Genetic Diversity of Staphylococcus aureus Strains from a Tertiary Care Hospital in Rawalpindi, Pakistan
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Identification
2.2. Antibiotic Susceptibility Testing
2.3. Molecular Characterization
3. Results
3.1. Bacterial Isolation and Identification
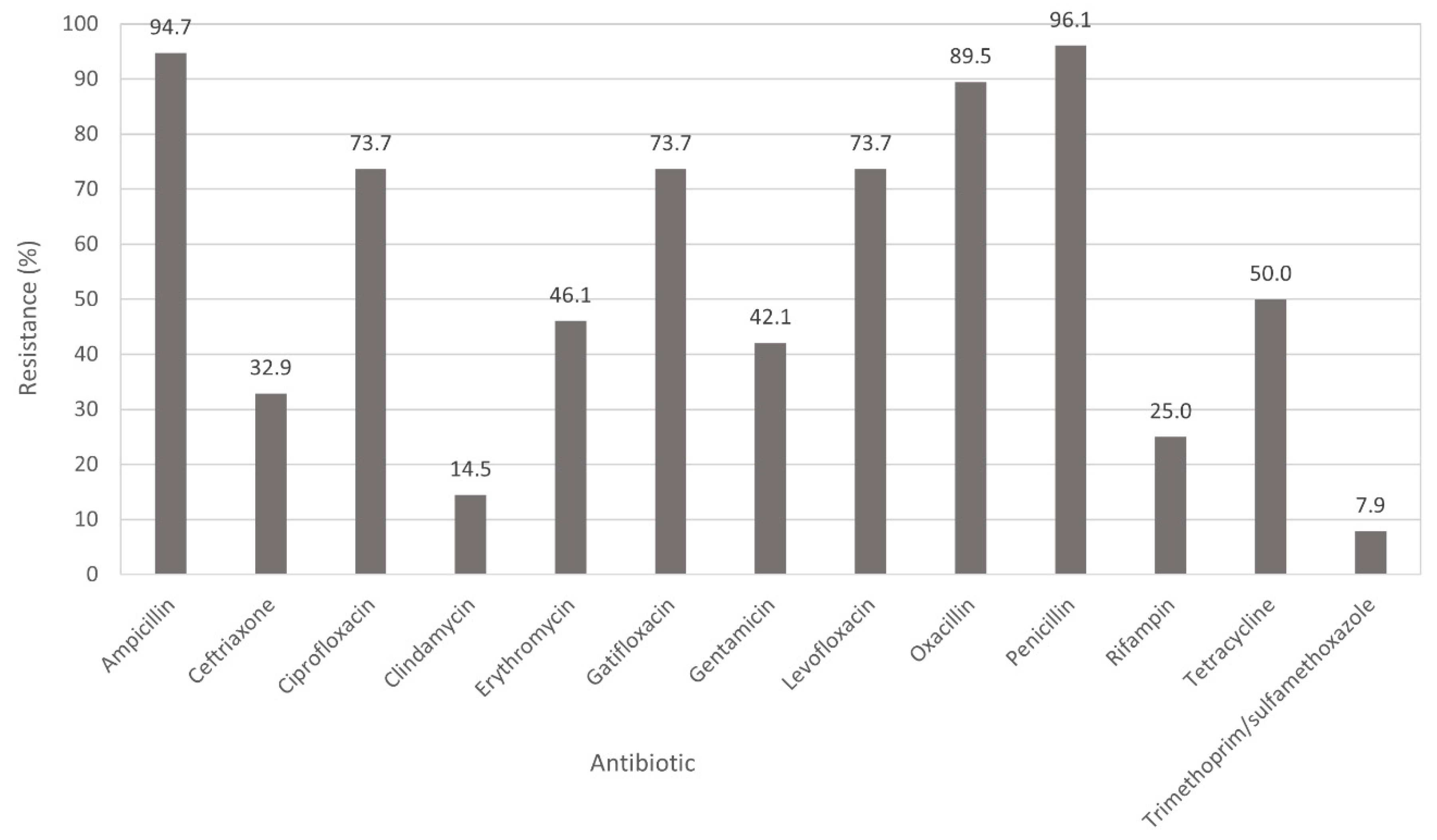
3.2. Antimicrobial Susceptibility Testing
3.3. Frequency of Antibiotic Resistance Genes
3.4. Molecular Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gould, D.; Chamberlaine, A. Staphylococcus aureus: A review of the literature. J. Clin. Nurs. 1995, 4, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Williams, R.E.; Jevons, M.P.; Shooter, R.A.; Hunter, C.J.; Girling, J.A.; Griffiths, J.D.; Taylor, G.W. Nasal staphylococci and sepsis in hospital patients. Br. Med. J. 1959, 2, 658–662. [Google Scholar] [CrossRef] [PubMed]
- Omuse, G.; Van Zyl, K.N.; Hoek, K.; Abdulgader, S.; Kariuki, S.; Whitelaw, A.; Revathi, G. Molecular characterization of Staphylococcus aureus isolates from various healthcare institutions in Nairobi, Kenya: A cross sectional study. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 51. [Google Scholar] [CrossRef]
- Choudhury, R.; Panda, S.; Singh, D.V. Emergence and dissemination of antibiotic resistance: A global problem. Indian J. Med. Microbiol. 2012, 30, 384–390. [Google Scholar] [CrossRef]
- Sergelidis, D.; Angelidis, A.S. Methicillin-resistant Staphylococcus aureus: A controversial food-borne pathogen. Lett. Appl. Microbiol. 2017, 64, 409–418. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar]
- Schentag, J.J.; Hyatt, J.M.; Carr, J.R.; Paladino, J.A.; Birmingham, M.C.; Zimmer, G.S.; Cumbo, T.J. Genesis of methicillin-resistant Staphylococcus aureus (MRSA), how treatment of MRSA infections has selected for vancomycin-resistant Enterococcus faecium, and the importance of antibiotic management and infection control. Clin. Infect. Dis. 1998, 26, 1204–1214. [Google Scholar] [CrossRef]
- Cosgrove, S.E.; Qi, Y.; Kaye, K.S.; Harbarth, S.; Karchmer, A.W.; Carmeli, Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: Mortality, length of stay, and hospital charges. Infect. Control. Hosp. Epidemiol. 2005, 26, 166–174. [Google Scholar] [CrossRef]
- DeLeo, F.R.; Otto, M.; Kreiswirth, B.N.; Chambers, H.F. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 2010, 375, 1557–1568. [Google Scholar] [CrossRef]
- Stapleton, P.D.; Taylor, P.W. Methicillin resistance in Staphylococcus aureus: Mechanisms and modulation. Sci. Prog. 2002, 85 Pt 1, 57–72. [Google Scholar] [CrossRef]
- Rukan, M.; Jamil, H.; Bokhari, H.A.; Khattak, A.A.; Khan, A.N.; Ullah, Z.; Hussain, S.; Syed, M.A. Nasal carriage of highly resistant methicillin resistant Staphylococcus aureus (MRSA) strains by hospital staff in Hazara region of Pakistan. J. Pak. Med. Assoc. 2021, 71, 47–50. [Google Scholar] [PubMed]
- Khan, A.A.; Ali, A.; Tharmalingam, N.; Mylonakis, E.; Zahra, R. First report of mecC gene in clinical methicillin resistant S. aureus (MRSA) from tertiary care hospital Islamabad, Pakistan. J. Infect. Public Health 2020, 13, 1501–1507. [Google Scholar] [CrossRef]
- Taj, Y.; Abdullah, F.E.; Kazmi, S.U. Current pattern of antibiotic resistance in Staphylococcus aureus clinical isolates and the emergence of vancomycin resistance. J. Coll. Physicians Surg. Pak. 2010, 20, 728–732. [Google Scholar]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, S.Z.; Ahmed, S. Prevalence of meticillin resistance among Staphylococcus aureus isolates in Pakistan and its clinical outcome. J. Hosp. Infect. 2007, 67, 101–102. [Google Scholar] [CrossRef]
- Jamil, B.; Gawlik, D.; Syed, M.A.; Shah, A.A.; Abbasi, S.A.; Muller, E.; Reissig, A.; Ehricht, R.; Monecke, S. Hospital-acquired methicillin-resistant Staphylococcus aureus (MRSA) from Pakistan: Molecular characterisation by microarray technology. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Madzgalla, S.; Syed, M.A.; Khan, M.A.; Rehman, S.S.; Muller, E.; Reissig, A.; Ehricht, R.; Monecke, S. Molecular characterization of Staphylococcus aureus isolates causing skin and soft tissue infections in patients from Malakand, Pakistan. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1541–1547. [Google Scholar] [CrossRef]
- Poulsen, A.B.; Skov, R.; Pallesen, L.V. Detection of methicillin resistance in coagulase-negative staphylococci and in staphylococci directly from simulated blood cultures using the EVIGENE MRSA Detection Kit. J. Antimicrob. Chemother. 2003, 51, 419–421. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Volume CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Seventeenth Informational Supplement, 28th ed.; Volume CLSI Supplement M100-S17 No. 1; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2007. [Google Scholar]
- Strommenger, B.; Kettlitz, C.; Werner, G.; Witte, W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 4089–4094. [Google Scholar] [CrossRef]
- McDougal, L.K.; Steward, C.D.; Killgore, G.E.; Chaitram, J.M.; McAllister, S.K.; Tenover, F.C. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J. Clin. Microbiol. 2003, 41, 5113–5120. [Google Scholar] [CrossRef]
- Oliveira, D.C.; de Lencastre, H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents. Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Strommenger, B.; Braulke, C.; Heuck, D.; Schmidt, C.; Pasemann, B.; Nubel, U.; Witte, W. spa Typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J. Clin. Microbiol. 2008, 46, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, D.; Claus, H.; Witte, W.; Rothganger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Qasim, M.; Rahman, H.; Khan, J.; Haroon, M.; Muhammad, N.; Khan, A.; Muhammad, N. High frequency of methicillin-resistant Staphylococcus aureus in Peshawar Region of Pakistan. Springerplus 2016, 5, 600. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, T.; Muhammad, I.N.; Khan, M.N.; Naz, S.; Bashir, L.; Sarosh, N.; Masood, R.; Ali, A.; Fatima, S.; Naqvi, T. MRSA: Prevalence and susceptibility pattern in health care setups of Karachi. Pak. J. Pharm. Sci. 2017, 30 (Suppl. S6), 2417–2421. [Google Scholar]
- Atif, M.; Azeem, M.; Saqib, A.; Scahill, S. Investigation of antimicrobial use at a tertiary care hospital in Southern Punjab, Pakistan using WHO methodology. Antimicrob. Resist. Infect. Control. 2017, 6, 41. [Google Scholar] [CrossRef]
- Hanif, A.; Ashar, S.M.; Rabnawaz, R.; Yasmeen, S. Self-medication of antibiotics among the students of Hamdard University, Pakistan. J. Public Health Dev. Ctries 2016, 2, 145–148. [Google Scholar]
- Asghar, S.; Atif, M.; Mushtaq, I.; Malik, I.; Hayat, K.; Babar, Z.U. Factors associated with inappropriate dispensing of antibiotics among non-pharmacist pharmacy workers. Res. Social Adm Pharm 2020, 16, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Hakim, S.T.; Arshed, S.; Iqbal, M.; Javaid, S. Vancomycin sensitivity of Staphylococcus aureus isolates from hospital patients in Karachi, Pakistan. Libyan J. Med. 2007, 2, 176–179. [Google Scholar] [CrossRef]
- Brohi, N.A.; Noor, A.A. Frequency of the occurrence of Methicillin-resistant Staphylococcus aureus (MRSA) infections in Hyderabad, Pakistan. Pak. J. Anal. Environ. Chem. 2017, 18, 84–90. [Google Scholar] [CrossRef]
- Bukhari, S.Z.; Ahmed, S.; Zia, N. Antimicrobial susceptibility pattern of Staphylococcus aureus on clinical isolates and efficacy of laboratory tests to diagnose MRSA: A multi-centre study. J. Ayub Med. Coll. Abbottabad 2011, 23, 139–142. [Google Scholar]
- Khan, Z.; Ahmed, N.; Rehman, A.; Khan, F.U.; Karatas, Y. Utilization pattern of antibiotics and patient care indicators in the teaching hospitals, Islamabad, Pakistan. SN Comp. Clin. Med. 2019, 1, 812–816. [Google Scholar] [CrossRef]
- Atif, M.; Azeem, M.; Sarwar, M.R.; Shahid, S.; Javaid, S.; Ikram, H.; Baig, U.; Scahill, S. WHO/INRUD prescribing indicators and prescribing trends of antibiotics in the Accident and Emergency Department of Bahawal Victoria Hospital, Pakistan. Springerplus 2016, 5, 1928. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Shehzad, A.; Shehzad, O.; Al-Suhaimi, E.A. Inpatient antibiotics pharmacology and physiological use in Hayatabad medical complex, Pakistan. Int. J. Physiol. Pathophysiol. Pharmacol. 2013, 5, 120–127. [Google Scholar] [PubMed]
- Syed, M.A.; Shah, S.H.H.; Sherafzal, Y.; Shafi-Ur-Rehman, S.; Khan, M.A.; Barrett, J.B.; Woodley, T.A.; Jamil, B.; Abbasi, S.A.; Jackson, C.R. Detection and molecular characterization of methicillin-resistant Staphylococcus aureus from table eggs in Haripur, Pakistan. Foodborne Pathog. Dis. 2018, 15, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Arfat, Y.; Johnson, M.; Malik, S.A.; Morrisey, J.A.; Bayliss, C.D. Epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) isolates from Pakistan. African J. Microbiol. Res. 2013, 7, 568–576. [Google Scholar]
- Brennan, G.I.; Shore, A.C.; Corcoran, S.; Tecklenborg, S.; Coleman, D.C.; O’Connell, B. Emergence of hospital- and community-associated panton-valentine leukocidin-positive methicillin-resistant Staphylococcus aureus genotype ST772-MRSA-V in Ireland and detailed investigation of an ST772-MRSA-V cluster in a neonatal intensive care unit. J. Clin. Microbiol. 2012, 50, 841–847. [Google Scholar] [CrossRef]
- Rajan, V.; Schoenfelder, S.M.; Ziebuhr, W.; Gopal, S. Genotyping of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) in a tertiary care centre in Mysore, South India: ST2371-SCCmec IV emerges as the major clone. Infect. Genet. Evol. 2015, 34, 230–235. [Google Scholar] [CrossRef]
- Prabhakara, S.; Khedkar, S.; Loganathan, R.M.; Chandana, S.; Gowda, M.; Arakere, G.; Seshasayee, A.S. Draft genome sequence of Staphylococcus aureus 118 (ST772), a major disease clone from India. J. Bacteriol. 2012, 194, 3727–3728. [Google Scholar] [CrossRef]
- D’Souza, N.; Rodrigues, C.; Mehta, A. Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence type (ST) 22 and ST 772 in Mumbai, India. J. Clin. Microbiol. 2010, 48, 1806–1811. [Google Scholar] [CrossRef]
- Ho, W.Y.; Choo, Q.C.; Chew, C.H. Predominance of three closely related methicillin-resistant Staphylococcus aureus clones carrying a unique ccrC-positive SCCmec type III and the emergence of spa t304 and t690 SCCmec type IV pvl(+) MRSA isolates in Kinta Valley, Malaysia. Microb. Drug. Resist. 2017, 23, 215–223. [Google Scholar] [CrossRef]
- Tang, Y.T.; Cao, R.; Xiao, N.; Li, Z.S.; Wang, R.; Zou, J.M.; Pei, J. Molecular epidemiology and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates in Xiangyang, China. J. Glob. Antimicrob. Resist. 2018, 12, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Di Gregorio, S.; Haim, M.S.; Vielma Vallenilla, J.; Cohen, V.; Rago, L.; Gulone, L.; Aanensen, D.M.; Argimon, S.; Mollerach, M. Genomic epidemiology of CC30 methicillin-resistant Staphylococcus aureus strains from Argentina reveals four major clades with distinctive genetic features. mSphere 2021, 6, e01297-20. [Google Scholar] [CrossRef]
- Ishihara, K.; Saito, M.; Shimokubo, N.; Muramatsu, Y.; Maetani, S.; Tamura, Y. Methicillin-resistant Staphylococcus aureus carriage among veterinary staff and dogs in private veterinary clinics in Hokkaido, Japan. Microbiol. Immunol. 2014, 58, 149–154. [Google Scholar] [CrossRef]
- Monecke, S.; Syed, M.A.; Khan, M.A.; Ahmed, S.; Tabassum, S.; Gawlik, D.; Muller, E.; Reissig, A.; Braun, S.D.; Ehricht, R. Genotyping of methicillin-resistant Staphylococcus aureus from sepsis patients in Pakistan and detection of antibodies against staphylococcal virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Deurenberg, R.H.; Vink, C.; Kalenic, S.; Friedrich, A.W.; Bruggeman, C.A.; Stobberingh, E.E. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2007, 13, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Shabir, S.; Hardy, K.J.; Abbasi, W.S.; McMurray, C.L.; Malik, S.A.; Wattal, C.; Hawkey, P.M. Epidemiological typing of meticillin-resistant Staphylococcus aureus isolates from Pakistan and India. J. Med. Microbiol. 2010, 59 Pt 3, 330–337. [Google Scholar] [CrossRef][Green Version]


| No. Isolates | No. Antibiotic Classes | ||
|---|---|---|---|
| Resistance Pattern a | MRSA | MSSA | |
| AmpCefCipCliEryGatGenLevOxaPenRifTetTri | 1 | 0 | 7 |
| AmpCefCipCliEryGatGenLevOxaPenRifTet | 8 | 1 | 6 |
| AmpCefCipEryGatGenLevOxaPenTetTri | 0 | 1 | 6 |
| AmpCefCipEryGatGenOxaPenRifTet | 8 | 1 | 6 |
| AmpCefCipEryGatLevOxaPenTet | 1 | 0 | 4 |
| AmpCefCipEryGatLevOxaPen | 1 | 0 | 3 |
| AmpCefCipGatGenLevOxaPen | 2 | 0 | 3 |
| AmpCefCipGatLevOxaPen | 1 | 0 | 2 |
| AmpCipCliEryGatGenLevOxaPen | 1 | 0 | 4 |
| AmpCipEryGatGenLevOxaPen | 0 | 1 | 4 |
| AmpCipEryGatLevOxaPen | 5 | 0 | 3 |
| AmpCipEryGatLevOxaPenTet | 3 | 0 | 4 |
| AmpCipEryGatLevPenTet | 0 | 2 | 4 |
| AmpCipGatGenLevOxaPen | 6 | 0 | 3 |
| AmpCipGatGenLevOxaPenTet | 1 | 0 | 4 |
| AmpCipGatGenLevOxaPenTri | 1 | 0 | 4 |
| AmpCipGatLevOxaPenTet | 2 | 0 | 3 |
| AmpCipGatLevOxaPen | 7 | 0 | 2 |
| AmpEryOxaPen | 1 | 0 | 2 |
| AmpOxaPen | 8 | 0 | 1 |
| AmpOxaPenTetTri | 1 | 0 | 3 |
| AmpOxaPenTet | 4 | 0 | 2 |
| AmpPenTet | 2 | 1 | 2 |
| AmpPen | 1 | 0 | 1 |
| CipGatLevOxaPen | 1 | 0 | 2 |
| CipGatLevOxa | 0 | 1 | 2 |
| Pan-susceptible | 1 | 1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syed, M.A.; Jamil, B.; Ramadan, H.; Rukan, M.; Ali, S.; Abbasi, S.A.; Woodley, T.A.; Jackson, C.R. Genetic Diversity of Staphylococcus aureus Strains from a Tertiary Care Hospital in Rawalpindi, Pakistan. Microorganisms 2021, 9, 2301. https://doi.org/10.3390/microorganisms9112301
Syed MA, Jamil B, Ramadan H, Rukan M, Ali S, Abbasi SA, Woodley TA, Jackson CR. Genetic Diversity of Staphylococcus aureus Strains from a Tertiary Care Hospital in Rawalpindi, Pakistan. Microorganisms. 2021; 9(11):2301. https://doi.org/10.3390/microorganisms9112301
Chicago/Turabian StyleSyed, Muhammad Ali, Bushra Jamil, Hazem Ramadan, Maria Rukan, Shahzad Ali, Shahid Ahmad Abbasi, Tiffanie A. Woodley, and Charlene R. Jackson. 2021. "Genetic Diversity of Staphylococcus aureus Strains from a Tertiary Care Hospital in Rawalpindi, Pakistan" Microorganisms 9, no. 11: 2301. https://doi.org/10.3390/microorganisms9112301
APA StyleSyed, M. A., Jamil, B., Ramadan, H., Rukan, M., Ali, S., Abbasi, S. A., Woodley, T. A., & Jackson, C. R. (2021). Genetic Diversity of Staphylococcus aureus Strains from a Tertiary Care Hospital in Rawalpindi, Pakistan. Microorganisms, 9(11), 2301. https://doi.org/10.3390/microorganisms9112301







