Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins
Abstract
1. Introduction
2. RiPPs, the Bacteriocins with Versatile Modes of Action
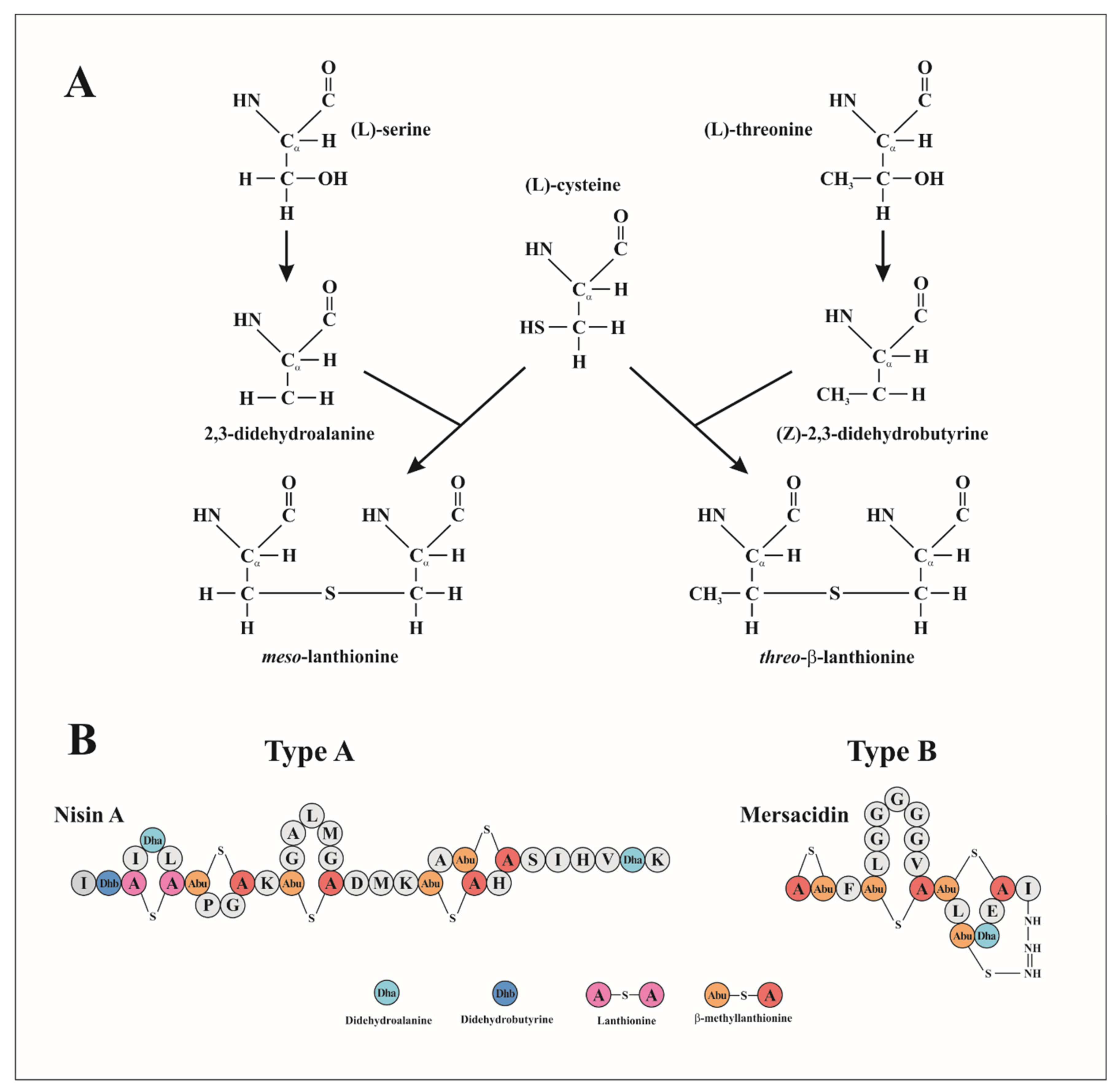
2.1. Class Ia: Lantibiotics
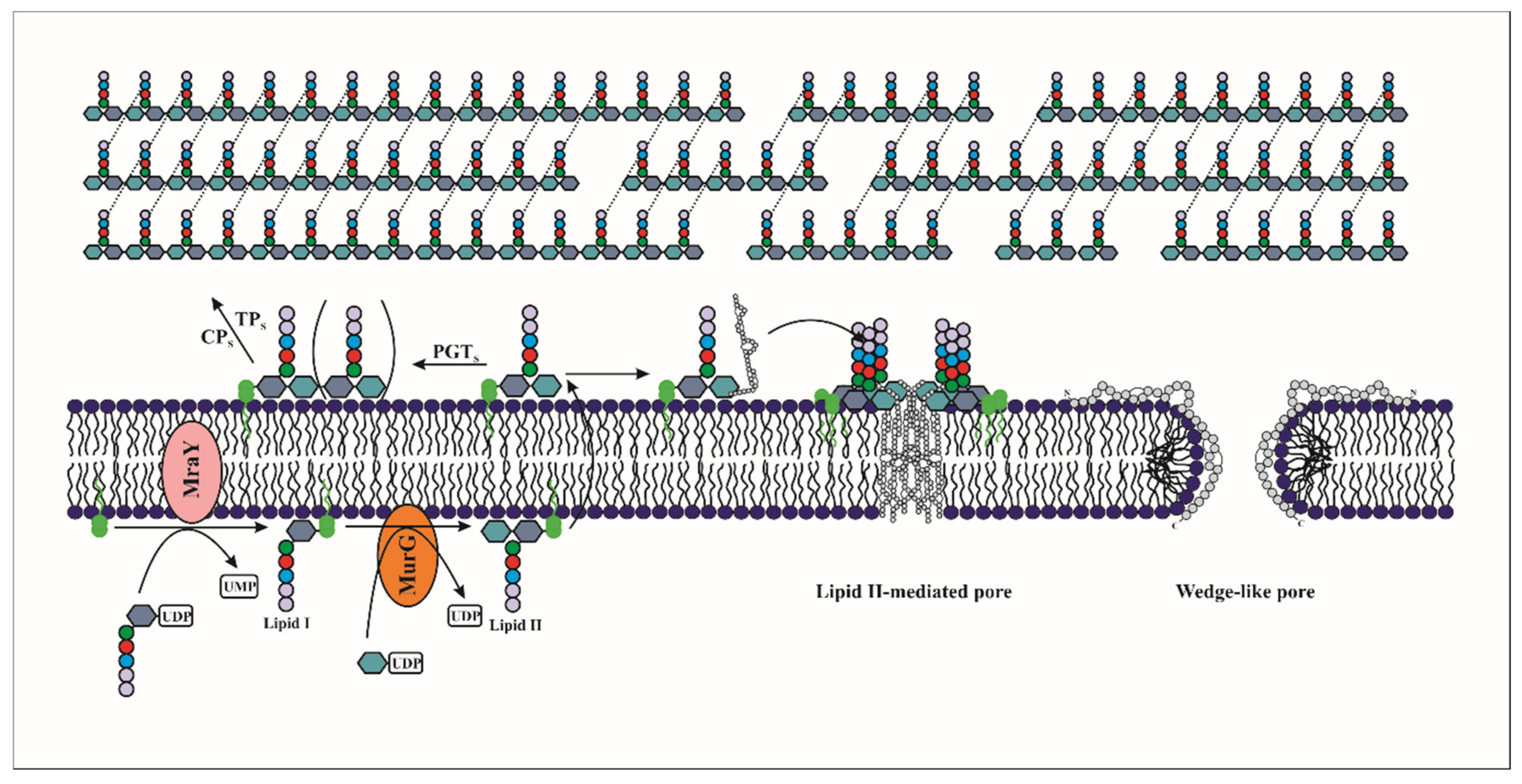
2.1.1. Mechanism of Action of Lantibiotics
2.1.2. Lantibiotic Immunity
2.2. Class Ib or Circular Bacteriocins
2.2.1. Mode of Action of Circular Bacteriocins
2.2.2. Immunity of Circular Bacteriocins
3. Class II, A Group with Not Only Pore Forming Bacteriocins
3.1. Class IIa Bacteriocins, or Pediocin-Like Bacteriocins (PLBs)
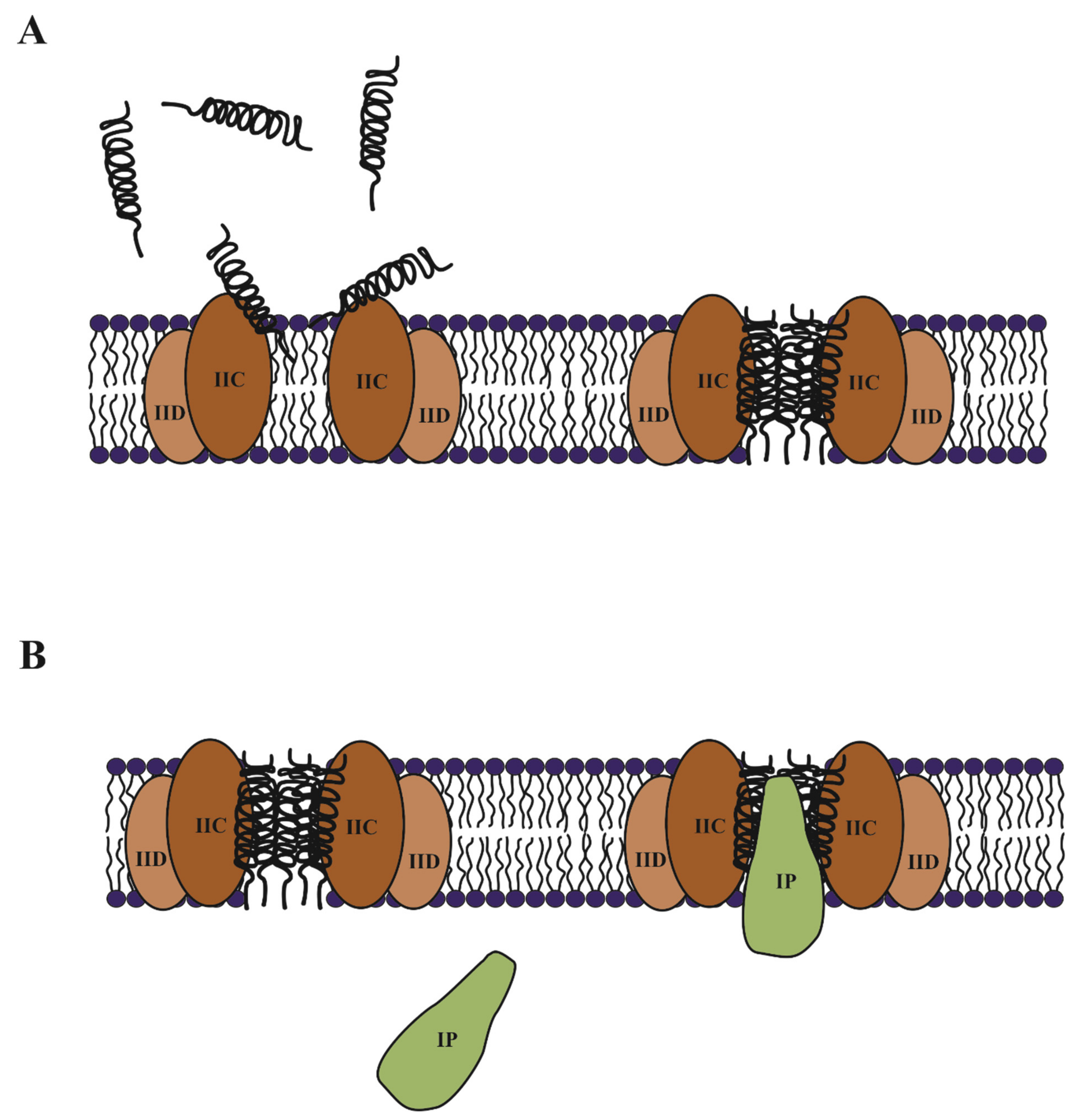
3.1.1. Mode of Action of Class IIa Bacteriocins
3.1.2. Immunity of Class IIa Bacteriocins
3.2. Class IIb Bacteriocins, or Two-Peptide Bacteriocins
3.2.1. Mode of Action of Class IIb Bacteriocins
3.2.2. Immunity of Class IIb Bacteriocins
3.3. Class IIc, Leaderless Bacteriocins
3.3.1. Mechanism of Action of Leaderless Bacteriocins
3.3.2. Immunity of Leaderless Bacteriocins
3.4. Class IId Bacteriocins, and Other Non-Pediocin-Like Single-Peptide Bacteriocins
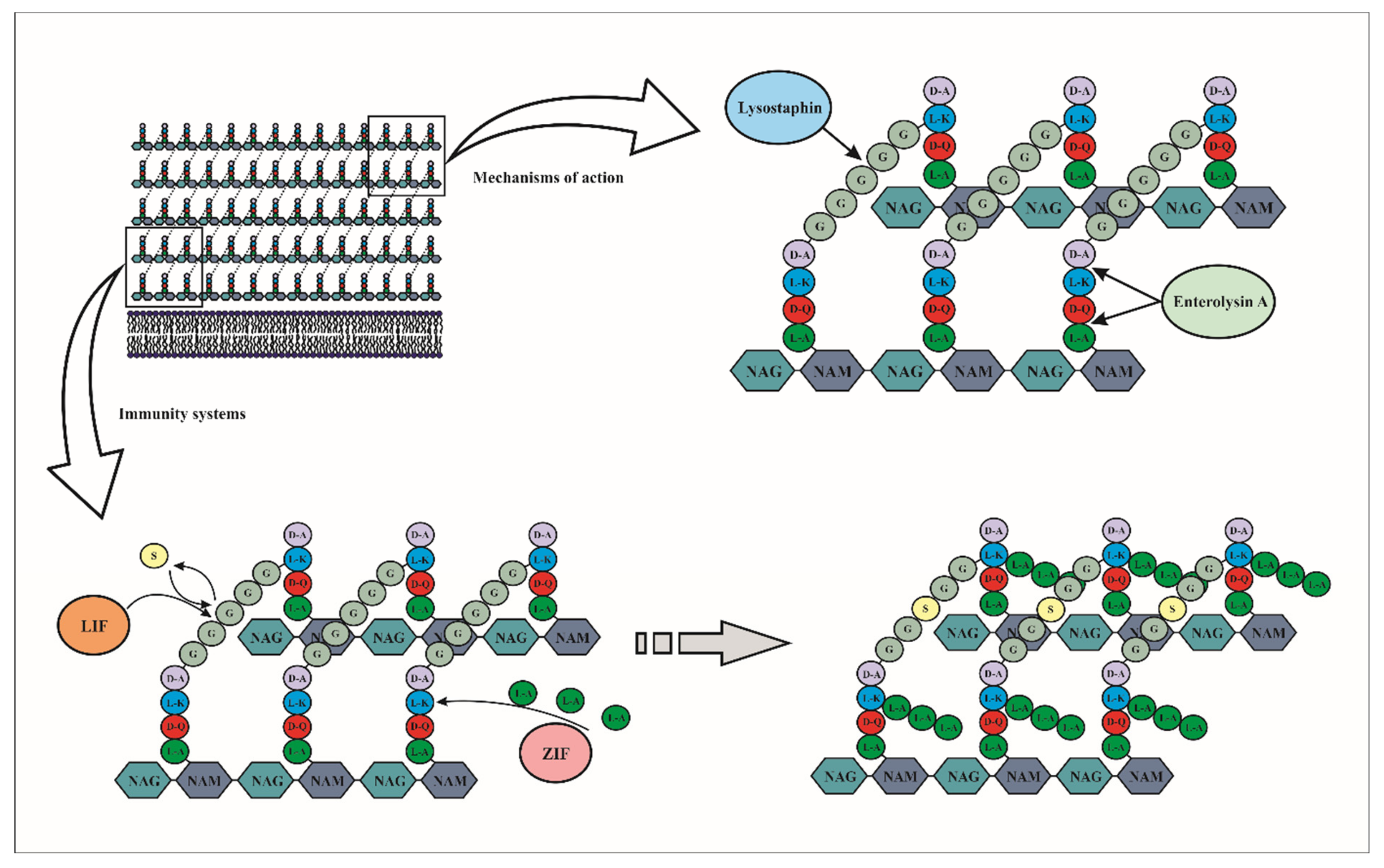
4. Class III, “Wall Breaker” Bacteriocins
4.1. Mode of Action of Class III Bacteriocins
4.2. Immunity Mechanism of Class III Bacteriocins
5. Concluding Remarks and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Drider, D.; Rebuffat, S. Prokaryotic Antimicrobial Peptides: From Genes to Applications; Springer Science & Business Media: New York, NY, USA, 2011; ISBN 978-1-4419-7692-5. [Google Scholar]
- Meade, E.; Slattery, M.A.; Garvey, M. Bacteriocins, Potent Antimicrobial Peptides and the Fight against Multi Drug Resistant Species: Resistance Is Futile? Antibiotics 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Settanni, L.; Corsetti, A. Application of Bacteriocins in Vegetable Food Biopreservation. Int. J. Food Microbiol. 2008, 121, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Siroli, L.; Camprini, L.; Pisano, M.B.; Patrignani, F.; Lanciotti, R. Volatile Molecule Profiles and Anti-Listeria Monocytogenes Activity of Nisin Producers Lactococcus Lactis Strains in Vegetable Drinks. Front. Microbiol. 2019, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Stern, N.J.; Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Pokhilenko, V.D.; Levchuk, V.P.; Svetoch, O.E.; Seal, B.S. Isolation of a Lactobacillus salivarius Strain and Purification of Its Bacteriocin, Which Is Inhibitory to Campylobacter Jejuni in the Chicken Gastrointestinal System. Antimicrob. Agents Chemother. 2006, 50, 3111–3116. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, S.; Kergourlay, G.; Dalgalarrondo, M.; Choiset, Y.; Ferchichi, M.; Prévost, H.; Pilet, M.-F.; Chobert, J.-M.; Manai, M.; Dousset, X. Purification and Characterization of a New Bacteriocin Active against Campylobacter Produced by Lactobacillus salivarius SMXD51. Food Microbiol. 2012, 32, 129–134. [Google Scholar] [CrossRef]
- Belguesmia, Y.; Bendjeddou, K.; Kempf, I.; Boukherroub, R.; Drider, D. Heterologous Biosynthesis of Five New Class II Bacteriocins from Lactobacillus paracasei CNCM I-5369 with Antagonistic Activity against Pathogenic Escherichia coli Strains. Front. Microbiol. 2020, 11, 1198. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Drider, D.; Bendali, F.; Naghmouchi, K.; Chikindas, M.L. Bacteriocins: Not Only Antibacterial Agents. Probiotics Antimicrob. Prot. 2016, 8, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M.T. Functions and Emerging Applications of Bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef]
- Klaenhammer, T.R. Genetics of Bacteriocins Produced by Lactic Acid Bacteria. FEMS Microbiol. Rev. 1993, 12, 39–85. [Google Scholar] [CrossRef]
- Drider, D.; Fimland, G.; Hechard, Y.; McMullen, L.M.; Prevost, H. The Continuing Story of Class IIa Bacteriocins. Microbiol. Mol. Biol. Rev. 2006, 70, 564–582. [Google Scholar] [CrossRef]
- Heng, N.C.K.; Tagg, J.R. What’s in a Name? Class Distinction for Bacteriocins. Nat. Rev. Microbiol. 2006, 4, 160. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L.M.T.; Dreyer, L.; Smith, C.; van Staden, A.D. A Review: The Fate of Bacteriocins in the Human Gastro-Intestinal Tract: Do They Cross the Gut–Blood Barrier? Front. Microbiol. 2018, 9, 2297. [Google Scholar] [CrossRef] [PubMed]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a New Generation of Antimicrobials: Toxicity Aspects and Regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef] [PubMed]
- Rebuffat, S. Bacteriocins from Gram-negative bacteria: A classification? In Prokaryotic Antimicrobial Peptides: From Genes to Applications; Drider, D., Rebuffat, S., Eds.; Springer: New York, NY, USA, 2011; pp. 55–72. ISBN 978-1-4419-7692-5. [Google Scholar]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Novel Bacteriocins from Lactic Acid Bacteria (LAB): Various Structures and Applications. Microb. Cell Fact. 2014, 13 (Suppl 1), S3. [Google Scholar] [CrossRef] [PubMed]
- Juturu, V.; Wu, J.C. Microbial Production of Bacteriocins: Latest Research Development and Applications. Biotechnol. Adv. 2018, 36, 2187–2200. [Google Scholar] [CrossRef]
- Daba, H.; Pandian, S.; Gosselin, J.F.; Simard, R.E.; Huang, J.; Lacroix, C. Detection and Activity of a Bacteriocin Produced by Leuconostoc Mesenteroides. Appl. Environ. Microbiol. 1991, 57, 3450–3455. [Google Scholar] [CrossRef]
- Hahn-Löbmann, S.; Stephan, A.; Schulz, S.; Schneider, T.; Shaverskyi, A.; Tusé, D.; Giritch, A.; Gleba, Y. Colicins and Salmocins—New Classes of Plant-Made Non-Antibiotic Food Antibacterials. Front. Plant Sci. 2019, 10, 437. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical Applications of Nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef]
- Masdea, L.; Kulik, E.M.; Hauser-Gerspach, I.; Ramseier, A.M.; Filippi, A.; Waltimo, T. Antimicrobial Activity of Streptococcus salivarius K12 on Bacteria Involved in Oral Malodour. Arch. Oral Biol. 2012, 57, 1041–1047. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Hale, J.D.F.; Heng, N.C.K.; Tagg, J.R. Developing Oral Probiotics from Streptococcus salivarius. Future Microbiol. 2012, 7, 1355–1371. [Google Scholar] [CrossRef]
- Srinivas, S.; Paul, D.C.; Colin, H. Lacticin 3147—Biosynthesis, Molecular Analysis, Immunity, Bioengineering and Applications. Curr. Protein Pept. Sci. 2012, 13, 193–204. [Google Scholar]
- Kitching, M.; Mathur, H.; Flynn, J.; Byrne, N.; Dillon, P.; Sayers, R.; Rea, M.C.; Hill, C.; Ross, R.P. A Live Bio-Therapeutic for Mastitis, Containing Lactococcus Lactis DPC3147 with Comparable Efficacy to Antibiotic Treatment. Front. Microbiol. 2019, 10, 2220. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hidalgo, M.; Montalbán-López, M.; Cebrián, R.; Valdivia, E.; Martínez-Bueno, M.; Maqueda, M. AS-48 Bacteriocin: Close to Perfection. Cell. Mol. Life Sci. 2011, 68, 2845–2857. [Google Scholar] [CrossRef]
- Montalbán-López, M.; Cebrián, R.; Galera, R.; Mingorance, L.; Martín-Platero, A.M.; Valdivia, E.; Martínez-Bueno, M.; Maqueda, M. Synergy of the Bacteriocin AS-48 and Antibiotics against Uropathogenic Enterococci. Antibiotics 2020, 9, 567. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Martínez, M.I.; Kok, J. Pediocin PA-1, a Wide-Spectrum Bacteriocin from Lactic Acid Bacteria. Crit. Rev. Food Sci. Nutr. 2002, 42, 91–121. [Google Scholar] [CrossRef] [PubMed]
- Araújo, C.; Muñoz-Atienza, E.; Poeta, P.; Igrejas, G.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Characterization of Pediococcus acidilactici Strains Isolated from Rainbow Trout (Oncorhynchus mykiss) Feed and Larvae: Safety, DNA Fingerprinting, and Bacteriocinogenicity. Dis. Aquat. Organ. 2016, 119, 129–143. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Lactobacillus Plantarum Isolated from Molasses Produces Bacteriocins Active against Gram-Negative Bacteria. Enzyme Microb. Technol. 2005, 36, 318–326. [Google Scholar] [CrossRef]
- Knoetze, H.; Todorov, S.D.; Dicks, L.M.T. A Class IIa Peptide from Enterococcus mundtii Inhibits Bacteria Associated with Otitis Media. Int. J. Antimicrob. Agents 2008, 31, 228–234. [Google Scholar] [CrossRef]
- Saeed, S.; Rasool, S.A.; Ahmad, S.; Zaidi, S.; Rehmani, S. Antiviral Activity of Staphylococcin 188: A Purified Bacteriocin like Inhibitory Substance Isolated from Staphylococcus aureus AB188. Res. J. Microbiol. 2007, 2, 796–806. [Google Scholar]
- Navarro, S.A.; Lanza, L.; Acuña, L.; Bellomio, A.; Chalón, M.C. Features and Applications of Ent35-MccV Hybrid Bacteriocin: Current State and Perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 6067–6077. [Google Scholar] [CrossRef]
- Todorov, S.D.; Wachsman, M.; Tomé, E.; Dousset, X.; Destro, M.T.; Dicks, L.M.T.; de Melo Franco, B.D.G.; Vaz-Velho, M.; Drider, D. Characterisation of an Antiviral Pediocin-like Bacteriocin Produced by Enterococcus faecium. Food Microbiol. 2010, 27, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Balgir, P.P.; Mittu, B.; Kumar, B.; Garg, N. Biomedical Applications of Fermenticin HV6b Isolated from Lactobacillus Fermentum HV6b MTCC10770. Biomed. Res. Int. 2013, 2013, e168438. [Google Scholar] [CrossRef] [PubMed]
- Villarante, K.I.; Elegado, F.B.; Iwatani, S.; Zendo, T.; Sonomoto, K.; de Guzman, E.E. Purification, Characterization and in Vitro Cytotoxicity of the Bacteriocin from Pediococcus Acidilactici K2a2-3 against Human Colon Adenocarcinoma (HT29) and Human Cervical Carcinoma (HeLa) Cells. World J. Microbiol. Biotechnol. 2011, 27, 975–980. [Google Scholar] [CrossRef]
- Belguesmia, Y.; Choiset, Y.; Rabesona, H.; Baudy-Floc’h, M.; Blay, G.L.; Haertlé, T.; Chobert, J.-M. Antifungal Properties of Durancins Isolated from Enterococcus Durans A5-11 and of Its Synthetic Fragments. Lett. Appl. Microbiol. 2013, 56, 237–244. [Google Scholar] [CrossRef]
- Heeney, D.D.; Yarov-Yarovoy, V.; Marco, M.L. Sensitivity to the Two Peptide Bacteriocin Plantaricin EF Is Dependent on CorC, a Membrane-Bound, Magnesium/Cobalt Efflux Protein. MicrobiologyOpen 2019, 8, e827. [Google Scholar] [CrossRef]
- Kadouri, D.E.; To, K.; Shanks, R.M.Q.; Doi, Y. Predatory Bacteria: A Potential Ally against Multidrug-Resistant Gram-Negative Pathogens. PLoS ONE 2013, 8, e63397. [Google Scholar] [CrossRef]
- Kang, B.S.; Seo, J.-G.; Lee, G.-S.; Kim, J.-H.; Kim, S.Y.; Han, Y.W.; Kang, H.; Kim, H.O.; Rhee, J.H.; Chung, M.-J.; et al. Antimicrobial Activity of Enterocins from Enterococcus faecalis SL-5 against Propionibacterium acnes, the Causative Agent in Acne Vulgaris, and Its Therapeutic Effect. J. Microbiol. 2009, 47, 101–109. [Google Scholar] [CrossRef]
- Jayakumar, J.; Kumar, V.A.; Biswas, L.; Biswas, R. Therapeutic Applications of Lysostaphin against Staphylococcus aureus. J. Appl. Microbiol. 2020, 131, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.; Huang, J.; Li, G.; Zhang, J.; Sun, Y.; Zeng, D.; Bao, W.; Zhong, J.; Huang, Q. Clinical Efficacy of Intravaginal Recombinant Lysostaphin Administration on Endometritis in Sows. Vet. Med. Sci. 2021, 7, 746–754. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, O.; Ross, R.P.; Hill, C. Lantibiotics: Structure, Biosynthesis and Mode of Action. FEMS Microbiol. Rev. 2001, 25, 285–308. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing Innate Immunity for Food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Rebuffat, S. The Manifold Roles of Microbial Ribosomal Peptide–Based Natural Products in Physiology and Ecology. J. Biol. Chem. 2020, 295, 34–54. [Google Scholar] [CrossRef]
- Willey, J.M.; van der Donk, W.A. Lantibiotics: Peptides of Diverse Structure and Function. Annu. Rev. Microbiol. 2007, 61, 477–501. [Google Scholar] [CrossRef]
- Daly, K.M.; Cotter, P.D.; Hill, C.; Ross, R.P. Lantibiotic Production by Pathogenic Microorganisms. Curr. Protein Pept. Sci. 2012, 13, 509–523. [Google Scholar] [CrossRef]
- Wilson-Stanford, S.; Smith, L. Commercial Development and Application of Type a Lantibiotics. Recent Pat. Anti-Infect. Drug Discov. 2011, 6, 175–185. [Google Scholar] [CrossRef]
- Van Kraaij, C.; Breukink, E.; Noordermeer, M.A.; Demel, R.A.; Siezen, R.J.; Kuipers, O.P.; de Kruijff, B. Pore Formation by Nisin Involves Translocation of Its C-Terminal Part across the Membrane. Biochemistry 1998, 37, 16033–16040. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, I.; Breukink, E.; van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; de Kruijff, B.; Sahl, H.G. Specific Binding of Nisin to the Peptidoglycan Precursor Lipid II Combines Pore Formation and Inhibition of Cell Wall Biosynthesis for Potent Antibiotic Activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, M.J.; Kok, J.; Venema, G.; Holo, H.; Nes, I.F.; Konings, W.N.; Abee, T. The Bacteriocin Lactococcin a Specifically Increases Permeability of Lactococcal Cytoplasmic Membranes in a Voltage-Independent, Protein-Mediated Manner. J. Bacteriol. 1991, 173, 7934–7941. [Google Scholar] [CrossRef]
- Montville, T.J.; Bruno, M.E. Evidence That Dissipation of Proton Motive Force Is a Common Mechanism of Action for Bacteriocins and Other Antimicrobial Proteins. Int. J. Food Microbiol. 1994, 24, 53–74. [Google Scholar] [CrossRef]
- Farha, M.A.; Verschoor, C.P.; Bowdish, D.; Brown, E.D. Collapsing the Proton Motive Force to Identify Synergistic Combinations against Staphylococcus aureus. Chem. Biol. 2013, 20, 1168–1178. [Google Scholar] [CrossRef]
- Kordel, M.; Schüller, F.; Sahl, H.G. Interaction of the Pore Forming-Peptide Antibiotics Pep 5, Nisin and Subtilin with Non-Energized Liposomes. FEBS Lett. 1989, 244, 99–102. [Google Scholar] [CrossRef]
- Moll, G.N.; Roberts, G.C.K.; Konings, W.N.; Driessen, A.J.M. Mechanism of Lantibiotic-Induced Pore-Formation. Antonie Leeuwenhoek 1996, 69, 185–191. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, R.E.; Ferretti, J.J.; Hynes, W.L. Nucleotide Sequence of the Streptococcin A-FF22 Lantibiotic Regulon: Model for Production of the Lantibiotic SA-FF22 by Strains of Streptococcus pyogenes. FEMS Microbiol. Lett. 1999, 175, 171–177. [Google Scholar] [CrossRef]
- Breukink, E.; Ganz, P.; de Kruijff, B.; Seelig, J. Binding of Nisin Z to Bilayer Vesicles as Determined with Isothermal Titration Calorimetry. Biochemistry 2000, 39, 10247–10254. [Google Scholar] [CrossRef]
- Giffard, C.J.; Dodd, H.M.; Horn, N.; Ladha, S.; Mackie, A.R.; Parr, A.; Gasson, M.J.; Sanders, D. Structure-Function Relations of Variant and Fragment Nisins Studied with Model Membrane Systems. Biochemistry 1997, 36, 3802–3810. [Google Scholar] [CrossRef] [PubMed]
- Héchard, Y.; Sahl, H.-G. Mode of Action of Modified and Unmodified Bacteriocins from Gram-Positive Bacteria. Biochimie 2002, 84, 545–557. [Google Scholar] [CrossRef]
- Driessen, A.J.; van den Hooven, H.W.; Kuiper, W.; van de Kamp, M.; Sahl, H.G.; Konings, R.N.; Konings, W.N. Mechanistic Studies of Lantibiotic-Induced Permeabilization of Phospholipid Vesicles. Biochemistry 1995, 34, 1606–1614. [Google Scholar] [CrossRef]
- Tolpekina, T.V.; den Otter, W.K.; Briels, W.J. Nucleation Free Energy of Pore Formation in an Amphiphilic Bilayer Studied by Molecular Dynamics Simulations. J. Chem. Phys. 2004, 121, 12060–12066. [Google Scholar] [CrossRef]
- Sahl, H.G.; Kordel, M.; Benz, R. Voltage-Dependent Depolarization of Bacterial Membranes and Artificial Lipid Bilayers by the Peptide Antibiotic Nisin. Arch. Microbiol. 1987, 149, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Garcerá, M.J.; Elferink, M.G.; Driessen, A.J.; Konings, W.N. In Vitro Pore-Forming Activity of the Lantibiotic Nisin. Role of Protonmotive Force and Lipid Composition. Eur. J. Biochem. 1993, 212, 417–422. [Google Scholar] [CrossRef]
- Breukink, E.; van Heusden, H.E.; Vollmerhaus, P.J.; Swiezewska, E.; Brunner, L.; Walker, S.; Heck, A.J.R.; de Kruijff, B. Lipid II Is an Intrinsic Component of the Pore Induced by Nisin in Bacterial Membranes. J. Biol. Chem. 2003, 278, 19898–19903. [Google Scholar] [CrossRef] [PubMed]
- Van Heusden, H.E.; de Kruijff, B.; Breukink, E. Lipid II Induces a Transmembrane Orientation of the Pore-Forming Peptide Lantibiotic Nisin. Biochemistry 2002, 41, 12171–12178. [Google Scholar] [CrossRef] [PubMed]
- Breukink, E.; Wiedemann, I.; van Kraaij, C.; Kuipers, O.P.; Sahl, H.G.; de Kruijff, B. Use of the Cell Wall Precursor Lipid II by a Pore-Forming Peptide Antibiotic. Science 1999, 286, 2361–2364. [Google Scholar] [CrossRef]
- Hsu, S.-T.D.; Breukink, E.; Tischenko, E.; Lutters, M.A.G.; de Kruijff, B.; Kaptein, R.; Bonvin, A.M.J.J.; van Nuland, N.A.J. The Nisin–Lipid II Complex Reveals a Pyrophosphate Cage That Provides a Blueprint for Novel Antibiotics. Nat. Struct. Mol. Biol. 2004, 11, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, I.; Benz, R.; Sahl, H.-G. Lipid II-Mediated Pore Formation by the Peptide Antibiotic Nisin: A Black Lipid Membrane Study. J. Bacteriol. 2004, 186, 3259–3261. [Google Scholar] [CrossRef]
- Hasper, H.E.; Kramer, N.E.; Smith, J.L.; Hillman, J.D.; Zachariah, C.; Kuipers, O.P.; de Kruijff, B.; Breukink, E. An Alternative Bactericidal Mechanism of Action for Lantibiotic Peptides That Target Lipid II. Science 2006, 313, 1636–1637. [Google Scholar] [CrossRef]
- Brötz, H.; Bierbaum, G.; Leopold, K.; Reynolds, P.E.; Sahl, H.G. The Lantibiotic Mersacidin Inhibits Peptidoglycan Synthesis by Targeting Lipid II. Antimicrob. Agents Chemother. 1998, 42, 154–160. [Google Scholar] [CrossRef]
- Dickman, R.; Mitchell, S.A.; Figueiredo, A.M.; Hansen, D.F.; Tabor, A.B. Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B. J. Org. Chem. 2019, 84, 11493–11512. [Google Scholar] [CrossRef]
- Zimmermann, N.; Metzger, J.W.; Jung, G. The Tetracyclic Lantibiotic Actagardine. 1H-NMR and 13C-NMR Assignments and Revised Primary Structure. Eur. J. Biochem. 1995, 228, 786–797. [Google Scholar] [CrossRef]
- Bierbaum, G.; Sahl, H.G. Autolytic System of Staphylococcus Simulans 22: Influence of Cationic Peptides on Activity of N-Acetylmuramoyl-L-Alanine Amidase. J. Bacteriol. 1987, 169, 5452–5458. [Google Scholar] [CrossRef]
- Liu, W.; Hansen, J.N. The Antimicrobial Effect of a Structural Variant of Subtilin against Outgrowing Bacillus Cereus T Spores and Vegetative Cells Occurs by Different Mechanisms. Appl. Environ. Microbiol. 1993, 59, 648–651. [Google Scholar] [CrossRef]
- Barbour, A.; Wescombe, P.; Smith, L. Evolution of Lantibiotic Salivaricins: New Weapons to Fight Infectious Diseases. Trends Microbiol. 2020, 28, 578–593. [Google Scholar] [CrossRef] [PubMed]
- Draper, L.A.; Ross, R.P.; Hill, C.; Cotter, P.D. Lantibiotic Immunity. Curr. Protein Pept. Sci. 2008, 9, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Hacker, C.; Christ, N.A.; Duchardt-Ferner, E.; Korn, S.; Göbl, C.; Berninger, L.; Düsterhus, S.; Hellmich, U.A.; Madl, T.; Kötter, P.; et al. The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin. J. Biol. Chem. 2015, 290, 28869–28886. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.; Heinzmann, S.; Solovieva, I.; Entian, K.-D. Function of Lactococcus Lactis Nisin Immunity Genes NisI and NisFEG after Coordinated Expression in the Surrogate Host Bacillus subtilis. J. Biol. Chem. 2003, 278, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Ra, R.; Beerthuyzen, M.M.; de Vos, W.M.; Saris, P.E.J.; Kuipers, O.P. Effects of Gene Disruptions in the Nisin Gene Cluster of Lactococcus lactis on Nisin Production and Producer Immunity. Microbiology 1999, 145 Pt 5, 1227–1233. [Google Scholar] [CrossRef]
- Siegers, K.; Entian, K.D. Genes Involved in Immunity to the Lantibiotic Nisin Produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 1995, 61, 1082–1089. [Google Scholar] [CrossRef]
- Koponen, O.; Takala, T.M.; Saarela, U.; Qiao, M.; Saris, P.E.J. Distribution of the NisI Immunity Protein and Enhancement of Nisin Activity by the Lipid-Free NisI. FEMS Microbiol. Lett. 2004, 231, 85–90. [Google Scholar] [CrossRef]
- AlKhatib, Z.; Lagedroste, M.; Zaschke, J.; Wagner, M.; Abts, A.; Fey, I.; Kleinschrodt, D.; Smits, S.H.J. The C-Terminus of Nisin Is Important for the ABC Transporter NisFEG to Confer Immunity in Lactococcus lactis. MicrobiologyOpen 2014, 3, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Li, R.; Liu, F.; Xu, H.; Li, B.; Yuan, Y.; Saris, P.E.J.; Qiao, M. Mu Insertion in FeuD Triggers the Increase in Nisin Immunity in Lactococcus lactis subsp. lactis N8. J. Appl. Microbiol. 2016, 120, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; Bayjanov, J.; Renckens, B.; Wels, M.; van Hijum, S.A.F.T.; Molenaar, D.; van Hylckama Vlieg, J.E.T. Complete Genome Sequence of Lactococcus lactis subsp. lactis KF147, a Plant-Associated Lactic Acid Bacterium. J. Bacteriol. 2010, 192, 2649–2650. [Google Scholar] [CrossRef]
- Halami, P.M.; Stein, T.; Chandrashekar, A.; Entian, K.-D. Maturation and Processing of SpaI, the Lipoprotein Involved in Subtilin Immunity in Bacillus subtilis ATCC 6633. Microbiol. Res. 2010, 165, 183–189. [Google Scholar] [CrossRef]
- Geiger, C.; Korn, S.M.; Häsler, M.; Peetz, O.; Martin, J.; Kötter, P.; Morgner, N.; Entian, K.-D. LanI-Mediated Lantibiotic Immunity in Bacillus subtilis: Functional Analysis. Appl. Environ. Microbiol. 2019, 85, e00534-19. [Google Scholar] [CrossRef]
- Christ, N.A.; Bochmann, S.; Gottstein, D.; Duchardt-Ferner, E.; Hellmich, U.A.; Düsterhus, S.; Kötter, P.; Güntert, P.; Entian, K.-D.; Wöhnert, J. The First Structure of a Lantibiotic Immunity Protein, SpaI from Bacillus subtilis, Reveals a Novel Fold. J. Biol. Chem. 2012, 287, 35286–35298. [Google Scholar] [CrossRef]
- Khosa, S.; Lagedroste, M.; Smits, S.H.J. Protein Defense Systems against the Lantibiotic Nisin: Function of the Immunity Protein NisI and the Resistance Protein NSR. Front. Microbiol. 2016, 7, 504. [Google Scholar] [CrossRef]
- Jeong, J.H.; Ha, S.C. Crystal Structure of NisI in a Lipid-Free Form, the Nisin Immunity Protein, from Lactococcus lactis. Antimicrob. Agents Chemother. 2018, 62, e01966-17. [Google Scholar] [CrossRef]
- Hoffmann, A.; Schneider, T.; Pag, U.; Sahl, H.-G. Localization and Functional Analysis of PepI, the Immunity Peptide of Pep5-Producing Staphylococcus epidermidis Strain 5. Appl. Environ. Microbiol. 2004, 70, 3263–3271. [Google Scholar] [CrossRef]
- Van Belkum, M.J.; Martin-Visscher, L.A.; Vederas, J.C. Structure and Genetics of Circular Bacteriocins. Trends Microbiol. 2011, 19, 411–418. [Google Scholar] [CrossRef]
- Maqueda, M.; Gálvez, A.; Bueno, M.M.; Sanchez-Barrena, M.J.; González, C.; Albert, A.; Rico, M.; Valdivia, E. Peptide AS-48: Prototype of a New Class of Cyclic Bacteriocins. Curr. Protein Pept. Sci. 2004, 5, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Maqueda, M.; Sánchez-Hidalgo, M.; Fernández, M.; Montalbán-López, M.; Valdivia, E.; Martínez-Bueno, M. Genetic Features of Circular Bacteriocins Produced by Gram-Positive Bacteria. FEMS Microbiol. Rev. 2008, 32, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsen, C.; Brede, D.A.; Nes, I.F.; Diep, D.B. Circular Bacteriocins: Biosynthesis and Mode of Action. Appl. Environ. Microbiol. 2014, 80, 6854–6862. [Google Scholar] [CrossRef]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Circular and Leaderless Bacteriocins: Biosynthesis, Mode of Action, Applications, and Prospects. Front. Microbiol. 2018, 9, 2085. [Google Scholar] [CrossRef] [PubMed]
- Gálvez, A.; Maqueda, M.; Valdivia, E.; Quesada, A.; Montoya, E. Characterization and Partial Purification of a Broad Spectrum Antibiotic AS-48 Produced by Streptococcus faecalis. Can. J. Microbiol. 1986, 32, 765–771. [Google Scholar] [CrossRef]
- Sánchez-Barrena, M.J.; Martınez-Ripoll, M.; Gálvez, A.; Valdivia, E.; Maqueda, M.; Cruz, V.; Albert, A. Structure of Bacteriocin AS-48: From Soluble State to Membrane Bound State. J. Mol. Biol. 2003, 334, 541–549. [Google Scholar] [CrossRef]
- Cebrián, R.; Martínez-Bueno, M.; Valdivia, E.; Albert, A.; Maqueda, M.; Sánchez-Barrena, M.J. The Bacteriocin AS-48 Requires Dimer Dissociation Followed by Hydrophobic Interactions with the Membrane for Antibacterial Activity. J. Struct. Biol. 2015, 190, 162–172. [Google Scholar] [CrossRef]
- Gálvez, A.; Maqueda, M.; Martínez-Bueno, M.; Valdivia, E. Permeation of Bacterial Cells, Permeation of Cytoplasmic and Artificial Membrane Vesicles, and Channel Formation on Lipid Bilayers by Peptide Antibiotic AS-48. J. Bacteriol. 1991, 173, 886–892. [Google Scholar] [CrossRef]
- Cruz, V.L.; Ramos, J.; Melo, M.N.; Martinez-Salazar, J. Bacteriocin AS-48 Binding to Model Membranes and Pore Formation as Revealed by Coarse-Grained Simulations. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2524–2531. [Google Scholar] [CrossRef]
- Gabrielsen, C.; Brede, D.A.; Hernández, P.E.; Nes, I.F.; Diep, D.B. The Maltose ABC Transporter in Lactococcus lactis Facilitates High-Level Sensitivity to the Circular Bacteriocin Garvicin ML. Antimicrob. Agents Chemother. 2012, 56, 2908–2915. [Google Scholar] [CrossRef]
- Gor, M.-C.; Vezina, B.; McMahon, R.M.; King, G.J.; Panjikar, S.; Rehm, B.H.A.; Martin, J.L.; Smith, A.T. Crystal Structure and Site-Directed Mutagenesis of Circular Bacteriocin Plantacyclin B21AG Reveals Cationic and Aromatic Residues Important for Antimicrobial Activity. Sci. Rep. 2020, 10, 17398. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Bueno, M.; Maqueda, M.; Gálvez, A.; Samyn, B.; Van Beeumen, J.; Coyette, J.; Valdivia, E. Determination of the Gene Sequence and the Molecular Structure of the Enterococcal Peptide Antibiotic AS-48. J. Bacteriol. 1994, 176, 6334–6339. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Valdivia, E.; Martínez-Bueno, M.; Fernández, M.; Soler-González, A.S.; Ramírez-Rodrigo, H.; Maqueda, M. Characterization of a New Operon, as-48EFGH, from the as-48 Gene Cluster Involved in Immunity to Enterocin AS-48. Appl. Environ. Microbiol. 2003, 69, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Cebrián, R.; Rodríguez-Ruano, S.; Martínez-Bueno, M.; Valdivia, E.; Maqueda, M.; Montalbán-López, M. Analysis of the Promoters Involved in Enterocin AS-48 Expression. PLoS ONE 2014, 9, e90603. [Google Scholar] [CrossRef]
- Martínez-Bueno, M.; Valdivia, E.; Gálvez, A.; Coyette, J.; Maqueda, M. Analysis of the Gene Cluster Involved in Production and Immunity of the Peptide Antibiotic AS-48 in Enterococcus faecalis. Mol. Microbiol. 1998, 27, 347–358. [Google Scholar] [CrossRef]
- Fernández, M.; Sánchez-Hidalgo, M.; García-Quintáns, N.; Martínez-Bueno, M.; Valdivia, E.; López, P.; Maqueda, M. Processing of As-48ABC RNA in AS-48 Enterocin Production by Enterococcus faecalis. J. Bacteriol. 2008, 190, 240–250. [Google Scholar] [CrossRef]
- Kemperman, R.; Jonker, M.; Nauta, A.; Kuipers, O.P.; Kok, J. Functional Analysis of the Gene Cluster Involved in Production of the Bacteriocin Circularin A by Clostridium beijerinckii ATCC 25752. Appl. Environ. Microbiol. 2003, 69, 5839–5848. [Google Scholar] [CrossRef]
- Van Belkum, M.J.; Martin-Visscher, L.A.; Vederas, J.C. Cloning and Characterization of the Gene Cluster Involved in the Production of the Circular Bacteriocin Barnocyclin A. Probiotics Antimicrob. Proteins 2010, 2, 218–225. [Google Scholar] [CrossRef]
- Borrero, J.; Brede, D.A.; Skaugen, M.; Diep, D.B.; Herranz, C.; Nes, I.F.; Cintas, L.M.; Hernández, P.E. Characterization of Garvicin ML, a Novel Circular Bacteriocin Produced by Lactococcus garvieae DCC43, Isolated from Mallard Ducks (Anas platyrhynchos). Appl. Environ. Microbiol. 2011, 77, 369–373. [Google Scholar] [CrossRef]
- Kalmokoff, M.L.; Cyr, T.D.; Hefford, M.A.; Whitford, M.F.; Teather, R.M. Butyrivibriocin AR10, a New Cyclic Bacteriocin Produced by the Ruminal Anaerobe Butyrivibrio Fibrisolvens AR10: Characterization of the Gene and Peptide. Can. J. Microbiol. 2003, 49, 763–773. [Google Scholar] [CrossRef]
- Wirawan, R.E.; Swanson, K.M.; Kleffmann, T.; Jack, R.W.; Tagg, J.R. 2007 Uberolysin: A Novel Cyclic Bacteriocin Produced by Streptococcus uberis. Microbiology 2007, 153, 1619–1630. [Google Scholar] [CrossRef]
- Belguesmia, Y.; Naghmouchi, K.; Chihib, N.-E.; Drider, D. Class IIa bacteriocins: Current knowledge and perspectives. In Prokaryotic Antimicrobial Peptides: From Genes to Applications; Drider, D., Rebuffat, S., Eds.; Springer: New York, NY, USA, 2011; pp. 171–195. ISBN 978-1-4419-7692-5. [Google Scholar]
- Belguesmia, Y.; Madi, A.; Sperandio, D.; Merieau, A.; Feuilloley, M.; Prévost, H.; Drider, D.; Connil, N. Growing Insights into the Safety of Bacteriocins: The Case of Enterocin S37. Res. Microbiol. 2011, 162, 159–163. [Google Scholar] [CrossRef]
- Nissen-Meyer, J.; Rogne, P.; Oppegard, C.; Haugen, H.S.; Kristiansen, P.E. Structure-Function Relationships of the Non-Lanthionine-Containing Peptide (Class II) Bacteriocins Produced by Gram-Positive Bacteria. Curr. Pharm. Biotechnol. 2009, 10, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Chikindas, M.L.; García-Garcerá, M.J.; Driessen, A.J.; Ledeboer, A.M.; Nissen-Meyer, J.; Nes, I.F.; Abee, T.; Konings, W.N.; Venema, G. Pediocin PA-1, a Bacteriocin from Pediococcus Acidilactici PAC1.0, Forms Hydrophilic Pores in the Cytoplasmic Membrane of Target Cells. Appl. Environ. Microbiol. 1993, 59, 3577–3584. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ludescher, R.D.; Montville, T.J. Electrostatic Interactions, but Not the YGNGV Consensus Motif, Govern the Binding of Pediocin PA-1 and Its Fragments to Phospholipid Vesicles. Appl. Environ. Microbiol. 1997, 63, 4770–4777. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shapira, R.; Eisenstein, M.; Montville, T.J. Functional Characterization of Pediocin PA-1 Binding to Liposomes in the Absence of a Protein Receptor and Its Relationship to a Predicted Tertiary Structure. Appl. Environ. Microbiol. 1997, 63, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Fimland, G.; Jack, R.; Jung, G.; Nes, I.F.; Nissen-Meyer, J. The Bactericidal Activity of Pediocin SPA-1 Is Specifically Inhibited by a 15-Fmer Fragment That Spans the Bacteriocin from the Center toward the C Terminus. Appl. Environ. Microbiol. 1998, 64, 5057–5060. [Google Scholar] [CrossRef]
- Diep, D.B.; Skaugen, M.; Salehian, Z.; Holo, H.; Nes, I.F. Common Mechanisms of Target Cell Recognition and Immunity for Class II Bacteriocins. Proc. Natl. Acad. Sci. USA 2007, 104, 2384–2389. [Google Scholar] [CrossRef]
- Kjos, M.; Salehian, Z.; Nes, I.F.; Diep, D.B. An Extracellular Loop of the Mannose Phosphotransferase System Component IIC Is Responsible for Specific Targeting by Class IIa Bacteriocins. J. Bacteriol. Res. 2010, 192, 5906–5913. [Google Scholar] [CrossRef]
- Ríos Colombo, N.S.; Chalón, M.C.; Navarro, S.A.; Bellomio, A. Pediocin-like Bacteriocins: New Perspectives on Mechanism of Action and Immunity. Curr. Genet. 2018, 64, 345–351. [Google Scholar] [CrossRef]
- Barraza, D.E.; Colombo, N.S.R.; Galván, A.E.; Acuña, L.; Minahk, C.J.; Bellomio, A.; Chalón, M.C. New Insights into Enterocin CRL35: Mechanism of Action and Immunity Revealed by Heterologous Expression in Escherichia coli. Mol. Microbiol. 2017, 105, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, G.; Wang, C.; Ren, F.; Hao, Y. Both IIC and IID Components of Mannose Phosphotransferase System Are Involved in the Specific Recognition between Immunity Protein PedB and Bacteriocin-Receptor Complex. PLoS ONE 2016, 11, e0164973. [Google Scholar] [CrossRef] [PubMed]
- Nissen-Meyer, J.; Oppegård, C.; Rogne, P.; Haugen, H.S.; Kristiansen, P.E. Structure and Mode-of-Action of the Two-Peptide (Class-IIb) Bacteriocins. Probiotics Antimicro. Prot. 2010, 2, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Senes, A.; Engel, D.E.; DeGrado, W.F. Folding of Helical Membrane Proteins: The Role of Polar, GxxxG-like and Proline Motifs. Curr. Opin. Struct. Biol. 2004, 14, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Kjos, M.; Oppegård, C.; Diep, D.B.; Nes, I.F.; Veening, J.-W.; Nissen-Meyer, J.; Kristensen, T. Sensitivity to the Two-Peptide Bacteriocin Lactococcin G Is Dependent on UppP, an Enzyme Involved in Cell-Wall Synthesis. Mol. Microbiol. 2014, 92, 1177–1187. [Google Scholar] [CrossRef]
- Oppegård, C.; Emanuelsen, L.; Thorbek, L.; Fimland, G.; Nissen-Meyer, J. The Lactococcin G Immunity Protein Recognizes Specific Regions in Both Peptides Constituting the Two-Peptide Bacteriocin Lactococcin G. Appl. Environ. Microbiol. 2010, 76, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, N.; Zendo, T.; Koga, S.; Shigeri, Y.; Sonomoto, K. 2015 Molecular Characterization of the Genes Involved in the Secretion and Immunity of Lactococcin Q, a Two-Peptide Bacteriocin Produced by Lactococcus lactis QU 4. Microbiology 2015, 161, 2069–2078. [Google Scholar] [CrossRef]
- Kjos, M.; Snipen, L.; Salehian, Z.; Nes, I.F.; Diep, D.B. The Abi Proteins and Their Involvement in Bacteriocin Self-Immunity. J. Bacteriol. 2010, 192, 2068–2076. [Google Scholar] [CrossRef]
- Britton, A.P.; Ende SR van, d.e.r.; Belkum MJ, v.a.n.; Martin-Visscher, L.A. The Membrane Topology of Immunity Proteins for the Two-Peptide Bacteriocins Carnobacteriocin XY, Lactococcin G, and Lactococcin MN Shows Structural Diversity. MicrobiologyOpen 2020, 9, e00957. [Google Scholar] [CrossRef]
- Yoneyama, F.; Imura, Y.; Ohno, K.; Zendo, T.; Nakayama, J.; Matsuzaki, K.; Sonomoto, K. Peptide-Lipid Huge Toroidal Pore, a New Antimicrobial Mechanism Mediated by a Lactococcal Bacteriocin, Lacticin Q. Antimicrob. Agents Chemother. 2009, 53, 3211–3217. [Google Scholar] [CrossRef]
- Netz, D.J.; Bastos, M.D.; Sahl, H.G. Mode of Action of the Antimicrobial Peptide Aureocin A53 from Staphylococcus aureus. Appl. Environ. Microbiol. 2002, 68, 5274–5280. [Google Scholar] [CrossRef]
- Uzelac, G.; Kojic, M.; Lozo, J.; Aleksandrzak-Piekarczyk, T.; Gabrielsen, C.; Kristensen, T.; Nes, I.F.; Diep, D.B.; Topisirovic, L. A Zn-Dependent Metallopeptidase Is Responsible for Sensitivity to LsbB, a Class II Leaderless Bacteriocin of Lactococcus lactis subsp. lactis BGMN1-5. J. Bacteriol. Res. 2013, 195, 5614–5621. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, J.D.; Coelho, M.L.; Ceotto, H.; Potter, A.; Fleming, L.R.; Salehian, Z.; Nes, I.F.; Bastos, M.D. Genes Involved in Immunity to and Secretion of Aureocin A53, an Atypical Class II Bacteriocin Produced by Staphylococcus aureus A53. J. Bacteriol. 2012, 194, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.L.V.; Coutinho, B.G.; Cabral da Silva Santos, O.; Nes, I.F.; Bastos, M. Immunity to the Staphylococcus aureus Leaderless Four-Peptide Bacteriocin Aureocin A70 Is Conferred by AurI, an Integral Membrane Protein. Res. Microbiol. 2014, 165, 50–59. [Google Scholar] [CrossRef]
- Iwatani, S.; Yoneyama, F.; Miyashita, S.; Zendo, T.; Nakayama, J.; Sonomoto, K. Identification of the Genes Involved in the Secretion and Self-Immunity of Lacticin Q, an Unmodified Leaderless Bacteriocin from Lactococcus lactis QU 5. Microbiology 2012, 158, 2927–2935. [Google Scholar] [CrossRef]
- Holo, H.; Nilssen, O.; Nes, I.F. Lactococcin A, a New Bacteriocin from Lactococcus lactis subsp. cremoris: Isolation and Characterization of the Protein and Its Gene. J. Bacteriol. 1991, 173, 3879–3887. [Google Scholar] [CrossRef]
- Martínez, B.; Böttiger, T.; Schneider, T.; Rodríguez, A.; Sahl, H.-G.; Wiedemann, I. Specific Interaction of the Unmodified Bacteriocin Lactococcin 972 with the Cell Wall Precursor Lipid II. Appl. Environ. Microbiol. 2008, 74, 4666–4670. [Google Scholar] [CrossRef]
- Venema, K.; Haverkort, R.E.; Abee, T.; Haandrikman, A.J.; Leenhouts, K.J.; de Leij, L.; Venema, G.; Kok, J. Mode of Action of LciA, the Lactococcin a Immunity Protein. Mol. Microbiol. 1994, 14, 521–532. [Google Scholar] [CrossRef]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a Cell Wall-Degrading Bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.J.; Rawlings, N.D. Families and Clans of Cysteine Peptidases. Perspect. Drug Discov. Des. 1996, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Vilas, A. Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Formatex Research Center: Norristown, PA, USA, 2010; ISBN 978-84-614-6195-0. [Google Scholar]
- Kim, S.J.; Chang, J.; Singh, M. Peptidoglycan Architecture of Gram-Positive Bacteria by Solid-State NMR. Biochim. Biophys. Acta 2015, 1848, 350–362. [Google Scholar] [CrossRef]
- Khan, H.; Flint, S.H.; Yu, P.-L. Determination of the Mode of Action of Enterolysin A, Produced by Enterococcus faecalis B9510. J. Appl. Microbiol. 2013, 115, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Bastos, M.D.; Coutinho, B.G.; Coelho, M.L. Lysostaphin: A Staphylococcal Bacteriolysin with Potential Clinical Applications. Pharmaceuticals 2010, 3, 1139–1161. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, X.; Zhang, X.; Wu, H.; Zou, Y.; Li, P.; Sun, C.; Xu, W.; Liu, F.; Wang, D. Class III Bacteriocin Helveticin-M Causes Sublethal Damage on Target Cells through Impairment of Cell Wall and Membrane. J. Ind. Microbiol. Biotechnol. 2018, 45, 213–227. [Google Scholar] [CrossRef]
- Gargis, S.R.; Heath, H.E.; LeBlanc, P.A.; Dekker, L.; Simmonds, R.S.; Sloan, G.L. Inhibition of the Activity of Both Domains of Lysostaphin through Peptidoglycan Modification by the Lysostaphin Immunity Protein. Appl. Environ. Microbiol. 2010, 76, 6944–6946. [Google Scholar] [CrossRef][Green Version]
- Gargis, S.R.; Gargis, A.S.; Heath, H.E.; Heath, L.S.; LeBlanc, P.A.; Senn, M.M.; Berger-Bächi, B.; Simmonds, R.S.; Sloan, G.L. Zif, the Zoocin a Immunity Factor, Is a FemABX-like Immunity Protein with a Novel Mode of Action. Appl. Environ. Microbiol. 2009, 75, 6205–6210. [Google Scholar] [CrossRef]
- Joerger, M.C.; Klaenhammer, T.R. Characterization and Purification of Helveticin J and Evidence for a Chromosomally Determined Bacteriocin Produced by Lactobacillus helveticus 481. J. Bacteriol. 1986, 167, 439–446. [Google Scholar] [CrossRef]
- Collins, F.W.J.; O’Connor, P.M.; O’Sullivan, O.; Gómez-Sala, B.; Rea, M.C.; Hill, C.; Ross, R.P. Bacteriocin Gene-Trait Matching across the Complete Lactobacillus Pan-Genome. Sci. Rep. 2017, 7, 3481. [Google Scholar] [CrossRef]
- Gálvez, A.; Abriouel, H.; López, R.L.; Ben Omar, N. Bacteriocin-Based Strategies for Food Biopreservation. Int. J. Food Microbiol. 2007, 120, 51–70. [Google Scholar] [CrossRef]
- Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Dusemund, B.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gundert-Remy, U.; Kuhnle, G.G.; et al. Safety of Nisin (E 234) as a Food Additive in the Light of New Toxicological Data and the Proposed Extension of Use. EFSA J. 2017, 15, e05063. [Google Scholar] [CrossRef] [PubMed]
- Faustman, C.; Aaron, D.; Negowetti, N.; Leib, E.B. Ten Years Post-GAO Assessment, FDA Remains Uninformed of Potentially Harmful GRAS Substances in Foods. Crit. Rev. Food Sci. Nutr. 2021, 61, 1260–1268. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. An audience with … Jim O’Neill. Nat. Rev. Drug Discov. 2016, 15, 526. [Google Scholar] [CrossRef]
- Mesa-Pereira, B.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Heterologous Expression of Biopreservative Bacteriocins with a View to Low Cost Production. Front. Microbiol. 2018, 9, 1654. [Google Scholar] [CrossRef]
- Hols, P.; Ledesma-García, L.; Gabant, P.; Mignolet, J. Mobilization of Microbiota Commensals and Their Bacteriocins for Therapeutics. Trends Microbiol. 2019, 27, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Drider, D. Gut Microbiota Is an Important Source of Bacteriocins and Their In Situ Expression Can Be Explored for Treatment of Bacterial Infections. Probiotics Antimicrob. Proteins 2021. [Google Scholar] [CrossRef]
- Zgheib, H.; Drider, D.; Belguesmia, Y. Broadening and Enhancing Bacteriocins Activities by Association with Bioactive Substances. Int. J. Environ. Res. Public Health 2020, 17, 7835. [Google Scholar] [CrossRef]


| Class | Bacteriocin | Producer Bacteria | Applications | Reference |
|---|---|---|---|---|
| Class I Lantibiotic | Nisin | Lactococcus lactis spp. | Food industry: natural preservative, inhibits most of the food-borne pathogens | [18] |
| Human health: infection diseases, oral care, cancer therapy | [22] | |||
| GP15cin | Lactobacillus plantarum GP15 | Patented bacteriocin for control of unwanted bacteria in ethanol fermentation plants | [8] | |
| Salivaricin A2/B | Streptococcus salivarius K12 | Active against Halitosis producing bacteria and Streptococcus pyogenes, which causes pharyngitis | [23,24] | |
| Lacticin 3147 | Lactococcus lactis 3147 | Inhibits the growth of many Gram-positive food-borne pathogens | [25] | |
| Treatment for bovine mastitis | [26] | |||
| Class I cyclic | Enterocin AS-48 | Enterococcus faecalis subsp. liquefaciens | promising perspectives to be used as biopreservatives in food | [27] |
| Active against uropathogenic enterococci | [28] | |||
| Class IIa Pediocin-like | Pediocin PA-1 | Pediococcus acidilactici PAC1.0 | Food industry: natural preservative | [29] |
| Potential activity against fish pathogens | [30] | |||
| ST4V | Enterococcus mundtii ST4SA | antibacterial and antiviral activity | [31] | |
| effective against middle ear infections | [32] | |||
| Staphylococcin 188 | Staphylococcus aureu AB188 | Anti-enterococci and antiviral activity | [33] | |
| Enterocin CRL35 | Enterococcus faecium CRL35 | Anti-Gram-positive pathogens and anti-Gram-negative pathogens when hybridized to microcin V | [34] | |
| ST5H | Enterococcus faecium ST5Ha | Active against human food pathogens and virus | [35] | |
| Fermenticin HV6b | Lactobacillus fermentum HV6b MTCC | inhibits pathogens which causes vaginal infections in humans and has a spermicidal activity. | [36] | |
| Pediocin K2a2–3 | Pediococcus acidilactici K2a2–3 | Anti-cancer activity against HT-29 colon adenocarcinoma cells | [37] | |
| Class IIb two-peptide | Durancin A5-11a/b | Enterococcus durans A5-11 | Antifungical activity | [38] |
| Plantaricin EF | Lactobacillus plantarum NCIMB8826-R | Anti-inflammatory effect and reduction of weight in obese mice | [39] | |
| Abp118 | Lactobacillus salivarius UCC118 | Enhancement of the bacteria probiotic effect and modulation of the mouse and pig intestinal microbiota | [40] | |
| Class IIc Leaderless | Enterocin SL-5 | Enterococcus faecalis SL-5 | Active against inflammatory acne lesions caused by Propionibacterium acnes | [41] |
| Class III | Lysostaphin | Staphylococcus simulans | Active against Staphylococcus aureus and other pathogens of endometritis sows | [42,43] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Ramos, A.; Madi-Moussa, D.; Coucheney, F.; Drider, D. Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins. Microorganisms 2021, 9, 2107. https://doi.org/10.3390/microorganisms9102107
Pérez-Ramos A, Madi-Moussa D, Coucheney F, Drider D. Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins. Microorganisms. 2021; 9(10):2107. https://doi.org/10.3390/microorganisms9102107
Chicago/Turabian StylePérez-Ramos, Adrián, Désiré Madi-Moussa, Françoise Coucheney, and Djamel Drider. 2021. "Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins" Microorganisms 9, no. 10: 2107. https://doi.org/10.3390/microorganisms9102107
APA StylePérez-Ramos, A., Madi-Moussa, D., Coucheney, F., & Drider, D. (2021). Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins. Microorganisms, 9(10), 2107. https://doi.org/10.3390/microorganisms9102107







