Abstract
Staphylococcus xylosus forms biofilm embedded in an extracellular polymeric matrix. As extracellular DNA (eDNA) resulting from cell lysis has been found in several staphylococcal biofilms, we investigated S. xylosus biofilm in vitro by a microscopic approach and identified the mechanisms involved in cell lysis by a transcriptomic approach. Confocal laser scanning microscopy (CLSM) analyses of the biofilms, together with DNA staining and DNase treatment, revealed that eDNA constituted an important component of the matrix. This eDNA resulted from cell lysis by two mechanisms, overexpression of phage-related genes and of cidABC encoding a holin protein that is an effector of murein hydrolase activity. This lysis might furnish nutrients for the remaining cells as highlighted by genes overexpressed in nucleotide salvage, in amino sugar catabolism and in inorganic ion transports. Several genes involved in DNA/RNA repair and genes encoding proteases and chaperones involved in protein turnover were up-regulated. Furthermore, S. xylosus perceived osmotic and oxidative stresses and responded by up-regulating genes involved in osmoprotectant synthesis and in detoxification. This study provides new insight into the physiology of S. xylosus in biofilm.
1. Introduction
Staphylococcus xylosus is a commensal species of the epithelium and mucous membranes of warm-blooded animals. It is frequently isolated from the skin of farm animals [1,2], hence its prevalence in foods of animal origin such as milk and milk products and fermented meat products [3,4]. Furthermore, S. xylosus colonizes the manufacturing environment of dry fermented sausage plants in relation to its ability to form biofilm [4,5]. Indeed, S. xylosus forms multilayered biofilm where cells are embedded in an extracellular polymeric matrix [5], which is a common trait in bacterial biofilms [6,7,8]. In Staphylococcus epidermidis and Staphylococcus aureus, one component of the matrix is designated as poly-N-acetylglucosamine polysaccharide (PNAG) whose production is governed by the ica operon [9,10]. In S. xylosus, however, exopolymer biosynthesis appears to be ica-independent and the composition of the polymer remains unknown [5,11]. Other surface components have been described as required for biofilm cohesion in S. aureus and S. epidermidis, namely, teichoic acids, the accumulation associated protein (Aap), and the biofilm-associated protein (Bap) [6,12,13,14,15]. In addition, the observation that certain strains of S. epidermidis and S. aureus form ica-independent biofilms has shown that extracellular DNA (eDNA) can serve as a natural glue connecting neighboring cells to each other [9,16,17,18]. In fact, eDNA is a component of the biofilm matrix of many bacterial species [19,20,21].
eDNA released from cells could result from autolysis. In S. epidermidis and S. aureus, eDNA in biofilm was released through the activity of the prominent murein hydrolase Atl [22,23]. In these two species, a two-component regulatory system LytSR affects murein hydrolase activity and autolysis [24]. LytSR regulates the expression of the lrgAB operon, which, together with the cidABC operon, has been shown to be a regulator in the control of cell death and lysis during biofilm development [25,26]. The cidA gene encodes a putative holin protein that is an effector of murein hydrolase activity and cell lysis [27], while lrgA encodes a putative antiholin that is an inhibitor of these processes [25]. Furthermore, in S. aureus CidR enhances cidABC, lrgAB, and alsSD (encoding proteins involved in acetoin production) expression in response to carbohydrate metabolism [28].
Other known mechanisms of eDNA release include phage-mediated cell death. Phage release has been observed in biofilms of both Gram-negative and Gram-positive bacteria [29,30,31]. In Pseudomonas aeruginosa, the phage Pf4 mediates the formation of small-colony variants in biofilms [29]. Lysogenic S. aureus cells in planktonic and biofilm cultures release phages into their surroundings; two morphologically distinct phages are observed [31]. The resulting lysis of cells in biofilm might promote the persistence of the remaining cells by furnishing nutrients.
In S. xylosus, genes encoding the holin/antiholin system and genes encoding phage proteins are highly overexpressed during growth in a meat model [32]. Moreover, a phage capsid protein is overexpressed in S. xylosus cultivated in biofilm compared to planktonic culture [11]. These results highlight that cell lysis could result in the release of DNA. Thus, the aim of this study was first to check if eDNA is released during biofilm formation by a microscopic approach and then to identify the mechanisms involved in cell lysis by a transcriptomic approach.
2. Materials and Methods
2.1. Bacterial Strain and Biofilm Formation
S. xylosus strain C2a expressing cyan fluorescent (C2a-B2) was used [33]. This strain contains the erythromycin-resistance plasmid pJEBAN2. The strain C2a-B2 was pre-cultivated in Brain Heart Infusion (BHI, Becton, Dickinson and Company, Le Pont de Claix, France) with 10 µg/mL erythromycin at 30 °C, with stirring at 170 rpm for 24 h. Bacterial concentration was then measured by determining the optical density at 600 nm (OD600) and appropriate dilution was prepared. The strain was then inoculated at 106 CFU/mL in BHI in the Lab-Tek chamber slide system (1 chamber borosilicate cover glass system, NUNC 15536, 8.6 cm2) and incubated at 30 °C for 9, 24 and 48 h in a humid chamber. After incubation, the supernatant in the Lab-Tek chambers was removed, the adhered cells were washed twice with tryptone salt and then were detached by scratching in tryptone salt. The bacterial population of biofilm after 9, 24 and 48 h were determined by 10-fold serial dilutions on BHI agar plates and enumerated after 24-h culture at 30 °C. In parallel, the cells detached by scratching in tryptone salt were pelleted by centrifugation for 2 min at 4500× g and at 4 °C and the pellet was immediately frozen in liquid nitrogen and stored at −80 °C before extraction of RNA. Three independent experiments were performed.
2.2. Confocal Laser Scanning Microscopy Analyses of the Biofilms
S. xylosus C2a-B2 biofilms in the Lab-Tek chamber slide system were analyzed by confocal laser scanning microscopy (CLSM) at the three times of incubation: 9, 24 and 48 h. To test for the presence of eDNA in the biofilm, DNase was used under two conditions. First, after incubation at 9, 24 and 48 h and removal of the supernatants, the biofilms were overlaid with DNase solution (100 U, Roche, Mannheim, Germany) for 15 min. Second, DNase (25 U, Roche) was added at the same time as inoculation of the biofilm and the incubation lasted 9, 24 and 48 h. eDNA was stained with 1 µM TOTO-3-iodide (Thermo Fisher Scientific, Molecular probes, IllKirch-Graffenstaden, France) at all incubation times and in all conditions (treated or not with DNase).
After incubation of the biofilms treated or not by DNase, the supernatants in the Lab-Tek chambers were removed and the adhered cells, after washing, were treated with CitiFluor™ (75% AF1 + 25% AF3, UKC Chem. Lab Canterbury, UK) as previously described [33]. The biofilms were observed with a LEICA SP5 CLSM (Leica Microsystems, Nanterre, France, objective x63) at λex = 458 nm to observe the fluorescence of the strain C2a-B2 and at λex = 633 nm to observe DNA stained by TOTO-3. Horizontal cross-sections were acquired consecutively along the z-axis using a scanning step size of 1 μm, defining the so-called stacks, to cover the whole biofilm height of one randomly chosen (x, y) coordinate as described [33]. Images were acquired for three biofilm replicates per sampling time. Three-dimensional projections of biofilm structure were reconstructed using the Easy 3D function of the IMARIS 7.0 software (Bitplane, Zurich, Switzerland). Quantitative structural parameters of the biofilms, such as biovolume, substratum coverage, roughness and thickness were calculated using the software COMSTAT 1 [34] under MATLAB.
2.3. Phage Induction and Transmission Electron Microscopy
S. xylosus C2a-B2 strain was grown for 6 h in BHI up to OD600: 0.3–0.4 and was then treated with mitomycin C (2 μg/mL, Sigma-Aldrich, Saint-Quentin-Fallavier, France) to induce prophages. The culture was incubated at 37 °C and 70 rpm for 6 h. Cells were harvested by centrifugation for 10 min at 4500× g. The resulting supernatant was filtered through a 0.45 µm membrane and was centrifuged for 30 min at 25,000× g and 4 °C. The pellet was used for transmission electron microscopy. The phages were pre-fixed in 0.5% (w/v) glutaraldehyde, stained with 0.5% (w/v) uranyl acetate, and examined with a Hitachi H-7650 at a magnification of 120,000-fold.
2.4. RNA Extraction, Labeling and Microarray Analyses
For RNA extraction from S. xylosus biofilms after 9, 24 or 48 h of incubation, cell pellets were thawed on ice and resuspended in 500 μL of ice-cold Tris-EDTA buffer. Samples were transferred to tubes containing 600 mg of zirconia-silica beads (0.1 mm diameter, BioSpec Products, Bartlesville, OK, USA), 500 μL of acid phenol, 50 µL of sodium dodecyl sulfate (10%) and 3.5 μL of β-Mercaptoethanol. Cells were disrupted using a FastPrep® (MP Biomedicals, Illkirch-Graffenstaden, France). After the addition of 200 μL of chloroform and centrifugation, the aqueous phase containing RNA was collected and purified with the Nucleospin RNA II kit (Macherey Nagel, Hoerdt, France) according to the manufacturer’s instructions. A supplementary treatment was performed with Turbo DNase (Ambion, Austin, TX, USA) to remove residual DNA. The absence of DNA contamination was checked by PCR targeting the tuf gene. Total RNA was quantified using a NanoDrop 1000 and RNA quality was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. RNA samples were stored at −80 °C. They were reverse-transcribed to cDNA and labeled as previously described [35]. A complete description of the array developed for S. xylosus C2a is available at the NCBI Gene Expression Omnibus (GEO) database under platform accession number GPL19201 and the complete genome of S. xylosus C2a is available under accession number LN554884. Significant differences in the probe set intensities between the different conditions were identified using a linear model with an empirical Bayes method using all information probes to moderate the standard errors of the estimated log-fold changes [36]. The probabilities were corrected by the Benjamini–Hochberg procedure to control the false-discovery rate (FDR) with a p value cut-off of 0.05. All probes with an FDR ≤ 0.05 are considered to be differentially expressed. Finally, a gene was considered to be differentially expressed if more than 50% of the corresponding probes were differentially expressed and if the ratio of expression was ≥2 or ≤0.5.
The targeted genes for qPCR and primer sequences used to validate microarray data are listed in Supplementary Table S1. The analyses were performed on the same samples of RNA as used for the microarray experiments. The relative fold change of gene expression, using measured tuf housekeeping gene expression, was determined by the 2−ΔΔCt method [37].
3. Results and Discussion
3.1. Evidence of Extracellular DNA by CLSM Analyses
After 9 h, the population in biofilm was 7.2 ± 0.3 log CFU/cm2, a small increase to 7.9 ± 0.2 log CFU/cm2 was observed at 24 h and then the population remained stable at 8.2 ± 0.5 log CFU/cm2 at 48 h.
CLSM coupled with cell-impermeant DNA binding fluorescent stain such as TOTO-3 is a powerful tool to study biofilm matrices as it allows real-time visualization of living microorganisms [38]. Using this approach, we observed the architecture of the S. xylosus biofilm by recording the fluorescence of the labeled strain S. xylosus C2a-B2 (green) and the stained eDNA by TOTO-3 (red) during the 48 h of incubation (Figure 1A). The structural parameters, biovolume, average thickness, roughness and coverage were extracted from confocal images and are represented in Figure 2. S. xylosus-B2 produced a rough biofilm composed of aggregates covering 37% of the surface with a maximum thickness of 10.8 µm and an average one of 4.4 µm at 9 h of incubation. Then, a flat thick biofilm was observed from 24 h of incubation up to 48 h. The biovolume and the average thickness of the biofilm increased strongly after 9 h of incubation, while the roughness decreased (Figure 2, C2a control). An increasing amount of eDNA was measured in the S. xylosus biofilm during the incubation period, this increase being particularly noticeable between 24 and 48 h, as evidenced by high values of biovolume, thickness and coverage (Figure 1A and Figure 2, eDNA control). Such release of eDNA has been seen in numerous bacteria grown in biofilms such as S. aureus, S. epidermidis, Listeria monocytogenes, Pseudomonas aeruginosa, Bacillus cereus, and Burkholderia pseudomallei [18,19,20,21,39,40,41]. CLSM images revealed eDNA particularly at the base of the biofilm of S. xylosus as indicated by the red fluorescence on the bottom while the upper layer displayed green cells (Figure 1A). For B. pseudomallei, eDNA was also visualized essentially at the base of the biofilm [41]. For the latter species, as well as for S. aureus, eDNA was observed at the beginning of biofilm formation facilitating their adhesion to the support [16,41]. In our study, the first time of observation was 9 h and already a significant amount of eDNA was observed in the S. xylosus biofilm (Figure 1A). A previous study has shown that eDNA promoted adhesion of S. xylosus cells to hydrophobic surfaces [42].
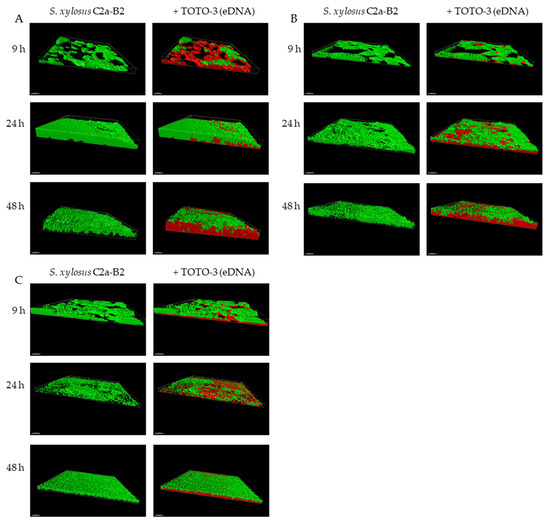
Figure 1.
3D projection of biofilms of S. xylosus C2a-B2 and DNA stained by TOTO-3 (A), after DNase treatment at the end of each incubation time (B), after incubation in the presence of DNase from the start (C), at three sampling times using Imaris software; Scale bar—30 µm.
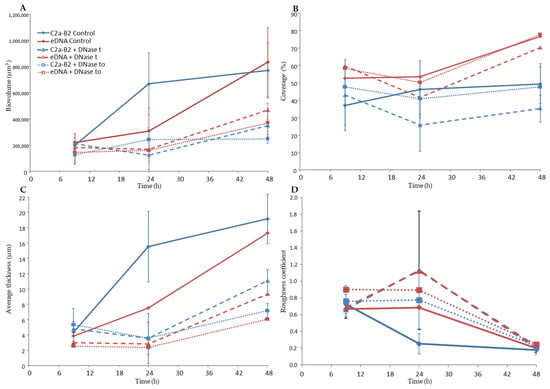
Figure 2.
Quantitative structural parameters of the biofilms of S. xylosus C2a-B2 and eDNA stained by TOTO-3 at three sampling times. (A) Biovolume, (B) Coverage, (C) Average thickness and (D) roughness coefficient. Biovolume C (µm3) represented the overall volume of cells in the observation field. Coverage (%) reflected the efficiency of substratum colonization by bacteria. The average thickness (μm) of biofilms was determined from the confocal stack images. Roughness coefficient provided a measure of variations in biofilm thickness and was an indicator of the superficial biofilm interface heterogeneity. Three independent experiments were performed per sampling time. C2a-B2 control (continuous blue curve) and eDNA control (continuous red curve): no treatment; C2a-B2 + DNase t (dashed blue curve) and eDNA + DNase t (dashed red curve): DNase treatment at the end of each incubation time; C2a-B2 + DNase to (dotted blue curve) and eDNA + DNase to (dotted red curve): after incubation in the presence of DNase from the start.
CLSM images revealed that DNase treatments of S. xylosus biofilm decreased eDNA, biovolume and average thickness of the biofilms during the incubation time (Figure 1B,C and, DNase t, DNase t0). The heterogeneity of the biofilms was increased in the presence of DNase as shown by higher roughness coefficient at 24 h of incubation. The use of DNase for biofilm removal is effective, but dependent on the age of the biofilm [21]. Under our conditions, a greater efficiency of DNase was observed on the biofilm of S. xylosus at 9 h compared to the following times of 24 and 48 h (Figure 1B,C). Similarly, young biofilms were easily removed, but DNase treatment was less effective on mature biofilm of S. epidermidis, P. aeruginosa and B. pseudomallei [19,22,41]. The addition of DNase at the start of the experiment did not inhibit biofilm formation by S. xylosus, but affected its biovolume and average thickness during incubation (Figure 1C and Figure 2, DNase t0). In fact, this treatment at t0 was more effective than that carried out at 48 h, in particular on the thickness of the biofilm at 48 h (Figure 2). DNase inhibited S. aureus biofilm formation, detached preformed biofilm and rendered preformed biofilm sensitive to detergent killing [9]. The release of DNA depends on the growing conditions. Under our conditions, the release of DNA by S. xylosus was observed during growth in BHI, a nutritionally rich medium. For L. monocytogenes, the DNase-sensitive biofilm was only observed in diluted medium with low ionic strength [40]. While for S. epidermidis, DNA release has been evidenced in the biofilm formed in whole blood-derived platelet concentrates [18].
3.2. Cell Lysis
A transcriptomic approach was used to determine the mechanism involved in the release of DNA. It revealed that 833 (338 down- and 495 up-regulated) genes were differentially expressed at 24 and 48 h, by comparison with 9 h used as reference (Supplementary Table S2). Notably, 458 genes were differentially expressed at the two times of incubation. These genes were classified into different functional categories: the most represented being metabolism (30%) followed by information storage and processing (10%), cellular processes (8%) and phage (7%). This analysis was validated by qPCR on selected genes and a good correlation was noted for the two times of incubation (24 h: r2 = 0.91, y = 1.205x − 0.0656; 48 h: r2 = 0.93, y = 1.1209x − 0.1745) (Supplementary Table S1).
The transcriptomic data revealed the overexpression of two mechanisms that could be involved in cell lysis, one implicating prophage genes and the other the cidABC genes (Table 1). Many prophage genes located in two distinct chromosomal loci were overexpressed (Table 1). The first region of about 15 kb (SXYL_01051-66) is poorly characterized and includes an integrase encoded by SXYL-1051 and several potential tail and host lysis-related genes. The second prophage region of 39.4 kb from SXYL_01727-88 displays the typical organization of the Siphoviridae phage genome, with typical functional modules of lysogeny, DNA metabolism, DNA packaging and head, tail and host lysis-related genes [43]. Temperate Siphoviridae are the main prophages found in coagulase-negative Staphylococcus genomes [44]. The gene SXYL_01783 encodes the phage antirepressor protein. In Salmonella phage P22, the antirepressor overcomes the repressor protein involved in the maintenance of lysogeny [45]. In the S. xylosus biofilm at both 24 and 48 h incubation, the lysogeny could switch to the lytic cycle. To determine if lytic production of phage particles can be induced from S. xylosus C2a, a planktonic culture was treated with mitomycin C Observation by transmission electron microscopy seemed to reveal only one type of phage with a shaped head and a long thin tail, as Siphoviridae (Figure 3). A spontaneous release of phages from S. aureus biofilm cells has already been observed [31]. These phages were detected over a period of 48–72 h in biofilm cultures. Likewise, lysogenic pneumococcal strains are able to release phage particles during biofilm development by spontaneous induction of prophage and hence release DNA [46]. In Shewanella oneidensis, prophage-mediated lysis results in DNA release and enhanced biofilm formation [47]. In this bacterium, induction of the Lambda So prophage occurs in RecA-dependent manner, involving oxidative stress-induced DNA damage as the major trigger. Iron and, in a minor way, H2O2 are involved in this oxidative stress [48]. For S. xylosus, the mechanism that could induce the release of phage particles in the biofilm remains unknown, but we observed that genes involved in iron uptake and response to oxidative stress were up-regulated, as discussed in Section 3.9 and Section 3.10.

Table 1.
Genes of S. xylosus discussed in this study, overexpressed at 24 h and/or 48 h compared to 9 h in biofilm.
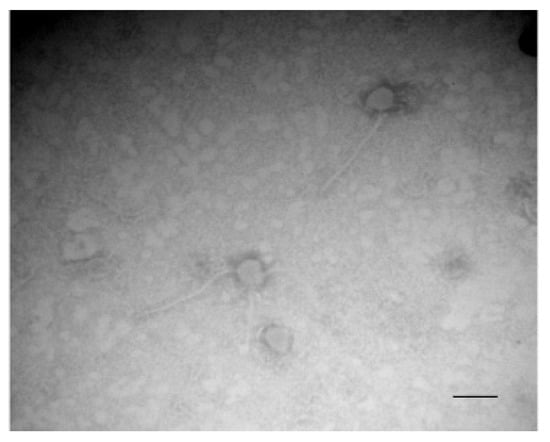
Figure 3.
Transmission electron microscopy of phages in the supernatant of S. xylosus after induction with mitomycin C. Bar, 100 nm. Magnification, ×120,000.
In addition to overexpression of genes encoding proteins from the two phage loci, the cidABC operon in sessile cells of S. xylosus was overexpressed particularly at 48 h (>10-fold) (Table 1). This operon has been widely studied in S. aureus [26]. CidA forms pores in the membrane allowing the murein hydrolase to reach and degrade peptidoglycan. CidB protein, like CidA, contains multiple predicted membrane-spanning domains, but its role is not yet established. CidC is a pyruvate oxidase that decarboxylates pyruvate to acetate. The cid operon plays a significant role during S. aureus biofilm development [27]. Thus, the biofilm produced by the S. aureus cidA mutant is more loosely compact and less adherent to the substrate. DNase treatment of this biofilm has a small effect, while it destabilizes the wild-type S. aureus biofilm revealing that DNA is released because of the cidA-mediated lysis of a subset of the bacterial population [27]. In our study, the overexpression of cidA from 24 h could have contributed to the lysis of S. xylosus and thus the release of DNA. Death and lysis in staphylococcal biofilms are under the control of a regulatory network such as CidR, which induces cidABC, lrgAB and alsSD transcription in response to the accumulation of intracellular pyruvate or acetate [24,26]. In our conditions, cidABC, as already mentioned, was overexpressed and lrgAB was not modulated. The alsSD operon is not present in the genome of S. xylosus, but ilvNB encoding an acetolactate synthase and budA encoding an acetolactate decarboxylase leading to acetoin were overexpressed (Table 1). In S. aureus biofilm, acetate derived from CidC activity potentiates cell death by a mechanism dependent on intracellular acidification and respiratory inhibition and AlsSD counters CidC by diverting carbon flux towards neutral rather than acidic byproducts, consuming protons in the process [49]. Based on our results, and as summarized in Figure 4 for S. xylosus, CidC, IlvNB, and BudA could modulate cell death to achieve optimal biofilm biomass. Lysis might be an advantage for the biofilm community because the remaining cells can gain nutrients from dead and lysed neighboring cells, as we will discuss in the section on metabolism.
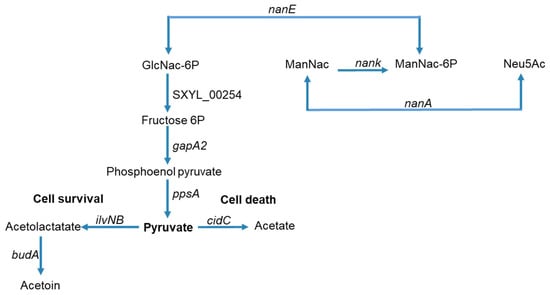
Figure 4.
Summary of carbohydrate and amino sugar catabolism in S. xylosus biofilm showing the up-regulated genes (the level of expression of these genes and the corresponding enzymes are given in Table 1). GlcNac-6P = N-acyl-d-glucosamine 6-phosphate, ManNac = N-acyl-d-mannosamine, ManNac-6P = N-acyl-d-mannosamine 6-phosphate, Neu5Ac = N-acetyl neuraminate.
3.3. Slow Cellular Process in the Mature Biofilm
The physiology of S. xylosus biofilm, like S. aureus and S. epidermis biofilms, is characterized by a general down-regulation of active cell processes such as protein, DNA, and cell wall syntheses that is typical of slow-growing cells [50]. However, several genes involved in the DNA machinery were up-regulated (Table 1). Thirteen genes encoding transcriptional regulators orchestrating gene activity and 12 genes involved in translation and ribosomal biogenesis were up-regulated. It is noteworthy that, walK was up-regulated (Table 1). It encodes a member of the two-component regulatory system WalRK. In S. aureus, WalRK positively controls biofilm formation [51] and is essential for cell viability mainly by controlling the transcription of cell wall lysing enzymes [52]. WalKR in S. aureus activated the transcription of nine genes involved in the different steps of cell wall turnover (lytM, atlA, isaA, sceD, ssaA, and four ssaA-related genes) [51]. In our study, only the isaA gene, and two sceD genes were up-regulated (Table 1).
3.4. DNA/RNA, Protein Repair Systems
It is worthy of mention that, 10 genes encoding proteins involved in DNA/RNA repair were up-regulated in S. xylosus in biofilm (Table 1). Among them, dinB encodes DNA polymerase IV, which is up-regulated during the SOS response to DNA damage in Escherichia coli and contributes to spontaneous mutation in slow-growing or non-growing cells [53]. The recF gene, which was up-regulated in our conditions, encodes RecF, which is required for DNA replication and for daughter strand gap repair [54]. This gene is also up-regulated in S. epidermidis in biofilm [55]. The gene tag encoding DNA-3-methyladenine glycosylase was up-regulated in S. xylosus; this enzyme is a base excision repair glycosylase that recognizes and excises a variety of alkylated bases from DNA [56]. The gene SXYL_01206 encoding the RadC family protein was up-regulated in S. xylosus. This protein is involved in DNA replication and repair [57]. The gene rnr encoding a ribonuclease R was also up-regulated. In E. coli, this RNase acts over a range of substrates, such as, ribosomal, transfer, messenger, and small non-coding RNAs [58]. The UvrABC repair system, encoded by uvrABC, which was overexpressed in our conditions, catalyzes the recognition and processing of DNA lesions. Endonuclease III is a ubiquitous DNA repair enzyme that repairs oxidized pyrimidine base lesions in DNA. It is encoded by the SXYL_02241 gene, which was up-regulated in our conditions.
Bacterial cells are equipped to adapt to various environmental conditions. They have developed a general response, like heat shock proteins, chaperones and ATP-dependent proteases, to deal with damaged proteins. In our study, 10 genes encoding proteases and chaperones involved in protein turnover were up-regulated (Table 1). S. xylosus overexpressed ctsR and the clpC, clpB, dnaK and groES genes. These genes are identified in S. aureus as belonging to the CtsR regulon [59]. Moreover, dnaK belongs to a cluster, which includes grpE, dnaJ and hcrA encoding a transcriptional regulator. All these genes were up-regulated in S. xylosus in biofilm. In S. aureus, the dnaK and groES operons also belong to the HrcA regulon embedded within the CtsR regulon, which controls HrcA synthesis [59]. A similar transcriptional regulation could occur in S. xylosus as revealed by the network of genes up-regulated under our conditions. The gene mecA up-regulated by S. xylosus encodes MecA, which enables the recognition and targeting of unfolded and aggregated proteins to the ClpC protease. Finally, the gene SXL_00505, up-regulated, encodes a YidC/Oxa1 family membrane protein insertase. These family members can function depending on the context as insertases, chaperones, and assembly factors for transmembrane proteins [60].
3.5. Pyrimidine and Purine Salvage
Uracil arising from cell lysis could be used as pyrimidine source. It can be taken up by the pyrP encoded permease, which is a part of the pyr operon including pyrCBPR, all of which was highly up-regulated at 48 h of incubation in S. xylosus (Table 1). PyrR can act as a phosphoribosyltransferase leading to UMP and is a regulatory protein of the pyr operon in Bacillus subtilis [61]. The genes pyrCB, carAB, and glnA2 genes, which were all up-regulated, could also contribute to the synthesis of UMP from glutamate/glutamine as already described for S. xylosus [62] (Table 1). As for S. xylosus, the genes encoding pyrimidine were strongly up-regulated in S. aureus in biofilm compared to planktonic cultures [63]. The nrdEF genes up-regulated in S. xylosus in biofilm could participate in the synthesis of pyrimidine deoxynucleotides. Three genes, including the repressor purR, which is involved in purine synthesis were up-regulated (Table 1). In B. subtilis, PurR regulated the 12-gene pur operon required for de novo synthesis of purine from the IMP pathway [61]. Such regulation could happen in S. xylosus; this operon is also down-regulated in S. aureus grown in biofilm [63].
3.6. Amino Sugar Catabolism
One of the main features of S. xylosus in sessile conditions was its potential to use amino sugars released from the cell wall of lysed bacteria as a carbon source. These amino sugars are catabolized via the amino sugar pathway, for which a cluster of four genes was up-regulated (nan, SXYL_00403-406, Table 1 and Figure 4). A complete nan system was defined as one that minimally includes orthologues of genes encoding NanA, NanE, and NanK [64]. This is the case with S. xylosus, which also comprises a transcriptional regulator of the RpiR family (Table 1). The nan systems of E. coli and S. aureus also include a regulator, albeit very different from each other and not characterized for S. aureus [64]. This nan pathway led to N-acetyl-glucosamine-6-phosphate, which could be further catabolized to pyruvate via several steps (Figure 4). In addition, in S. xylosus in biofilm, four genes involved in the pentose and glucuronate pathways were overexpressed (Table 1). Pyruvate could be catabolized by enzymes encoded by cidC, budA and ilvNB as described above (Figure 4) but all other genes involved in its catabolism and in the TCA cycle were down-regulated (Table S2). Finally, two cydBA genes encoding cytochrome oxidase, which generates a proton motive force, were overexpressed (Table 1). In E. coli, cytochrome bd oxidase was expressed under O2-limited conditions [65]. This suggests that S. xylosus embedded in an eDNA matrix as observed by microscopy seems to perceive an anaerobic environment as revealed by the transcriptomic data related to the catabolism of sugars.
3.7. Amino Acid Synthesis
Amino acids could be released during cell lysis. Indeed, two clusters of genes (SXYL_00264-66; SXYL_00661-65) coding for ABC type amino acid transporters and one gene encoding an ammonia permease were up-regulated in S. xylosus in biofilm, particularly at 48 h of incubation (Table 1). One of these clusters was involved in the transport of glutamate, a key component as the main nitrogen donor. In addition, glutamate could be synthesized by two pathways. One could involve alpha ketoglutarate and glutamine catabolized by glutamate synthase encoded by gltDB, which was overexpressed, together with gltC coding for a transcriptional activator (Table 1). In B. subtilis, the main regulatory role of GltC appears to be the prevention of a cycle of simultaneous glutamate synthesis and degradation [66]. The other pathway could involve the degradation of histidine with the overexpression of hutH and hutUI encoding histidine catabolic enzymes (Table 1). Then glutamate could be catabolized to glutamine-by-glutamine synthases encoded by glnA1, which was overexpressed only at 48 h of incubation, and glnA2, which was overexpressed at 24 and 48 h (Table 1). The glnA1 gene is in an operon with glnR encoding a repressor. In B. subtilis, GlnR represses the gln operon in the presence of glutamine. This repression is considered as a fine-tuning mechanism of gln expression [66]. Finally, glutamine could be involved in the synthesis of pyrimidine, as described in Section 3.5.
Almost all genes involved in the synthesis of branched-chain amino acids were up-regulated in S. xylosus in biofilm, especially at 48 h of incubation (cluster SXYl_00867-74, Table 1). Several genes involved in the synthesis of glycine, and of cysteine/methionine and alanine/lysine were up-regulated. Noteworthily, alr2 was highly overexpressed. It encodes an alanine racemase, which furnishes d-alanine for the synthesis of the cell wall. Surprisingly, the gene ldhD encoding a d-lactate dehydrogenase was highly up-regulated (>10-fold) both at 24 and 48 h. d-lactate instead of the usual d-alanine could be introduced in peptidoglycan, as already reported for Staphylococcus haemolyticus resistant to vancomycin [67].
3.8. Cofactor, Vitamin Synthesis
S. xylosus in biofilm overexpressed the cluster rib involved in riboflavin synthesis (Table 1). It also overexpressed a cluster of two genes encoding energy-coupling factor transporters (Table 1). These transporters mediate the uptake of essential vitamins and metal ions in many prokaryotes, in particular for those bacteria lacking the pathways for folate, biotin, and thiamine biosynthesis [68]. Biotin and thiamine are two vitamins essential for S. xylosus growth [69]. Seven other overexpressed genes are involved in the synthesis of porphyrin, cofactor and folate (Table 1).
3.9. Inorganic Ion Transport
In our conditions, three genes of the cluster pstSCAB involved in the import of inorganic phosphate were up-regulated at 48 h of incubation (Table 1). In S. aureus, the two-component system PhoPR is required for the expression of pstSCAB and is necessary for its growth under phosphate limiting conditions [70]. In our conditions, the genes encoding this system were down-regulated (Table S2). In S. xylosus biofilm, phosphate seems not to be limiting, as DNA was released in the growth medium.
Iron, an essential cofactor for several enzymes, is complexed with different proteins and its concentration is finely regulated. Fur (ferric uptake regulator) is involved in iron homeostasis and, is a repressor of three iron-acquiring systems (hts, sst, fhu) in S. xylosus [36,62]. In biofilm, fur was up-regulated, and consequently, these three systems were under-expressed (Table S2). Two clusters of genes, sitABC and SXYL_00561-63, were overexpressed at 24 and 48 h of incubation. The cluster sitABC, encoding an iron-regulated ABC transport involved in divalent metal uptake, was modulated in the presence of ferrous iron (FeSO4), while the cluster SXYL_00561-63 was highly up-regulated in the presence of ferritin in S. xylosus [71]. Note that, the gene dps encoding a Dps family protein was up-regulated. The crystal structure of Dps reveals structural homology with ferritins, a large family of iron storage proteins [72]. This led us to suppose that the cluster SXYL_00561-63 could be involved in the uptake of iron from Dps in S. xylosus in biofilm.
As mentioned for iron, all metal ions are required for biological reactions, but they are toxic in excess, and the intracellular availability of each is tightly regulated [73]. S. xylosus in biofilm up-regulated three genes involved in manganese acquisition and two clusters involved in metal efflux, one for zinc and cobalt and the other for copper (Table 1). Surplus of zinc and cobalt can be sensed by the regulator CzrA, encoded by czrA, which was highly up-regulated in our conditions, inducing the metal-efflux protein encoded by SXYL_00783. The copper chaperone CopZ, encoded by copZ, which was highly up-regulated in our conditions, could lead to the transcriptional de-repression of copA, which could result in the export of copper [73].
3.10. Response to Stress
The transcriptome profile of S. xylosus revealed the activation of stress-induced pathways within biofilm. The up-regulation of the cudTCAbetA cluster encoding a choline transporter (CudT), a regulator (CudC) and two enzymes (CudA, BetA) to form glycine betaine a powerful osmoprotectant indicated the perception of an osmotic stress (Table 1). In S. xylosus, the cudAbetA genes are up-regulated by choline and elevated NaCl concentrations [74]. In addition, the cluster mnhF2-A2 and three genes encoding Na+/H+ antiporter systems were up-regulated, particularly at 48 h of incubation. Similarly, the genes opuD encoding a glycine betaine transporter, prop a proline betaine transporter and mnhA a Na+/H+ antiporter unit are up-regulated in S. aureus in biofilm [75]. In addition, both genes encoding a glycine betaine transporter and a glycine betaine aldehyde dehydrogenase are up-regulated in S. epidermidis in biofilm [55].
S. xylosus also had to cope with oxidative stress, as shown by the up-regulation of seven genes involved in detoxification. Five of these genes (katB, trxB, ahpCF, SXL_00895) are under the control of the repressor PerR, whose gene is not modulated in our conditions (Table 1). These genes are overexpressed following nitrosative stress in a meat model [35]. In addition, katA was down-regulated, and sodA was not modulated in S. xylosus in biofilm, whereas these genes were found under-expressed in S. aureus in biofilm versus stationary growth phase [63]. A response to oxidative stress has already been reported for several biofilm-forming bacteria [76].
4. Conclusions
This study provides data on the physiology of S. xylosus in biofilm. It reveals that eDNA is a major component of the extracellular polymeric matrix and can be released by two mechanisms of cell lysis, lytic phage and the CidABC system. This lysis could furnish nutrients such as amino sugars, amino acids, nucleotides, and ions for the remaining cells. S. xylosus has developed defense mechanisms against osmotic and oxidant stresses. In addition, S. xylosus overexpressed several genes involved in DNA/RNA repair systems and in protein turnover. The ability of S. xylosus to form biofilm embedded in eDNA matrix allows it to colonize and survive in manufacturing environment.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9112192/s1, Table S1: Targeted genes of Staphylococcus xylosus for the validation of microarray data by qPCR: expression at 24 h and 48 h in biofilm compared with 9 h. Table S2: Total genes of Staphylococcus xylosus differentially expressed at 24 h and/or 48 h of incubation compared to 9 h in biofilm.
Author Contributions
Conceptualization, S.L. and R.T.; Data curation, P.M.; Formal analysis, S.L., P.M. and I.L.; Funding acquisition, S.L. and R.T.; Methodology, C.A., P.M. and I.L.; Supervision, S.L.; Writing—original draft, S.L. and R.T.; Writing—review & editing, S.L., I.L. and R.T. All authors have read and agreed to the published version of the manuscript.
Funding
The work was financially supported by the French National Research Agency (ANR) project NABAB (ANR-08-ALIA-11) and INRAE.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to David Marsh for correcting the English of our article.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Nagase, N.; Sasaki, A.; Yamashita, K.; Shimizu, A.; Wakita, Y.; Kitai, S.; Kawano, J. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 2002, 64, 245–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdier-Metz, I.; Gagne, G.; Bornes, S.; Monsallier, F.; Veisseire, P.; Delbès-Paus, C.; Montel, M.C. Cow teat skin, a potential source of diverse microbial populations for cheese production. Appl. Environ. Microbiol. 2012, 78, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Delbès, C.; Ali-Mandjee, L.; Montel, M.C. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 2007, 73, 1882–1891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leroy, S.; Giammarinaro, P.; Chacornac, J.P.; Lebert, I.; Talon, R. Biodiversity of indigenous staphylococci of naturally fermented dry sausages and manufacturing environments of small-scale processing units. Food Microbiol. 2010, 27, 249–301. [Google Scholar] [CrossRef] [PubMed]
- Planchon, S.; Gaillard-Martinie, B.; Dordet-Frisoni, E.; Bellon-Fontaine, M.N.; Leroy, S.; Labadie, J.; Hébraud, M.; Talon, R. Formation of biofilm by Staphylococcus xylosus. Int. J. Food Microbiol. 2006, 109, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Götz, F. Staphylococcus and biofilms. Mol. Microbiol. 2002, 43, 1367–1378. [Google Scholar] [CrossRef]
- Branda, S.S.; Vik, A.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 2005, 13, 20–26. [Google Scholar] [CrossRef]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 140, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Izano, E.A.; Amarante, M.A.; Kher, W.B.; Kaplan, J.B. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 2008, 74, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Planchon, S.; Desvaux, M.; Chafsey, I.; Chambon, C.; Leroy, S.; Hébraud, M.; Talon, R. Comparative subproteome analyses of planktonic and sessile Staphylococcus xylosus C2a: New insight in cell physiology of a coagulase-negative Staphylococcus in biofilm. J. Proteome Res. 2009, 8, 1797–1809. [Google Scholar] [CrossRef] [PubMed]
- Hussain, C.; Herrmann, M.; von Eiff, C.; Perdreau-Remington, F.; Peters, G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 1997, 65, 519–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cucarella, C.; Tormo, M.A.; Ubeda, C.; Trotonda, M.P.; Monzón, M.; Peris, C.; Amorena, B.; Lasa, I.; Penadès, J.R. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 2004, 72, 2177–2185. [Google Scholar] [CrossRef] [Green Version]
- Gross, M.; Cramton, S.E.; Götz, F.; Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 2001, 69, 3423–3426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadovskaya, I.; Vinogradov, E.; Li, J.; Jabbouri, S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus epidermidis RP62A, a reference biofilm-positive strain. Carbohydr. Res. 2004, 339, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Mann, E.E.; Rice, K.C.; Boles, B.R.; Endres, J.L.; Ranjit, D.; Chandramohan, L.; Tsang, L.H.; Smeltzer, M.S.; Horswill, A.R.; Bayles, K.W. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 2009, 4, e5822. [Google Scholar] [CrossRef] [Green Version]
- Kavanaugh, J.S.; Flack, C.E.; Lister, J.; Ricker, E.B.; Ibberson, C.B.; Jenul, C.; Moormeier, D.E.; Delmain, E.A.; Bayles, K.W.; Horswill, A.R. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio 2019, 10, e01137-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loza-Correa, M.; Ayala, J.A.; Perelman, I.; Hubbard, K.; Kalab, M.; Yi, Q.L.; Taha, M.; de Pedro, M.A.; Ramirez-Arcos, S. The peptidoglycan and biofilm matrix of Staphylococcus epidermidis undergo structural changes when exposed to human platelets. PLoS ONE 2019, 14, e0211132. [Google Scholar] [CrossRef] [PubMed]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef] [PubMed]
- Jakubovics, N.S.; Shields, R.C.; Rajarajan, N.; Burgess, J.G. Life after death: The critical role of extracellular DNA in microbial biofilms. Lett. Appl. Microbiol. 2013, 57, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Okshevsky, M.; Regina, V.R.; Meyer, R.L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef]
- Qin, Z.; Ou, Y.; Yang, L.; Zhu, Y.; Tolker-Nielsen, T.; Molin, S.; Qu, D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 2007, 153, 2083–2092. [Google Scholar] [CrossRef] [Green Version]
- Bose, J.L.; Lehman, M.K.; Fey, P.D.; Bayles, K.W. Contribution of the Staphylococcus aureus Atl AM and GL Murein Hydrolase Activities in Cell Division, Autolysis, and Biofilm Formation. PLoS ONE 2012, 7, e42244. [Google Scholar] [CrossRef]
- Sadykov, M.R.; Bayles, K.W. The control of death and lysis in staphylococcal biofilms: A coordination of physiological signals. Curr. Opin. Microbiol. 2012, 15, 211–215. [Google Scholar] [CrossRef] [Green Version]
- Groicher, K.H.; Firek, B.A.; Fujimoto, D.F.; Bayles, K.W. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 2000, 182, 1794–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, K.C.; Bayles, K.W. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 2008, 72, 85–109. [Google Scholar] [CrossRef] [Green Version]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.J.; Dunman, P.M.; Projan, S.J.; Bayles, K.W. Characterization of the Staphylococcus aureus CidR regulon: Elucidation of a novel role for acetoin metabolism in cell death and lysis. Mol. Microbiol. 2006, 60, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.S.; Givskovy, M.; Kjelleberg, S. Bacterial biofilms: Prokaryotic adventures in multicellularity. Curr. Opin. Microbiol. 2003, 6, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.S.; Lau, M.; Kjelleberg, S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004, 186, 8066–8073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resch, A.; Fehrenbacher, B.; Eisele, K.; Schaller, M.; Götz, F. Phage release from biofilm and planktonic Staphylococcus aureus cells. FEMS Microbiol. Lett. 2005, 252, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Vermassen, A.; Dordet-Frisoni, E.; de La Foye, A.; Micheau, P.; Laroute, V.; Leroy, S.; Talon, R. Adaptation of Staphylococcus xylosus to nutrients and osmotic stress in a salted meat model. Front. Microbiol. 2016, 7, 87. [Google Scholar] [CrossRef] [Green Version]
- Leroy, S.; Lebert, I.; Andant, C.; Talon, R. Interaction in dual species biofilms between Staphylococcus xylosus and Staphylococcus aureus. Int. J. Food Microbiol. 2020, 326, 108653. [Google Scholar] [CrossRef]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef] [Green Version]
- Vermassen, A.; de la Foye, A.; Loux, V.; Talon, R.; Leroy, S. Transcriptomic analysis of Staphylococcus xylosus in the presence of nitrate and nitrite in meat reveals its response to nitrosative stress. Front. Microbiol. 2014, 5, 691. [Google Scholar] [CrossRef] [Green Version]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schlafer, S.; Meyer, R.L. Confocal microscopy imaging of the biofilm matrix. J. Microbiol. Methods 2017, 138, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Vilain, S.; Pretorius, J.M.; Theron, J.; Brözel, V.S. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl. Environ. Microbiol. 2009, 75, 2861–2868. [Google Scholar] [CrossRef] [Green Version]
- Zetzmann, M.; Okshevsky, M.; Endres, J.; Sedlag, A.; Caccia, N.; Auchter, M.; Waidmann, M.S.; Desvaux, M.; Meyer, R.L.; Riedel, C.U. DNase-Sensitive and -Resistant Modes of Biofilm Formation by Listeria monocytogenes. Front. Microbiol. 2015, 6, 1428. [Google Scholar] [CrossRef] [Green Version]
- Pakkulnan, R.; Anutrakunchai, C.; Kanthawong, S.; Taweechaisupapong, S.; Chareonsudjai, P.; Chareonsudjai, S. Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS ONE 2019, 14, e0213288. [Google Scholar] [CrossRef]
- Regina, V.R.; Lokanathan, A.R.; Modrzyński, J.J.; Sutherland, D.S.; Meyer, R.L. Surface physicochemistry and ionic strength affects eDNA’s role in bacterial adhesion to abiotic surfaces. PLoS ONE 2014, 9, e105033. [Google Scholar] [CrossRef] [Green Version]
- Deghorain, M.; Bobay, L.M.; Smeesters, P.R.; Bousbata, S.; Vermeersch, M.; Perez-Morga, D.; Dreze, P.A.; Rocha, E.P.; Touchon, M.; Van Melderen, L. Characterization of Novel Phages Isolated in Coagulase-Negative Staphylococci Reveals Evolutionary Relationships with Staphylococcus aureus phages. J. Bacteriol. 2012, 194, 5829–5839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deghorain, M.; Van Melderen, L. The Staphylococci phages family: An overview. Viruses 2012, 4, 3316–3335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, M.; Truesdell, S.; Ramakrishnan, T.; Bronson, M.J. Dual control of lysogeny by bacteriophage P22: An antirepressor locus and its controlling elements. J. Mol. Biol. 1975, 91, 421–438. [Google Scholar] [CrossRef] [Green Version]
- Carrolo, M.; Frias, M.J.; Pinto, F.R.; Melo-Cristino, J.; Ramirez, M. Prophage Spontaneous Activation Promotes DNA Release Enhancing Biofilm Formation in Streptococcus pneumoniae. PLoS ONE 2010, 5, e15678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gödeke, J.; Paul, K.; Lassak, J.; Thormann, K.M. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2011, 5, 613–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binnenkade, L.; Teichmann, L.; Thormann, K.M. Iron triggers λSo prophage induction and release of extracellular DNA in Shewanella oneidensis MR-1biofilms. Appl. Environ. Microbiol. 2014, 80, 5304–5316. [Google Scholar] [CrossRef] [Green Version]
- Thomas, V.C.; Sadykov, M.R.; Chaudhari, S.S.; Jones, J.; Endres, J.L.; Widhelm, T.J.; Ahn, J.S.; Jawa, R.S.; Zimmerman, M.C.; Bayles, K.W. A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog. 2014, 10, e1004205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar] [CrossRef]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef] [Green Version]
- Gajdiss, M.; Monk, I.R.; Bertsche, U.; Kienemund, J.; Funk, T.; Dietrich, A.; Hort, M.; Sib, E.; Stinear, T.P.; Bierbaum, G. YycH and YycI Regulate Expression of Staphylococcus aureus Autolysins by Activation of WalRK Phosphorylation. Microorganisms 2020, 8, 870. [Google Scholar] [CrossRef]
- McKenzie, G.J.; Magner, D.B.; Lee, P.L.; Rosenberg, S.M. The dinB Operon and Spontaneous Mutation in Escherichia coli. J. Bacteriol. 2003, 185, 3972–3977. [Google Scholar] [CrossRef] [Green Version]
- Ayora, S.; Carrasco, B.; Cardenas, P.P.; Cesar, C.E.; Canas, C.; Yadav, T.; Marchisone, C.; Alonso, J.C. Double-strand break repair in bacteria: A view from Bacillus subtilis. FEMS Microbiol. Rev. 2011, 35, 1055–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Daniel, E.; Sturdevant, D.E.; Otto, M. Genome wide analysis of gene expression in Staphylococcus epidermidis biofilms: Insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 2005, 191, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Hollis, T.; Lau, A.; Ellenberger, T. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat. Res. DNA Repair 2000, 460, 201–210. [Google Scholar] [CrossRef]
- Iyer, L.M.; Zhang, D.; Rogozin, I.B.; Aravind, L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011, 39, 9473–9497. [Google Scholar] [CrossRef] [Green Version]
- Domingues, S.; Moreira, R.N.; Andrade, J.M.; Dos Santos, R.F.; Bárria, C.; Viegas, S.C.; Arraiano, C.M. The role of RNase R in trans-translation and ribosomal quality control. Biochimie 2015, 114, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Schuman, W.; Hecker, M.; Msadek, T. Regulation and function of heat-inducible genes in Bacillus subtilis. In Bacillus subtilis and Its Closest Relatives; American Society of Microbiology: Washington, DC, USA, 2002; pp. 359–368. [Google Scholar]
- Wang, P.; Dalbey, R.E. Inserting membrane proteins: The YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. et Biophys. Acta (BBA)—Biomembr. 2011, 1808, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Switzer, R.L.; Zalkin, H.; Saxild, H.H. Purine, Pyrimidine, and Pyridine Nucleotide Metabolism. In Bacillus subtilis and Its Closest Relatives; American Society of Microbiology: Washington, DC, USA, 2002; pp. 255–269. [Google Scholar]
- Leroy, S.; Vermassen, A.; Ras, G.; Talon, R. Insight into the genome of Staphylococcus xylosus, a ubiquitous species well adapted to meat products. Microorganisms 2017, 5, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beenken, K.E.; Dunman, P.M.; McAleese, F.; Macapagal, D.; Murphy, E.; Projan, S.J.; Blevins, J.S.; Smeltzer, M.S. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 2004, 186, 4665–4684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vimr, E.R.; Kalivoda, K.A.; Deszo, E.L.; Steenbergen, S.M. Diversity of Microbial Sialic Acid Metabolism. Microbiol. Mol. Biol. Rev. 2004, 68, 132–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borisov, V.B.; Gennis, R.B.; Hemp, J.; Verkhovsky, M.I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 2011, 1807, 1398–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belitsky, B.R. Biosynthesis of amino acids of the glutamate and aspartate families, alanine, and polyamines. In Bacillus subtilis and Its Closest Relatives; American Society of Microbiology: Washington, DC, USA, 2002; pp. 203–231. [Google Scholar]
- Billot-Klein, D.; Gutmann, L.; Bryant, D.; Bell, D.; van Heijenoort, J.; Grewal, J.; Shlaes, D.M. Peptidoglycan synthesis and structure in Staphylococcus haemolyticus expressing increasing levels of resistance to glycopeptide antibiotics. J. Bacteriol. 1996, 178, 4696–4703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schleimer, N.; Kaspar, U.; Drescher, M.; Seggewiß, J.; von Eiff, C.; Proctor, R.A.; Peters, G.; Kriegeskorte, A.; Becker, K. The Energy-Coupling Factor Transporter Module EcfAA’T, a Novel Candidate for the Genetic Basis of Fatty Acid-Auxotrophic Small-Colony Variants of Staphylococcus aureus. Front. Microbiol. 2018, 9, 1863. [Google Scholar] [CrossRef] [PubMed]
- Fiegler, H.; Brückner, R. Identification of the serine acetyltransferase gene of Staphylococcus xylosus. FEMS Microbiol. Lett. 1997, 148, 181–187. [Google Scholar] [CrossRef]
- Kelliher, J.L.; Radin, J.N.; Kehl-Fie, T.E. PhoPR contributes to Staphylococcus aureus growth during phosphate starvation and pathogenesis in an environment-specific manner. Infect. Immun. 2018, 86, e00371-18. [Google Scholar] [CrossRef] [Green Version]
- Vermassen, A.; Talon, R.; Leroy, S. Ferritin, an iron source in meat for Staphylococcus xylosus? Int. J. Food Microbiol. 2016, 225, 20–26. [Google Scholar] [CrossRef]
- Haikarainen, T.; Papageorgiou, A.C. 2010. Dps-like proteins: Structural and functional insights into a versatile protein family. Cell. Mol. Life Sci. 2010, 67, 341–351. [Google Scholar] [CrossRef]
- Ma, Z.; Jacobsen, F.E.; Giedroc, D.P. Chemistry Controls Bacterial Metal Homeostasis. Chem. Rev. 2009, 109, 4644–4681. [Google Scholar] [CrossRef] [Green Version]
- Rosenstein, R.; Futter-Bryniok, D.; Götz, F. The choline-converting pathway in Staphylococcus xylosus C2a: Genetic and physiological characterization. J. Bacteriol. 1999, 181, 2273–2278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resch, A.; Rosenstein, R.; Nerz, C.; Götz, F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 2005, 71, 2663–2676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beloin, C.; Ghigo, J.M. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005, 13, 16–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).