Serra da Estrela PDO Cheese Microbiome as Revealed by Next Generation Sequencing
Abstract
:1. Introduction
2. Materials and Methods
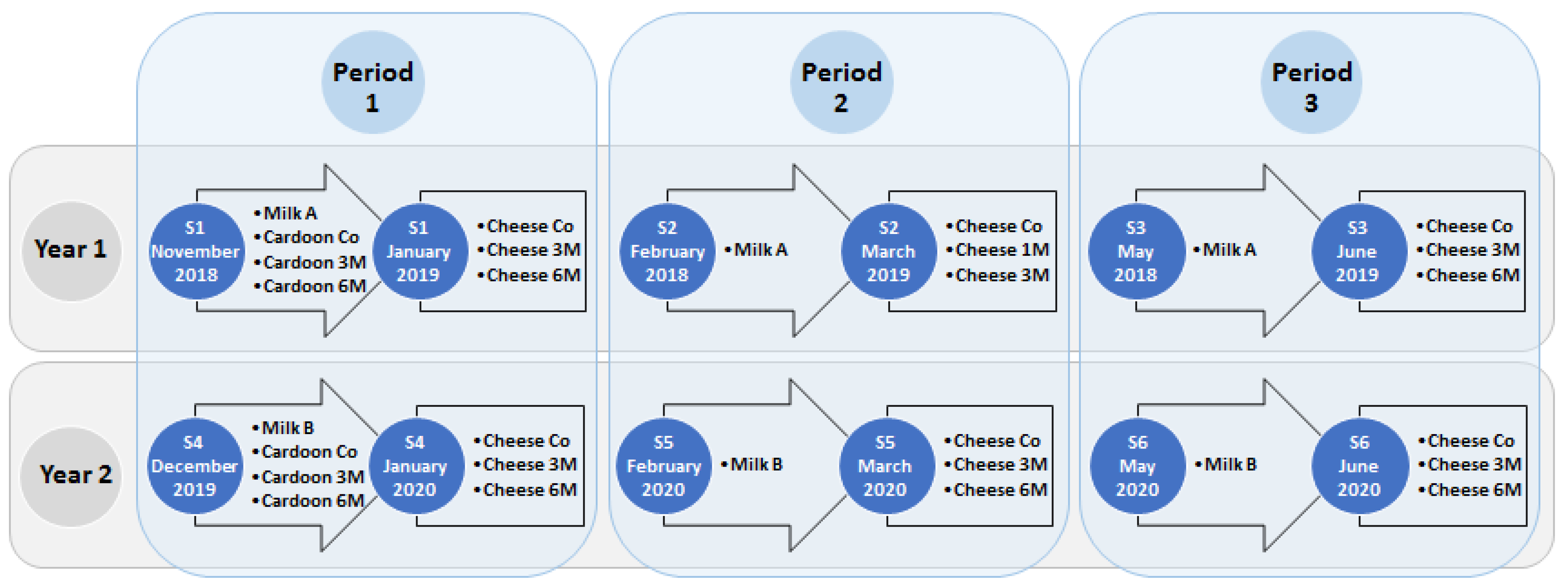
2.1. Cheese Manufacture and Sampling Strategy
2.2. Total DNA Extraction
2.3. Sequencing Preparation, Run and Processing
2.4. Bioinformatics and Statistical Analysis
2.5. Nucleotide Sequences Accession Number
3. Results and Discussion
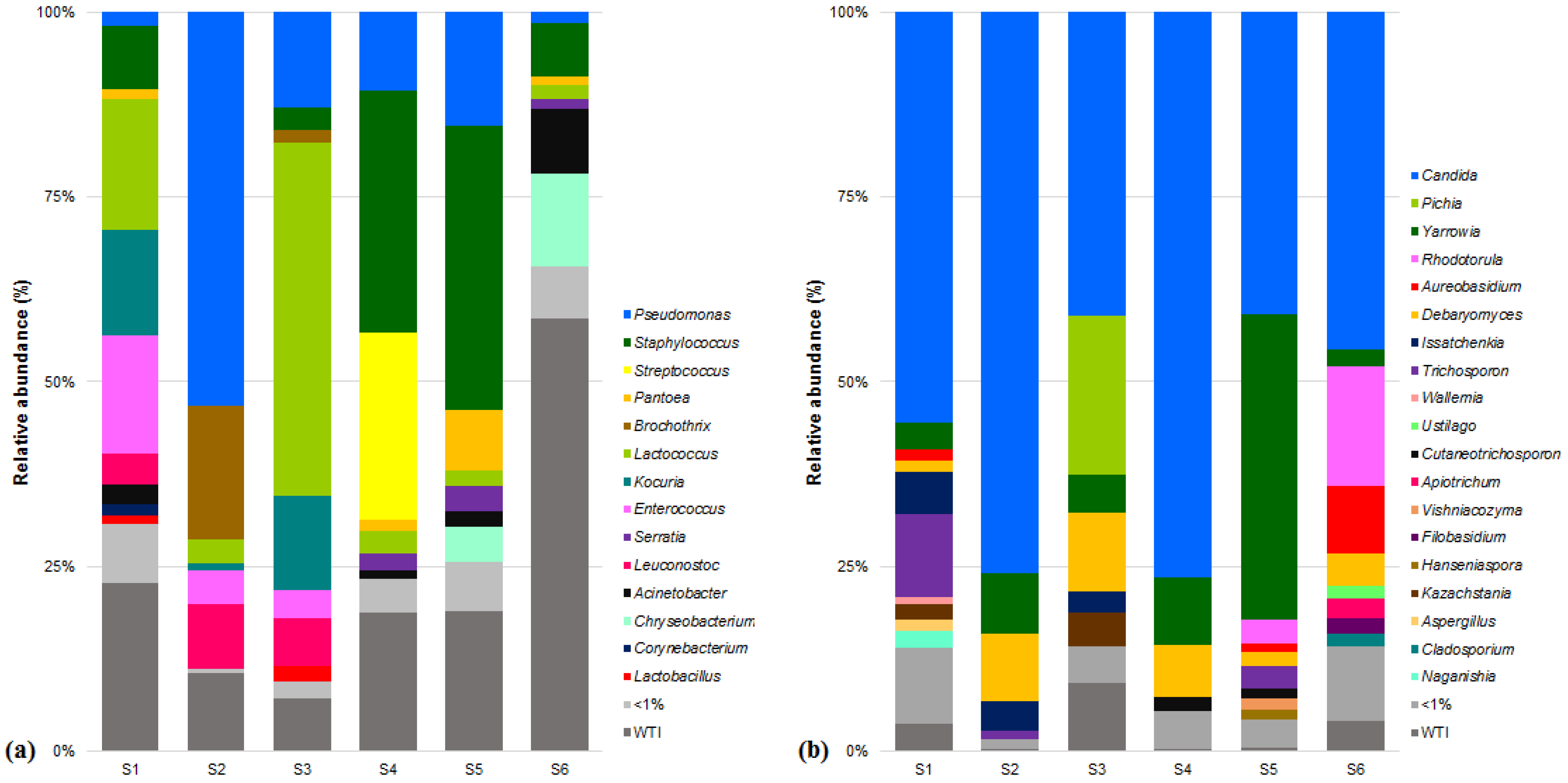
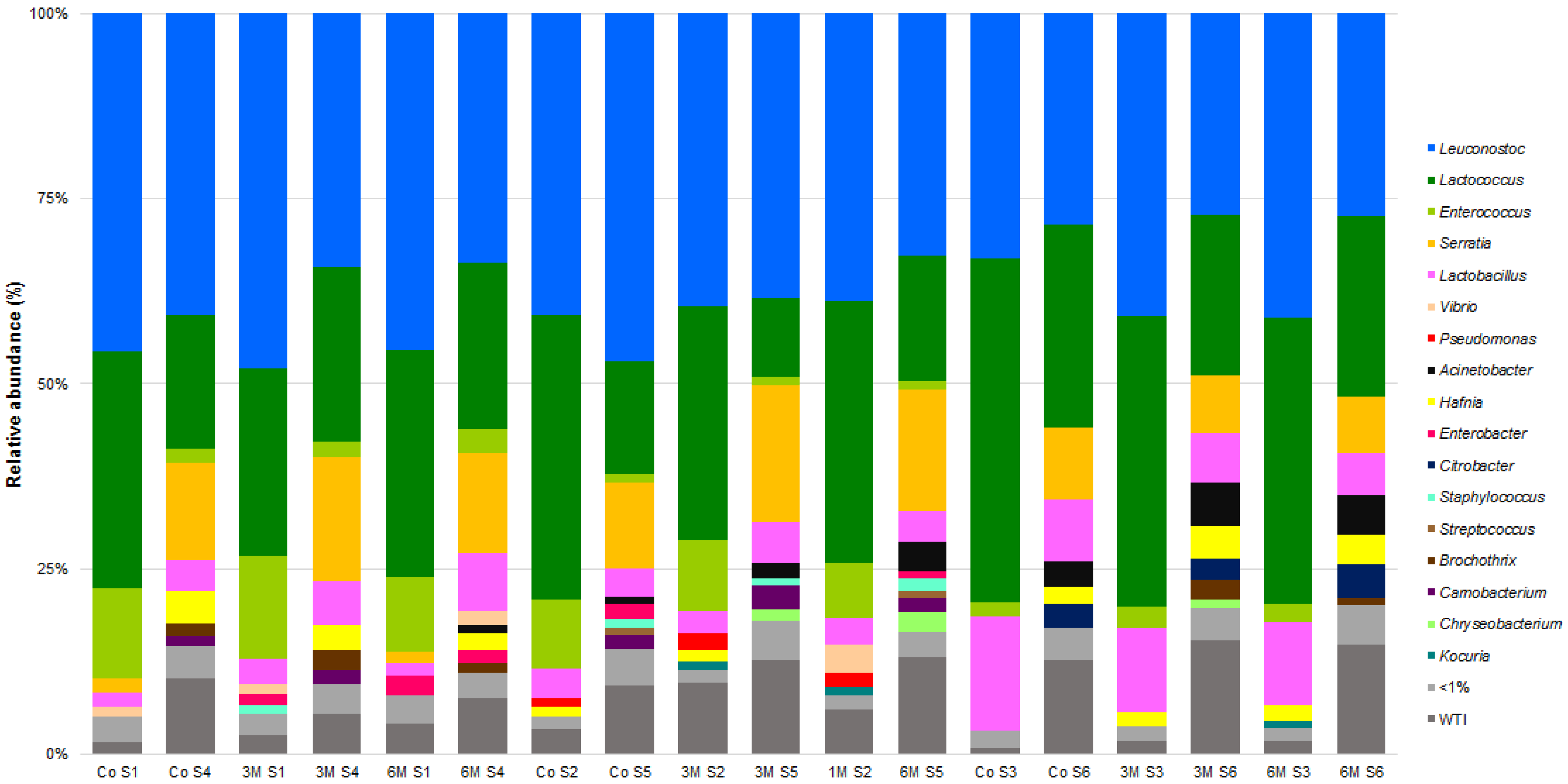
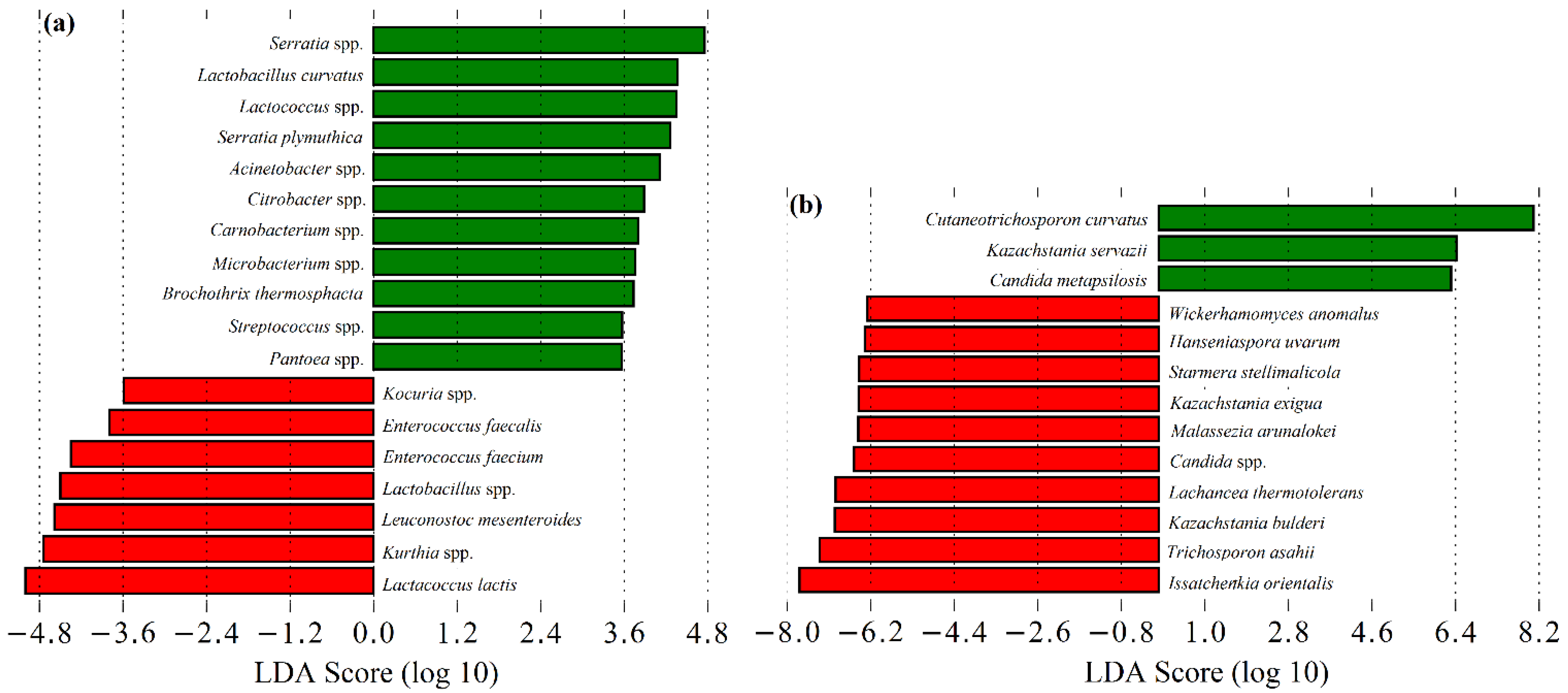
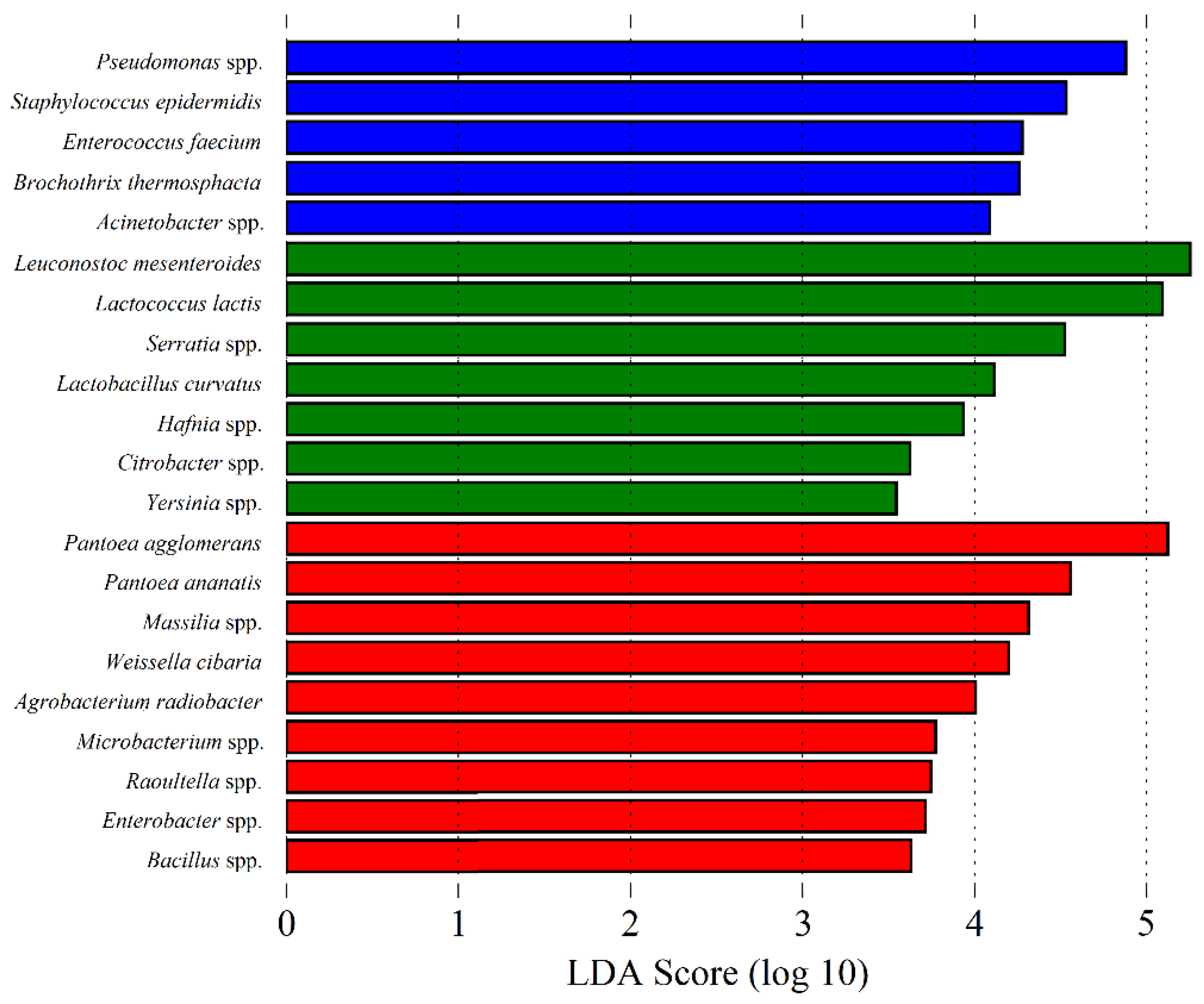
3.1. Microbial Community of Raw Ewes’ Milk
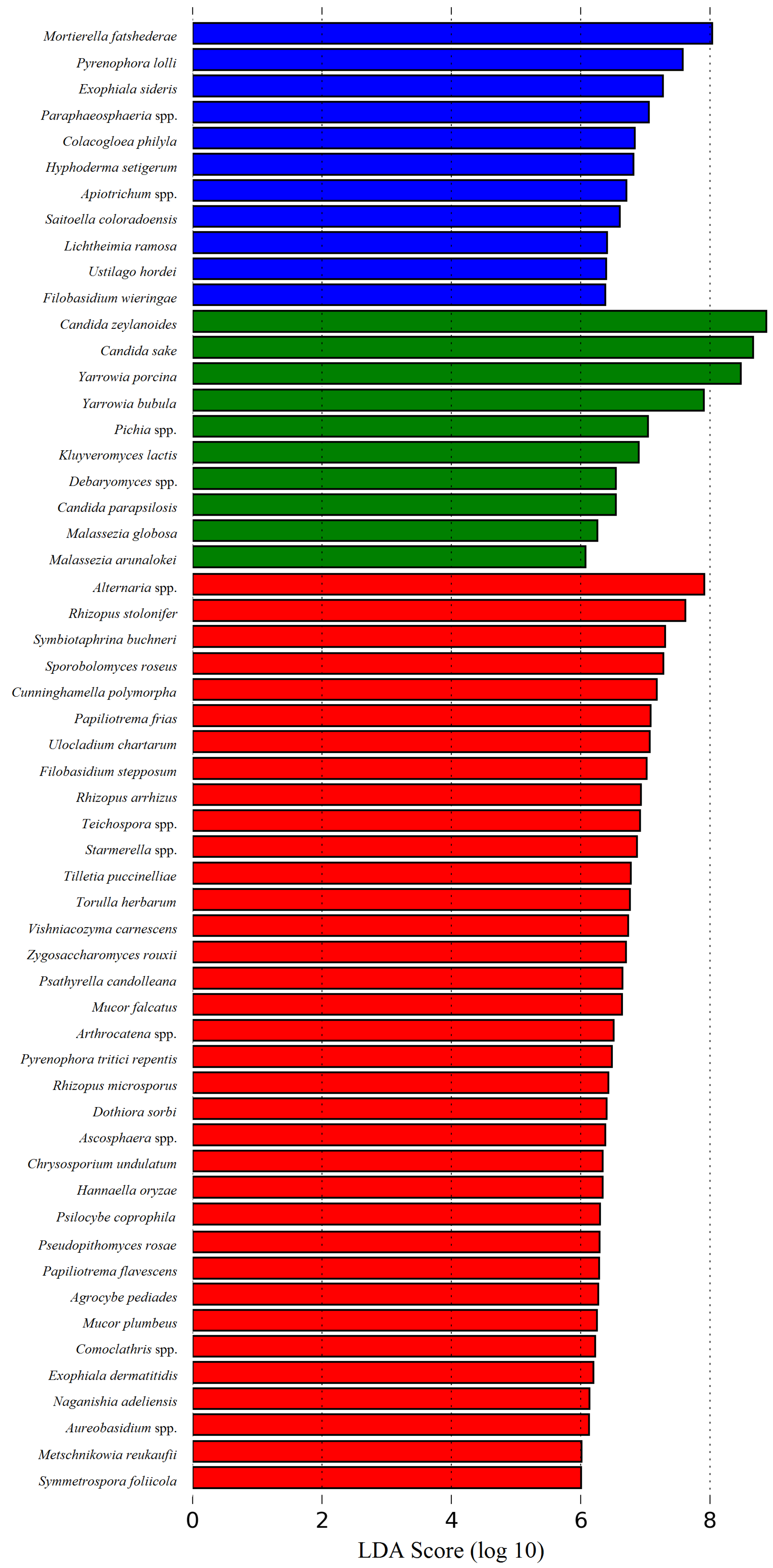
3.2. Microbial Community of Cardoon
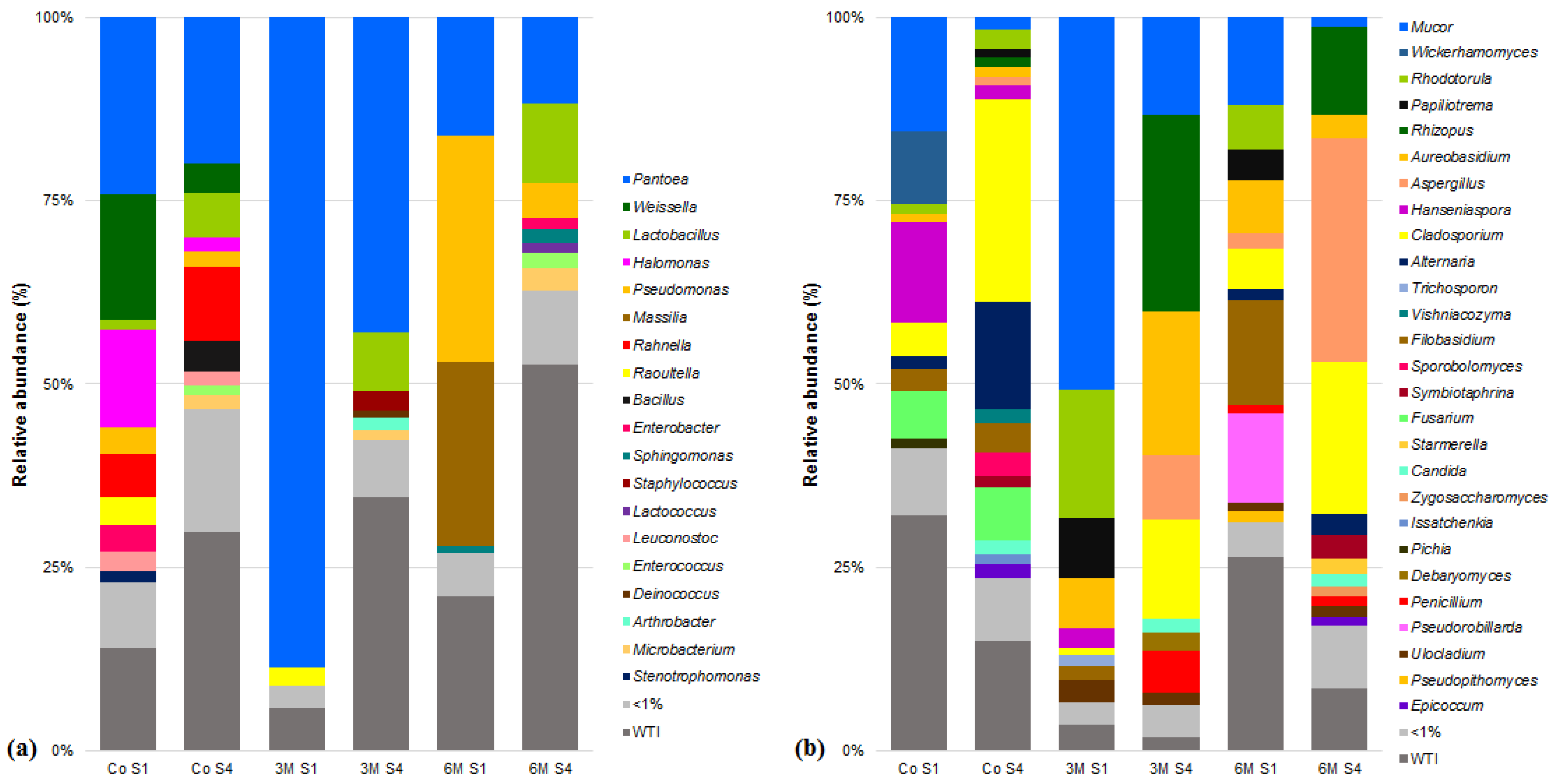
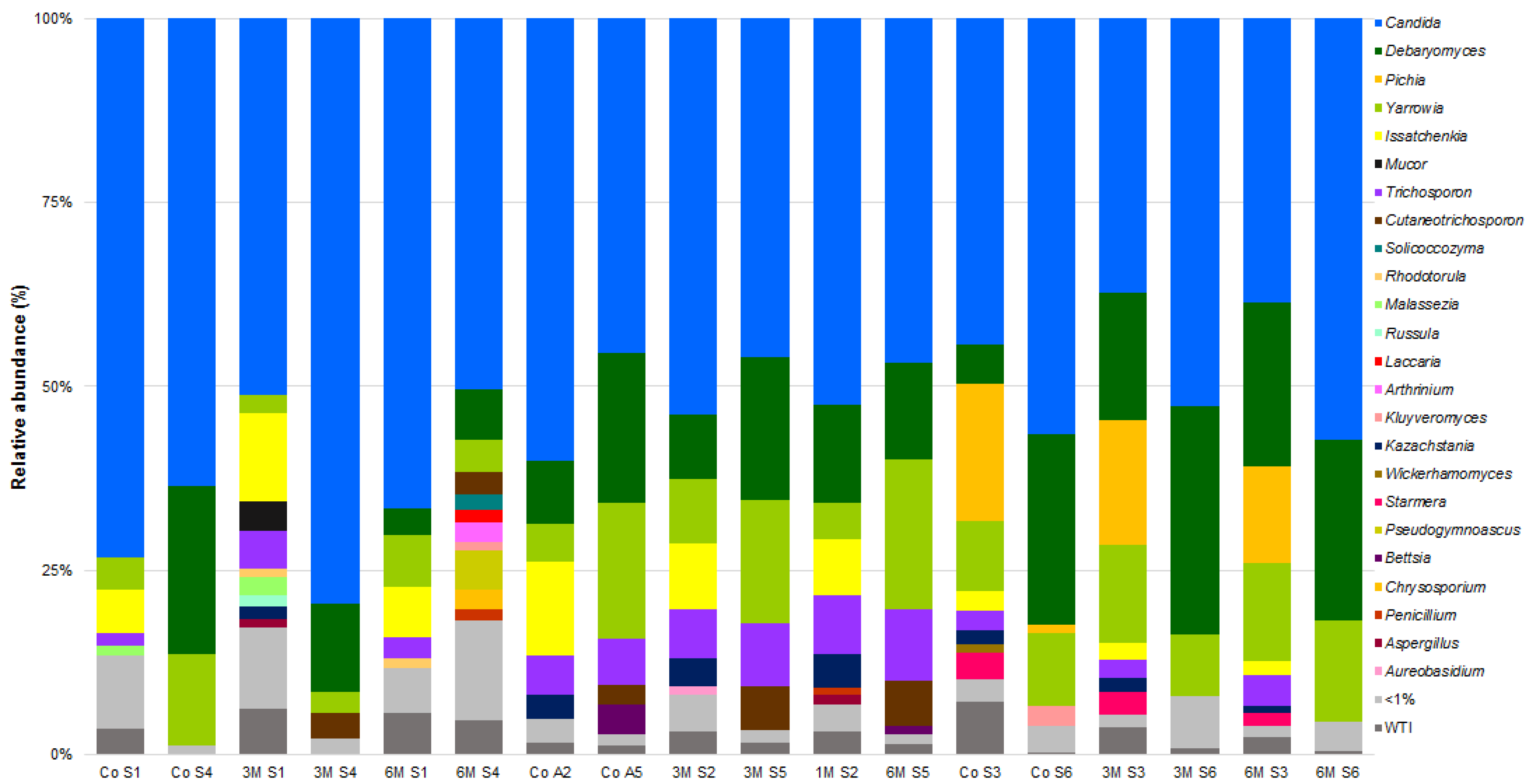
3.3. Microbial Community of Serra da Estrela Cheese
3.4. Matrices Taxa Variability and Core Community
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morales, M.; Bintsis, T.; Alichanidis, E.; Herian, K.; Jelen, P.; Hynes, E.R.; Perotti, M.C.; Bergamini, C.V.; Attard, E.; Grupetta, A.; et al. Soft cheeses (with rennet). In Global Cheesemaking Technology; Papademas, P., Bintsis, T., Eds.; Jonh Wiley & Sons, Ltd: Hoboken, NJ, USA, 2017; pp. 301–325. [Google Scholar] [CrossRef]
- Macedo, A.C.; Malcata, F.X.; Oliveira, J.C. The technology, Chemistry and Microbiology of Serra Cheese: A review. J. Dairy Sci. 1993, 76, 1725–1739. [Google Scholar] [CrossRef]
- Tavaria, F.K.; Malcata, F.X. On the microbiology of Serra da Estrela cheese: Geographical and chronological considerations. Food Microbiol. 2000, 17, 293–304. [Google Scholar] [CrossRef]
- Inácio, R.S.; Gomes, A.; Saraiva, J.A. Serra da Estrela cheese: A review. J. Food Process. Preserv. 2000, 44, e14412. [Google Scholar] [CrossRef]
- Tavaria, F.K.; Malcata, F.X. Microbiological Characterization of Serra da Estrela Cheese throughout Its Appellation d’Origine Protégée Region. J. Food Prot. 1998, 61, 601–607. [Google Scholar] [CrossRef]
- Macedo, A.C.; Malcata, F.X.; Hogg, T.A. Microbiological profile in Serra ewes’ cheese during ripening. J. Appl. Bacteriol. 1995, 79, 1–11. [Google Scholar] [CrossRef]
- Macedo, A.C.; Costa, M.L.; Malcata, F.X. Changes in the Microflora of Serra Cheese Evolution Throughout Ripening Time, Lactation Period and Axial Location. Int. Dairy J. 1996, 6, 79–94. [Google Scholar] [CrossRef]
- Dahl, S.; Tavaria, F.K.; Malcata, F.X. Relationships between flavor and microbiological profiles in Serra da Estrela cheese throughout ripening. Int. Dairy J. 2000, 10, 255–262. [Google Scholar] [CrossRef]
- Dalmasso, A.; Rio, M.; Civera, T.; Pattono, D.; Cardazzo, B.; Bottero, M.T. Characterization of microbiota in Plaisentif cheese by high–throughput sequencing. LWT Food Sci. Technol. 2016, 69, 490–496. [Google Scholar] [CrossRef]
- Park, W.; Yoo, J.; Oh, S.; Ham, J.; Jeong, S.; Kim, Y. Microbiological Characteristics of Gouda Cheese Manufactured with Pasteurized and Raw Milk during Ripening Using Next Generation Sequencing. Food Sci. Anim. Resour. 2019, 39, 585–600. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Pitino, I.; Ribbera, A.; Caggia, C. Pecorino Crotonese cheese: Study of bacterial population and flavour compounds. Food Microbiol. 2010, 27, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Filippis, F.; Parente, E.; Ercolini, D. Metagenomics insights into food fermentations. Microb. Biotechnol. 2017, 10, 91–102. [Google Scholar] [CrossRef]
- Irlinger, F.; Layec, S.; Hélinck, S.; Dugat-Bony, E. Cheese rind microbial communities: Diversity, composition and origin. FEMS Microbiol. Lett. 2015, 362, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonnala, B.R.; McSweeney, P.; Sheehan, J.J.; Cotter, P.D. Sequencing of the Cheese Microbiome and Its Relevance to Industry. Front. Microbiol. 2018, 9, 1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, O.; Cotter, P.D. Microbiota of Raw Milk and Raw Milk Cheeses. In Cheese—Chemistry, Physics and Microbiology, 4th ed.; McSweeney, P., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 301–316. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High–throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef] [Green Version]
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Quirasco, B.M. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 116–127. [Google Scholar] [CrossRef]
- Fuka, M.M.; Wallisch, S.; Engel, M.; Welzl, G.; Havranek, J.; Schloter, M. Dynamics of bacterial communities during the ripening process of different Croatian cheese types derived from raw ewe’s milk cheeses. PLoS ONE 2013, 8, e80734. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Mo, L.; Pan, L.; Hou, Q.; Li, C.; Darima, I.; Yu, J. Using PacBio sequencing to investigate the bacterial microbiota of traditional Buryatian cottage cheese and comparison with Italian and Kazakhstan artisanal cheeses. J. Dairy Sci. 2018, 101, 6885–6896. [Google Scholar] [CrossRef] [Green Version]
- Riquelme, C.; Câmara, S.; Dapkevicius, M.; Vinuesa, P.; Silva, C.; Malcata, F.X.; Rego, O.A. Characterization of the bacterial biodiversity in Pico cheese (an artisanal Azorean food). Int. J. Food Microbiol. 2015, 192, 86–94. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.; Benito, M.; Córdoba, M.; Alvarenga, N.; Herrera, S. Yeast community in traditional Portuguese Serpa cheese by culture–dependent and –independent DNA approaches. Int. J. Food Microbiol. 2017, 262, 63–70. [Google Scholar] [CrossRef]
- Barracosa, P.; Rosa, N.; Barros, M.; Pires, E. Selected Cardoon (Cynara cardunculus L.) Genotypes Suitable for PDO Cheeses in Mediterranean Regions. Chem. Biodivers. 2018, 15, e1800110. [Google Scholar] [CrossRef]
- Lima, S.F.; Bicalho, M.; Bicalho, R.C. Evaluation of milk sample fractions for characterization of milk microbiota from healthy and clinical mastitis cows. PLoS ONE 2018, 13, e0193671. [Google Scholar] [CrossRef] [Green Version]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next–generation sequencing–based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.; Vasco–Palacios, A.; Thu, P.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- 16S Metagenomic Sequencing Library Preparation. Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. 2013. Available online: https://emea.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 14 December 2020).
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, e00127-16. [Google Scholar] [CrossRef] [Green Version]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging Findings Background. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantos, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME release for Fungi. UNITE Community 2020. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Good, I.J. The population frequencies of species and the estimation of population parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Chao, A. Non–parametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Chao, A.; Lee, S.M. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 1992, 87, 210–217. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum–likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Jaccard, P. Nouvelles recherches sur la distribution floral. Bull. Soc. Vard. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Sørensen, T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on danish commons. Biol. Skr. 1948, 5, 1–34. [Google Scholar]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [Green Version]
- Kruskal, W.; Wallis, W. Use of Ranks in One–Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web–based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Montel, M.C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delavenne, E.; Mounier, J.; Asmani, K.; Jany, J.L.; Barbier, G.; Le Blay, G. Fungal diversity in cow, goat and ewe milk. Int. J. Food Microbiol. 2011, 151, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Blanco, C.; Gutiérrez-Gil, B.; Puente-Sánchez, F.; Marina, H.; Tamames, J.; Acedo, A.; Arranz, J.J. Microbiota characterization of sheep milk and its association with somatic cell count using 16s rRNA gene sequencing. J. Anim. Breed Genet. 2020, 137, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Fadda, M.E.; Mossa, V.; Pisano, M.B.; Deplano, M.; Cosentino, S. Occurrence and characterization of yeasts isolated from artisanal Fiore Sardo cheese. Int. J. Food Microbiol. 2004, 15, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Gaya, P.; Medina, M.; Nuñez, M. Enterobacteriaceae, coliforms, faecal coliforms and salmonellas in raw ewes’ milk. J. Appl. Bacteriol. 1987, 62, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Salmerón, J.; Vega, C.; Pérez-Elortondo, F.; Albisu, M.; Barrón, L. Effect of pasteurization and seasonal variations in the microflora of ewe’s milk for cheesemaking. Food Microbiol. 2002, 19, 167–174. [Google Scholar] [CrossRef]
- Spanamberg, A.; Fraga, C.; Ferreiro, L.; Aguinsky, M.; Sanches, E.; Roehe, C.; Lautert, C.; Santurio, J. Yeasts in the Raw Ewe’s Milk. Acta Sci. Vet. 2014, 42, 1236. [Google Scholar]
- Fröhlich-Wyder, M.T.; Arias-Roth, E.; Jakob, E. Cheese yeasts. Yeast 2019, 36, 129–141. [Google Scholar] [CrossRef]
- Akdouche, L.; Aissi, M.; Saadi, A. Prevalence and Identification of Yeasts Responsible for Mastitis in Dairy Cattle Farms in the Sidi Lahcene Region in the Wilaya of Sidi Bel abbes–Algeria. J. Adv. Dairy Res. 2018, 6, 206. [Google Scholar] [CrossRef]
- Zaragoza, C.S.; Olivares, R.A.; Watty, A.E.; Moctezuma, L.; Tanaca, L.V. Yeasts isolation from bovine mammary glands under different mastitis status in the Mexican High Plateu. Rev. Iberoam. Micol. 2011, 28, 79–82. [Google Scholar] [CrossRef]
- Specification book—Serra da Estrela cheese protected designation of origin. 2011. Available online: https://tradicional.dgadr.gov.pt/images/prod_imagens/queijos/docs/CE_Queijo_Serra.pdf (accessed on 15 March 2021).
- Callon, C.; Duthoit, F.; Delbès, C.; Ferrand, M.; Frileux, Y.; Crémoux, R.; Montel, M. Stability of microbial communities in goat milk during a lactation year: Molecular approaches. Syst. Appl. Microbiol. 2007, 30, 547–560. [Google Scholar] [CrossRef]
- Conceição, C.; Martins, P.; Alvarenga, N.; Dias, J.; Lamy, E.; Garrido, L.; Gomes, S.; Freitas, S.; Belo, A.; Brás, T.; et al. Cynara cardunculus: Use in Cheesemaking and Pharmaceutical Applications. In Technological Approaches for Novel Applications in Dairy Processing; Kuka, N., Ed.; IntechOpen: London, UK, 2018; pp. 73–107. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Salguero, J.; Sánchez, E.; Gómez, R.; Mata, C.; Vioque, M.; Tejada, L. Preliminary study of microbiological quality of cardoons of the genus Cynara L. used in manufacture of traditional cheeses. Milchwissenschaft 1999, 54, 688–689. [Google Scholar]
- Gómez, R.; Sánchez, E.; Vioque, M.; Ferreira, J.; Tejada, L.; Fernández-Salguero, J. Microbiological characteristics of ewes’ milk cheese manufactured using aqueous extracts of flowers from various species of cardoon Cynara L. Milchwissenschaft 2001, 56, 16–19. [Google Scholar]
- Ratão, I. Microbiological and Chemical Characterization of Traditional Cheese Made from Milk Produced by the Algarvian Goat Breed. Ph.D. Thesis, Cranfield University, Cranfield, UK, 2011. [Google Scholar]
- Bhadra, B.; Rao, R.S.; Singh, P.K.; Sarkar, P.K.; Shivaji, S. Yeasts and yeast–like fungi associated with tree bark: Diversity and identification of yeasts producing extracellular endoxylanases. Curr. Microbiol. 2008, 56, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Egbuta, M.; Mwanza, M.; Babalola, O. A Review of the Ubiquity of Ascomycetes Filamentous Fungi in Relation to Their Economic and Medical Importance. Adv. Microbiol. 2016, 6, 1140–1158. [Google Scholar] [CrossRef] [Green Version]
- Fox, P.F.; McSweeney, P. Cheese: An Overview. In Cheese—Chemistry, Physics and Microbiology, 4th ed.; Cotter, P.D., Everett, D.W., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 5–21. [Google Scholar] [CrossRef]
- Tuszyński, T.; Satora, P. Microbiological characteristics of the Węgierka Zwykła plum orchard in submontane region. Pol. J. Food Nutr. Sci. 2003, 53, 43–48. [Google Scholar]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Beresford, T.P. Microbiome Changes During Ripening. In Cheese—Chemistry, Physics and Microbiology, 4th ed.; Cotter, P.D., Everett, D.W., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 389–409. [Google Scholar] [CrossRef]
- Irlinger, F.; Helinck, S.; Jany, J.L. Secondary and Adjunct Cultures. In Cheese—Chemistry, Physics and Microbiology, 4th ed.; Cotter, P.D., Everett, D.W., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 273–300. [Google Scholar] [CrossRef]
- Picon, A. Cheese Microbial Ecology and Safety. In Global Cheesemaking Technology: Cheese Quality and Characteristics, 1st ed.; Papademas, P., Bintsis, T., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017; pp. 71–99. [Google Scholar] [CrossRef]
- Guerreiro, J. Molecular Methods for Authentication of Protected Denomination of Origin (PDO) Cheeses. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2006. [Google Scholar]
- Cardinali, F.; Ferrocino, I.; Milanović, V.; Belleggia, L.; Corvaglia, M.R.; Garofalo, C.; Foligni, R.; Mannozzi, C.; Mozzon, M.; Cocolin, L.; et al. Microbial communities and volatile profile of Queijo de Azeitão PDO cheese, a traditional Mediterranean thistle-curdled cheese from Portugal. Food Res. Int. 2021, 147, 110537. [Google Scholar] [CrossRef]
- Kačániová, M.; Terentjeva, M.; Kunová, S.; Haščík, P.; Kowalczewski, P.Ł.; Štefániková, J. Diversity of microbiota in Slovak summer ewes’ cheese “Bryndza”. Open Life Sci. 2021, 16, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, I.; Cagno, R.; Buchin, S.; Angelis, M.; Gobbetti, M. Spatial Distribution of the Metabolically Active Microbiota within Italian PDO Ewes’ Milk Cheeses. PLoS ONE 2016, 11, e0153213. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Dias, S.; Potes, M.E.; Marinho, A.; Malfeito-Ferreira, M.; Loureiro, V. Characterisation of yeast flora isolated from an artisanal Portuguese ewes’ cheese. Int. J. Food Microbiol. 2000, 60, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Spyrelli, E.; Stamatiou, A.; Tassou, C.; Nychas, G.; Doulgeraki, A. Microbiological and Metagenomic Analysis to Assess the Effect of Container Material on the Microbiota of Feta Cheese during Ripening. Fermentation 2020, 6, 12. [Google Scholar] [CrossRef] [Green Version]
- Padilla, B.; Manzanares, P.; Belloch, C. Yeast species and genetic heterogeneity within Debaryomyces hansenii along the ripening process of traditional ewes’ and goats’ cheeses. Food Microbiol. 2014, 38, 160–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavoie, K.; Touchette, M.; St-Gelais, D.; Labrie, S. Characterization of the fungal microflora in raw milk and specialty cheeses of the province of Quebec. Dairy Sci. Technol. 2012, 92, 455–468. [Google Scholar] [CrossRef] [Green Version]
- Soliman, N.; Aly, S. Occurrence and identification of yeast species isolated from Egyptian Karish cheese. J. Yeast Fungal Res. 2011, 2, 59–64. [Google Scholar]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Pathogens in Cheese and Foodborne Illnesses. In Fundamentals of Cheese Science; Fox, P.F., Guinee, T.P., Cogan, T.M., McSweeney, P.L.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 681–713. [Google Scholar] [CrossRef]
- Parente, E.; Cogan, T.M.; Powell, I.B. Starter Cultures: General Aspects. In Cheese—Chemistry, Physics and Microbiology, 4th ed.; Cotter, P.D., Everett, D.W., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 201–226. [Google Scholar]
- Cavanagh, D.; Fitzgerald, G.F.; McAuliffe, O. From field to fermentation: The origins of Lactococcus lactis and its domestication to the dairy environment. Food Microbiol. 2015, 47, 45–61. [Google Scholar] [CrossRef]
- Surber, G.; Schäper, C.; Wefers, D.; Rohm, H.; Jaros, D. Exopolysaccharides from Lactococcus lactis affect manufacture, texture and sensory properties of concentrated acid milk gel suspensions (fresh cheese). Int. Dairy J. 2021, 112, 104854. [Google Scholar] [CrossRef]
- McAuliffe, O. Genetics of Lactic Acid Bacteria. In Cheese—Chemistry, Physics and Microbiology, 4th ed.; Cotter, P.D., Everett, D.W., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 227–247. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Kapila, S.; Mani, J.; Malik, R.K. Comparison of susceptibility to opsonic killing by in vitro human immune response of Enterococcus strains isolated from dairy products, clinical samples and probiotic preparation. Int. J. Food Microbiol. 2009, 128, 513–515. [Google Scholar] [CrossRef]
- Nagy, E.; Dlauchy, D.; Medeiros, A.O.; Péter, G.; Rosa, C.A. Yarrowia porcina sp. nov. and Yarrowia bubula f.a. sp. nov., two yeast species from meat and river sediment. Antonie Van Leeuwenhoek 2014, 105, 697–707. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocha, R.; Vaz Velho, M.; Santos, J.; Fernandes, P. Serra da Estrela PDO Cheese Microbiome as Revealed by Next Generation Sequencing. Microorganisms 2021, 9, 2007. https://doi.org/10.3390/microorganisms9102007
Rocha R, Vaz Velho M, Santos J, Fernandes P. Serra da Estrela PDO Cheese Microbiome as Revealed by Next Generation Sequencing. Microorganisms. 2021; 9(10):2007. https://doi.org/10.3390/microorganisms9102007
Chicago/Turabian StyleRocha, Rui, Manuela Vaz Velho, Joana Santos, and Paulo Fernandes. 2021. "Serra da Estrela PDO Cheese Microbiome as Revealed by Next Generation Sequencing" Microorganisms 9, no. 10: 2007. https://doi.org/10.3390/microorganisms9102007
APA StyleRocha, R., Vaz Velho, M., Santos, J., & Fernandes, P. (2021). Serra da Estrela PDO Cheese Microbiome as Revealed by Next Generation Sequencing. Microorganisms, 9(10), 2007. https://doi.org/10.3390/microorganisms9102007






