Development of a Genoserotyping Method for Salmonella Infantis Detection on the Basis of Pangenome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Comparative Genome Analysis
2.3. Primer Design for a Diagnostic PCR Assay
2.4. Conventional Serotyping through Antisera Agglutination
2.5. PCR and Real-Time PCR
2.6. Evaluation of Sensitivity, Specificity, and Accuracy of the Diagnostic Assay
3. Results
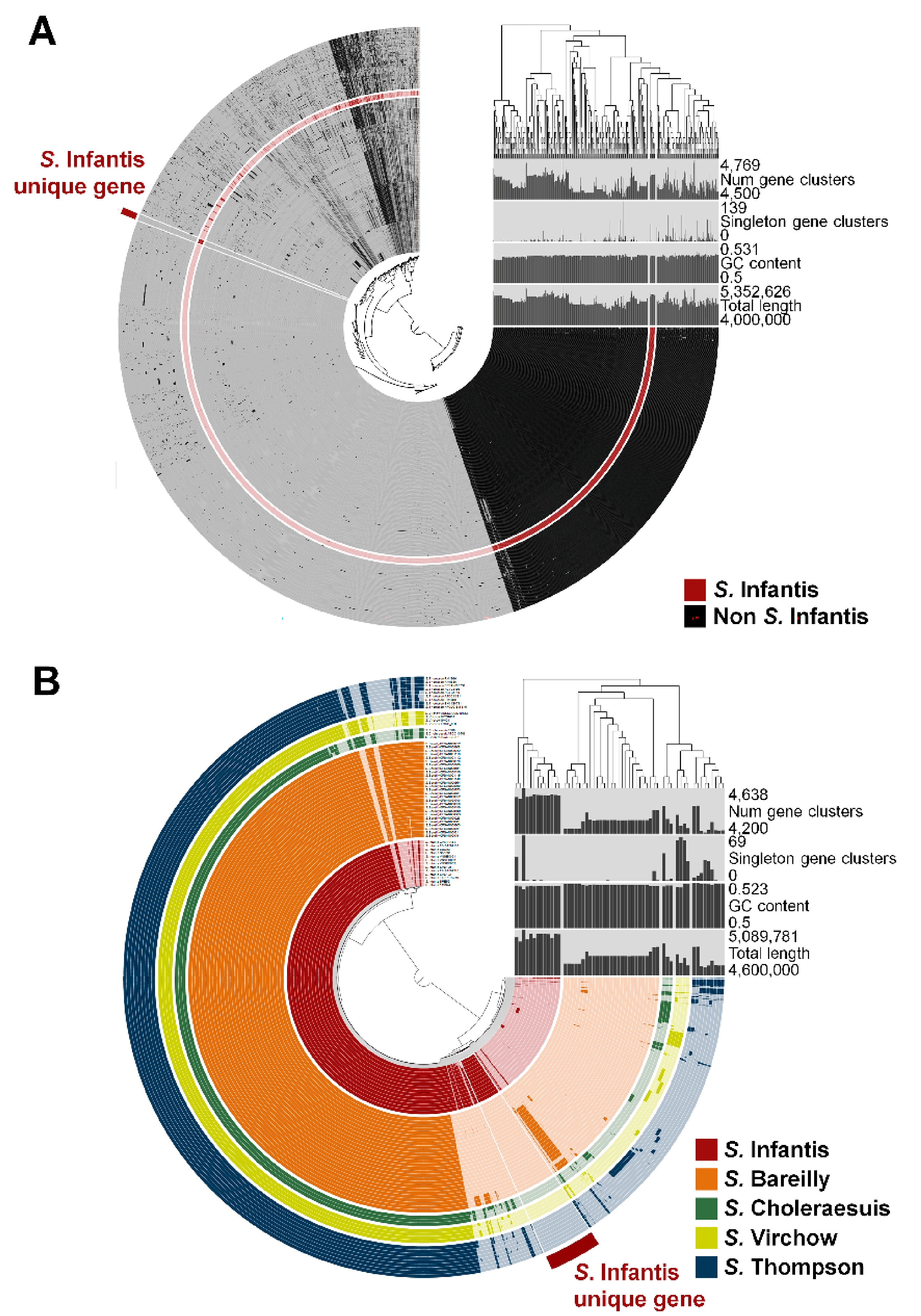
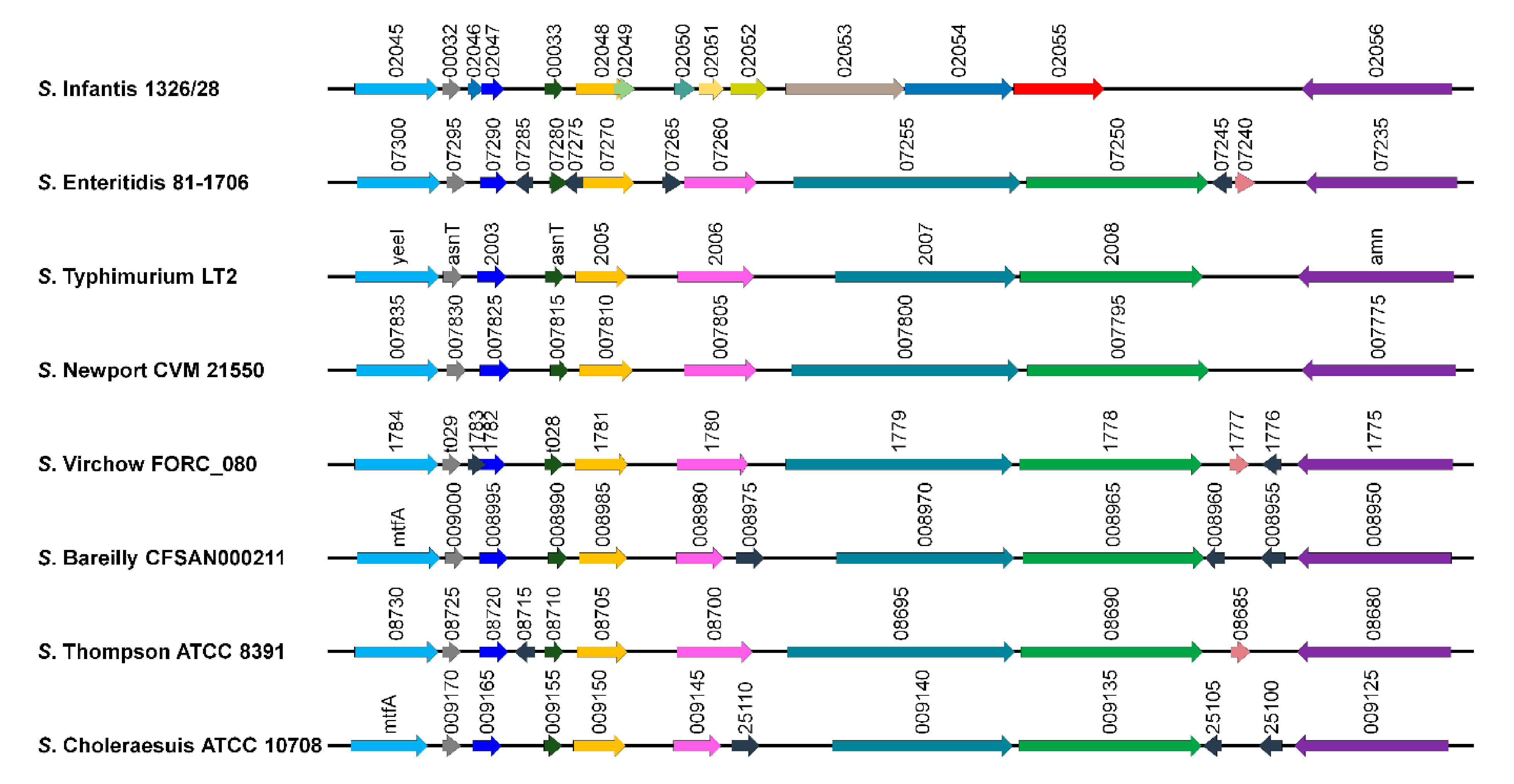
3.1. Identification of Gene Markers Specific to Serovar Infantis
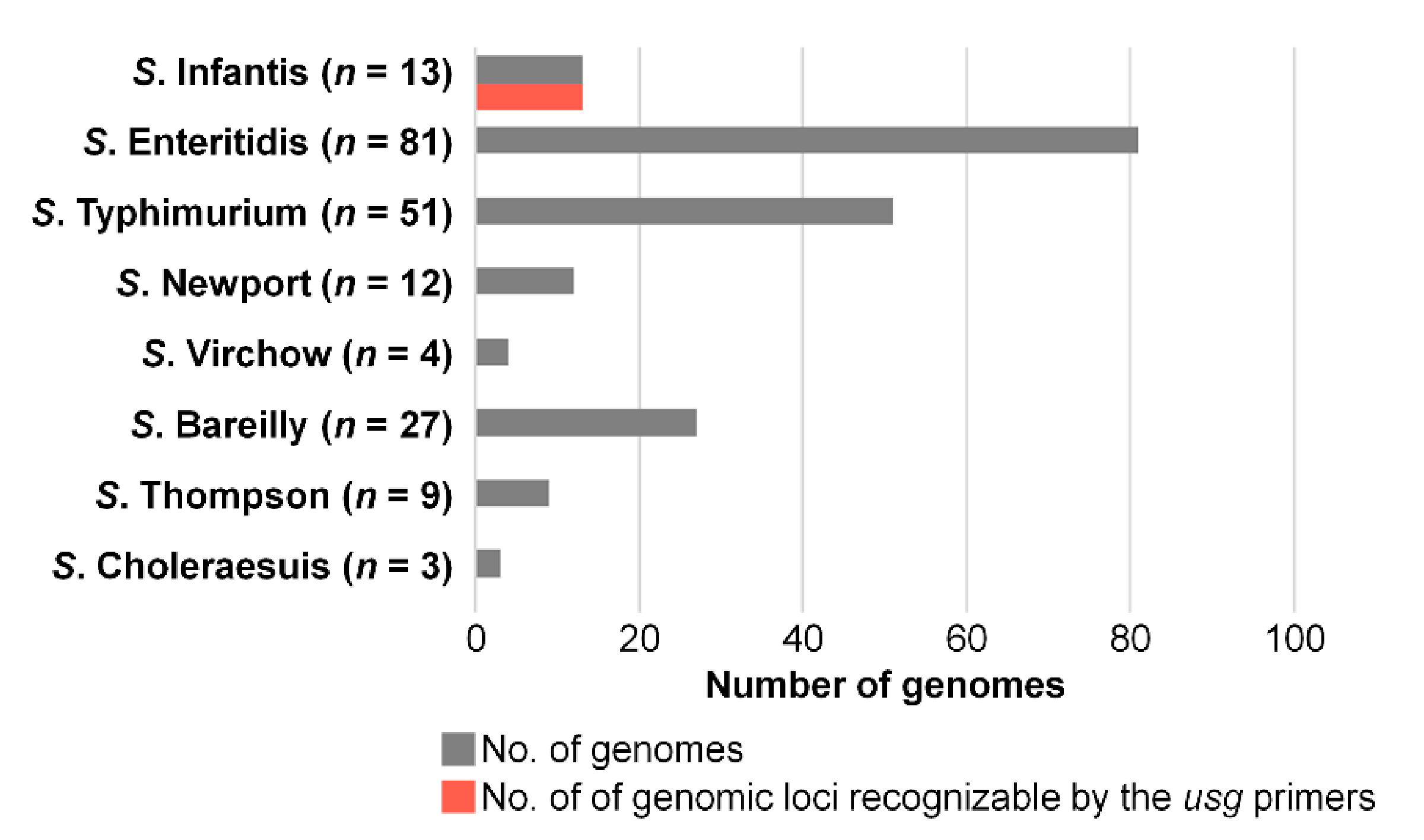
3.2. In Silico Prediction of Sensitivity and Specificity of the Candidate Gene Marker
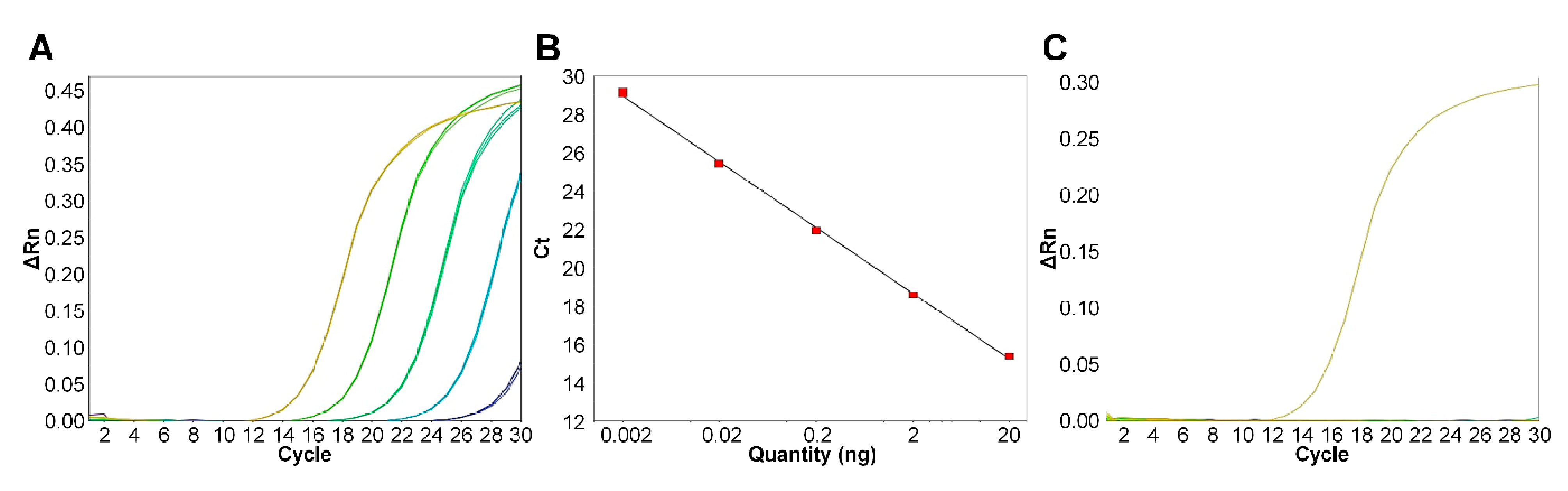
3.3. Efficacy Evaluation of the Diagnostic Primer Set in Real-Time PCR
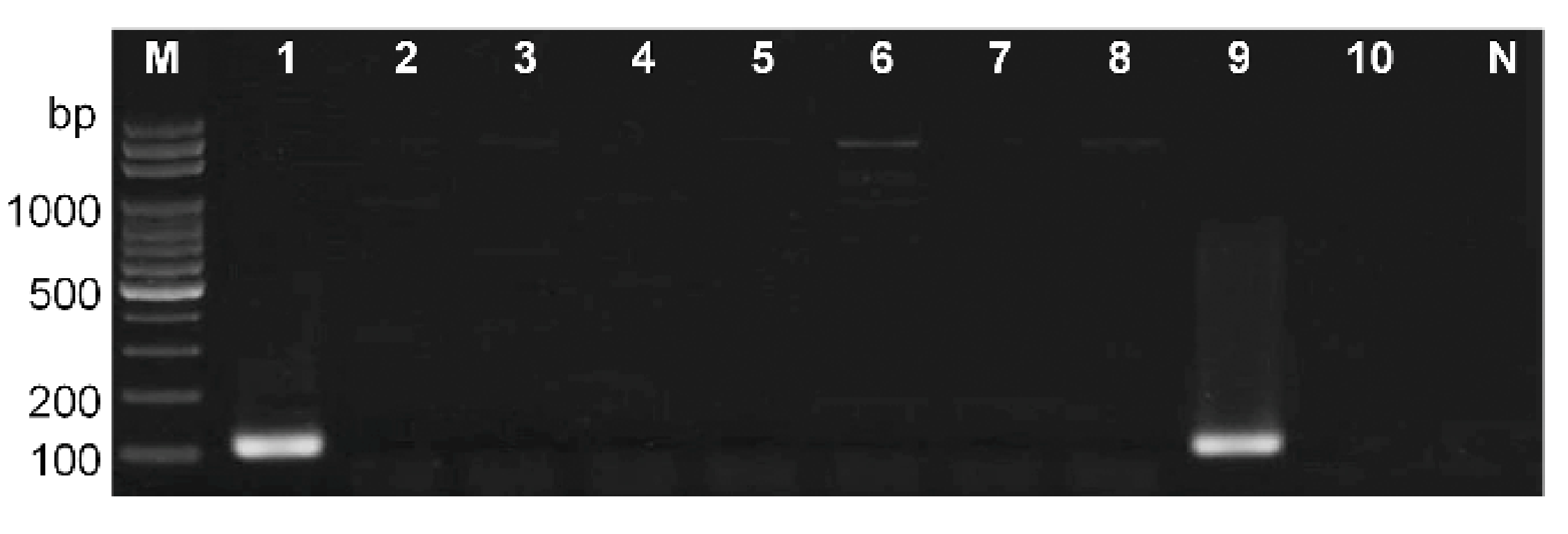
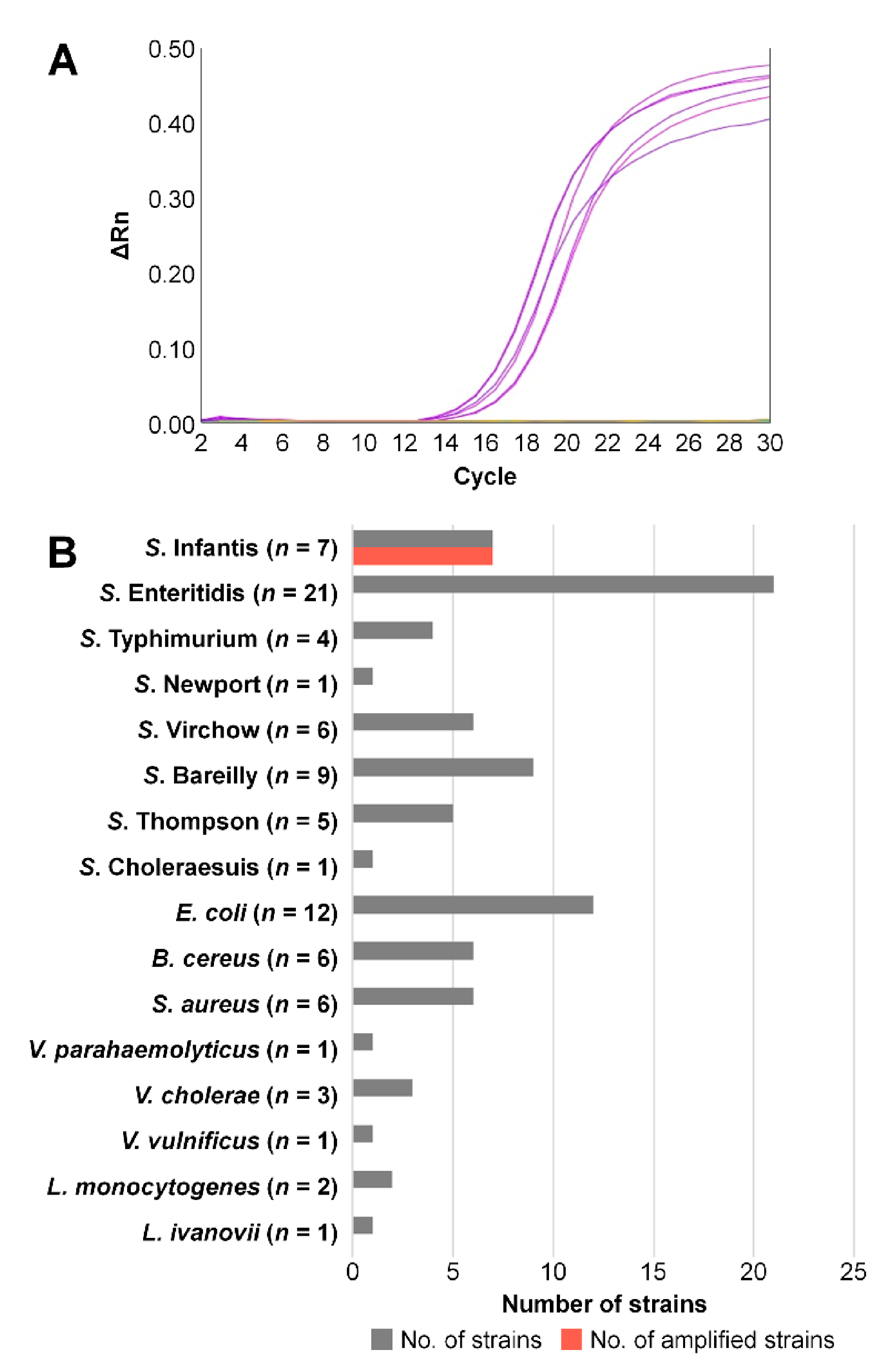
3.4. Validation of the Diagnostic PCR Method
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, H.M.; Wang, Y.; Su, L.H.; Chiu, C.H. Nontyphoid Salmonella infection: Microbiology, clinical features, and antimicrobial therapy. Pediatr. Neonatol. 2013, 54, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Crump, J.A.; Sjolund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef]
- Jones, T.F.; Ingram, L.A.; Cieslak, P.R.; Vugia, D.J.; Tobin-D’Angelo, M.; Hurd, S.; Medus, C.; Cronquist, A.; Angulo, F.J. Salmonellosis outcomes differ substantially by serotype. J. Infect. Dis. 2008, 198, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Griffin, P.M.; Cole, D.; Walsh, K.A.; Chai, S.J. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 1239–1244. [Google Scholar] [CrossRef]
- Miller, T.; Prager, R.; Rabsch, W.; Fehlhaber, K.; Voss, M. Epidemiological relationship between Salmonella Infantis isolates of human and broiler origin. Lohmann Inf. 2010, 45, 27–31. [Google Scholar]
- Nogrady, N.; Toth, A.; Kostyak, A.; Paszti, J.; Nagy, B. Emergence of multidrug-resistant clones of Salmonella Infantis in broiler chickens and humans in Hungary. J. Antimicrob. Chemother. 2007, 60, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Bassal, R.; Reisfeld, A.; Andorn, N.; Yishai, R.; Nissan, I.; Agmon, V.; Peled, N.; Block, C.; Keller, N.; Kenes, Y.; et al. Recent trends in the epidemiology of non-typhoidal Salmonella in Israel, 1999–2009. Epidemiol. Infect. 2012, 140, 1446–1453. [Google Scholar] [CrossRef]
- Kardos, G.; Farkas, T.; Antal, M.; Nogrady, N.; Kiss, I. Novel PCR assay for identification of Salmonella enterica serovar Infantis. Lett. Appl. Microbiol. 2007, 45, 421–425. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Mikoleit, M.; Carlson, V.P.; Karlsmose, S.; Vieira, A.R.; Jensen, A.B.; Seyfarth, A.M.; DeLong, S.M.; Weill, F.X.; Lo Fo Wong, D.M.; et al. WHO global salm-surv external quality assurance system for serotyping of Salmonella isolates from 2000 to 2007. J. Clin. Microbiol. 2009, 47, 2729–2736. [Google Scholar] [CrossRef]
- Guard, J.; Sanchez-Ingunza, R.; Morales, C.; Stewart, T.; Liljebjelke, K.; Van Kessel, J.; Ingram, K.; Jones, D.; Jackson, C.; Fedorka-Cray, P.; et al. Comparison of dkgB-linked intergenic sequence ribotyping to DNA microarray hybridization for assigning serotype to Salmonella enterica. FEMS Microbiol. Lett. 2012, 337, 61–72. [Google Scholar] [CrossRef][Green Version]
- Harbottle, H.; White, D.G.; McDermott, P.F.; Walker, R.D.; Zhao, S. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J. Clin. Microbiol. 2006, 44, 2449–2457. [Google Scholar] [CrossRef] [PubMed]
- Christensen, H.; Moller, P.L.; Vogensen, F.K.; Olsen, J.E. 16S to 23S rRNA spacer fragment length polymorphism of Salmonella enterica at subspecies and serotype levels. J. Appl. Microbiol. 2000, 89, 130–136. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Achtman, M.; Wain, J.; Weill, F.X.; Nair, S.; Zhou, Z.; Sangal, V.; Krauland, M.G.; Hale, J.L.; Harbottle, H.; Uesbeck, A.; et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012, 8, e1002776. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C.; Collins, M.; van Duyne, S.; Mikoleit, M.; Brown, T.; Fields, P. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 2007, 45, 3323–3334. [Google Scholar] [CrossRef] [PubMed]
- Leekitcharoenphon, P.; Nielsen, E.M.; Kaas, R.S.; Lund, O.; Aarestrup, F.M. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS ONE 2014, 9, e87991. [Google Scholar] [CrossRef]
- Ibrahim, G.M.; Morin, P.M. Salmonella serotyping using whole genome sequencing. Front. Microbiol. 2018, 9, 2993. [Google Scholar] [CrossRef]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.; Nash, J.H.; Taboada, E.N. The Salmonella in silico typing resource (SISTR): An open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef]
- Zhang, X.; Payne, M.; Wang, Q.; Sintchenko, V.; Lan, R. Highly sensitive and specific detection and serotyping of five prevalent Salmonella serovars by multiple cross-displacement amplification. J. Mol. Diagn. 2020, 22, 708–719. [Google Scholar] [CrossRef]
- Zou, Q.H.; Li, R.Q.; Liu, G.R.; Liu, S.L. Genotyping of Salmonella with lineage-specific genes: Correlation with serotyping. Int. J. Infect. Dis. 2016, 49, 134–140. [Google Scholar] [CrossRef][Green Version]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen, O.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Oberto, J. SyntTax: A web server linking synteny to prokaryotic taxonomy. BMC Bioinform. 2013, 14, 4. [Google Scholar] [CrossRef]
- Kamel, A.A.-E. Bioinformatic tools and guideline for PCR primer design. Afr. J. Biotechnol. 2003, 2, 91–95. [Google Scholar] [CrossRef]
- San Millan, R.M.; Martinez-Ballesteros, I.; Rementeria, A.; Garaizar, J.; Bikandi, J. Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Res. Notes 2013, 6, 513. [Google Scholar] [CrossRef] [PubMed]
- Grimont, P.A.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars. WHO Collab. Ref. Res. Salmonella 2007, 9, 1–166. Available online: https://www.pasteur.fr/sites/default/files/veng_0.pdf (accessed on 16 September 2019).
- Olasz, F.; Nagy, T.; Szabo, M.; Kiss, J.; Szmolka, A.; Barta, E.; van Tonder, A.; Thomson, N.; Barrow, P.; Nagy, B. Genome sequences of three Salmonella enterica subsp. enterica serovar Infantis strains from healthy broiler chicks in Hungary and in the United Kingdom. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Rabsch, W.; Hargis, B.M.; Tsolis, R.M.; Kingsley, R.A.; Hinz, K.H.; Tschape, H.; Baumler, A.J. Competitive exclusion of Salmonella Enteritidis by Salmonella Gallinarum in poultry. Emerg. Infect. Dis. 2000, 6, 443–448. [Google Scholar] [CrossRef]
- Alvarez, J.; Sota, M.; Vivanco, A.B.; Perales, I.; Cisterna, R.; Rementeria, A.; Garaizar, J. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J. Clin. Microbiol. 2004, 42, 1734–1738. [Google Scholar] [CrossRef]
- Heymans, R.; Vila, A.; van Heerwaarden, C.A.M.; Jansen, C.C.C.; Castelijn, G.A.A.; van der Voort, M.; Biesta-Peters, E.G. Rapid detection and differentiation of Salmonella species, Salmonella Typhimurium and Salmonella Enteritidis by multiplex quantitative PCR. PLoS ONE 2018, 13, e0206316. [Google Scholar] [CrossRef]
- Paiao, F.G.; Arisitides, L.G.; Murate, L.S.; Vilas-Boas, G.T.; Vilas-Boas, L.A.; Shimokomaki, M. Detection of Salmonella spp, Salmonella Enteritidis and Typhimurium in naturally infected broiler chickens by a multiplex PCR-based assay. Braz. J. Microbiol. 2013, 44, 37–41. [Google Scholar] [CrossRef]
- Chiang, Y.C.; Wang, H.H.; Ramireddy, L.; Chen, H.Y.; Shih, C.M.; Lin, C.K.; Tsen, H.Y. Designing a biochip following multiplex polymerase chain reaction for the detection of Salmonella serovars Typhimurium, Enteritidis, Infantis, Hadar, and Virchow in poultry products. J. Food Drug Anal. 2018, 26, 58–66. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.-M.; Baek, J.; Kim, E.; Kim, H.-B.; Ko, S.; Kim, D.; Yoon, H.; Kim, H.-Y. Development of a Genoserotyping Method for Salmonella Infantis Detection on the Basis of Pangenome Analysis. Microorganisms 2021, 9, 67. https://doi.org/10.3390/microorganisms9010067
Yang S-M, Baek J, Kim E, Kim H-B, Ko S, Kim D, Yoon H, Kim H-Y. Development of a Genoserotyping Method for Salmonella Infantis Detection on the Basis of Pangenome Analysis. Microorganisms. 2021; 9(1):67. https://doi.org/10.3390/microorganisms9010067
Chicago/Turabian StyleYang, Seung-Min, Jiwon Baek, Eiseul Kim, Hyeon-Be Kim, Seyoung Ko, Donghyuk Kim, Hyunjin Yoon, and Hae-Yeong Kim. 2021. "Development of a Genoserotyping Method for Salmonella Infantis Detection on the Basis of Pangenome Analysis" Microorganisms 9, no. 1: 67. https://doi.org/10.3390/microorganisms9010067
APA StyleYang, S.-M., Baek, J., Kim, E., Kim, H.-B., Ko, S., Kim, D., Yoon, H., & Kim, H.-Y. (2021). Development of a Genoserotyping Method for Salmonella Infantis Detection on the Basis of Pangenome Analysis. Microorganisms, 9(1), 67. https://doi.org/10.3390/microorganisms9010067





