
|---|---|---|---|
| FWD | CCCGAAAACGTGTCGCC | ||
| UL54 | REV | AAACGTTGACGCAGATACTGTAGC | 105 |
| PROBE (5 µM) | 6-FAM-TATCGTCAGCATCTGGTGC-BHQ-1 | ||
| FWD | AACTCAGCCTTCCCTAAGACCA | ||
| MIE | REV | GGGAGCACTGAGGCAAGTTC | 76 |
| PROBE (2 µM) | 6-FAM-CAATGGCTGCAGTCAGGCCATGG-TAMRA-6 | ||
| FWD | TGAGTATGCCTGCCGTGTGA | 105 | |
| B2M | REV | ACTCATACACAACTTTCAGCAGCTTAC | |
| PROBE (5 µM) | 6-FAM-CCATGTGACTTTGTCACAGCCCAAGATAGTT-TAMRA-6 | ||
| gB (UL55) | OUTER FWD | GAATRGCTGAYGGRTTGATCTTG | 590 |
| OUTER REV | GATCTCCTGGGATATACAGGACG | ||
| INNER FWD | GAGTTCCTTGAAGACCTCTAG | ||
| INNER REV | ACYTTCTGGGAAGCCTCGGAACG | 519 | |
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | |
|---|---|---|---|---|---|---|---|
| Age (years) | 33 | 57 | 62 | 55 | 56 | 34 | 42 |
| Male (M)/Female (F) | F | F | F | M | M | F | F |
| Ethnicity | Caucasian | Caucasian | Caucasian | Caucasian | Caucasian | Asian | Caucasian |
| RTR/Healthy | RTR | Healthy | Healthy | Healthy | RTR | Healthy | Healthy |
| Donor HCMV status | Negative | - | - | - | Positive | - | - |
| T-cell responses (EliSpot assay presented as cells producing interferon-γ/200,000 PBMC) a | |||||||
| HCMV lysate | 0 | 0 | 1 | 0 | 3 | 0 | 3 |
| IE-1 pooled peptides | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| pp65 pooled peptides | 0 | 0 | 1 | 0 | 1 | 0 | 2 |
| NK cells (flow cytometry) | |||||||
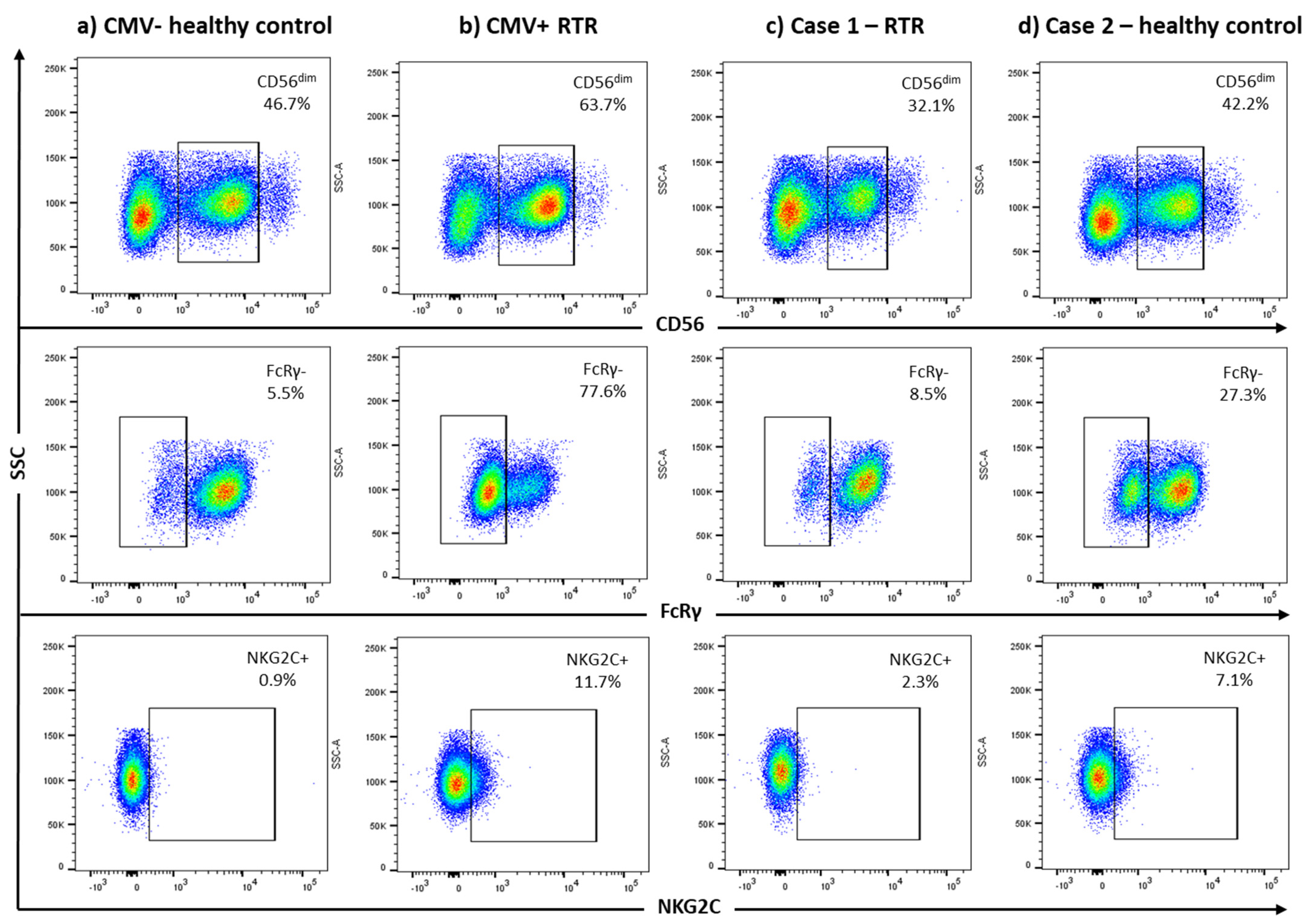
| FcRγ− (% CD3−CD56dim) | 8.5% | 27.3% | 2.7% | 3.9% | 8.8% | 4.1% | 13.8% |
| NKG2C+ (% CD3−CD56dim) | 2.3% | 7.1% | 4.3% | 1.8% | 2.1% | 2.8% | 2.8% |
| γδ T-cells (flow cytometry) | |||||||
| Vδ2− (% CD3+) | 0.19% | 0.5% | 2.2% | 0.1% | 0.4% | 0.7% | 1.2% |
| HCMV DNA/miRNA in saliva | |||||||
| HCMV DNA (UL54 qPCR) | Pos | Neg | Pos | Neg | Neg | Neg | Neg |
| HCMV DNA (UL55 nested PCR) | Pos | Pos | Pos | Neg | Neg | Neg | Neg |
| HCMV-encoded miRNA | Neg | miR-US5-2-3p | Neg | NT | NT | Neg | NT |
| Plasma HCMV vIL-10 (pg/mL) | |||||||
| 2006 | NA | NA | NA | NA | NA | 801 | NA |
| 2012 | <60 | NA | NA | 1840 | 945 | NA | NA |
| 2014 | <60 | <60 | <60 | 639 | <60 | 1440 | 166 |
| 2017 | NA | <60 | <60 | 916 | 704 | 1247 | 786 |
| 2021 | NA | NA | NA | NA | NA | 815 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waters, S.; Lee, S.; Irish, A.; Price, P. Challenging the Conventional Interpretation of HCMV Seronegativity. Microorganisms 2021, 9, 2382. https://doi.org/10.3390/microorganisms9112382
Waters S, Lee S, Irish A, Price P. Challenging the Conventional Interpretation of HCMV Seronegativity. Microorganisms. 2021; 9(11):2382. https://doi.org/10.3390/microorganisms9112382
Chicago/Turabian StyleWaters, Shelley, Silvia Lee, Ashley Irish, and Patricia Price. 2021. "Challenging the Conventional Interpretation of HCMV Seronegativity" Microorganisms 9, no. 11: 2382. https://doi.org/10.3390/microorganisms9112382
APA StyleWaters, S., Lee, S., Irish, A., & Price, P. (2021). Challenging the Conventional Interpretation of HCMV Seronegativity. Microorganisms, 9(11), 2382. https://doi.org/10.3390/microorganisms9112382







