Molecular Biology in the Improvement of Biological Nitrogen Fixation by Rhizobia and Extending the Scope to Cereals
Abstract
1. Introduction
2. Improvement of Rhizobial Strains
2.1. The Enhanced Efficiency of Nitrogen Fixation
2.2. Microsymbiont Competitiveness
2.3. Stress Tolerance
2.3.1. Drought and Salinity
2.3.2. Heat Stress
2.3.3. Metal Toxicity
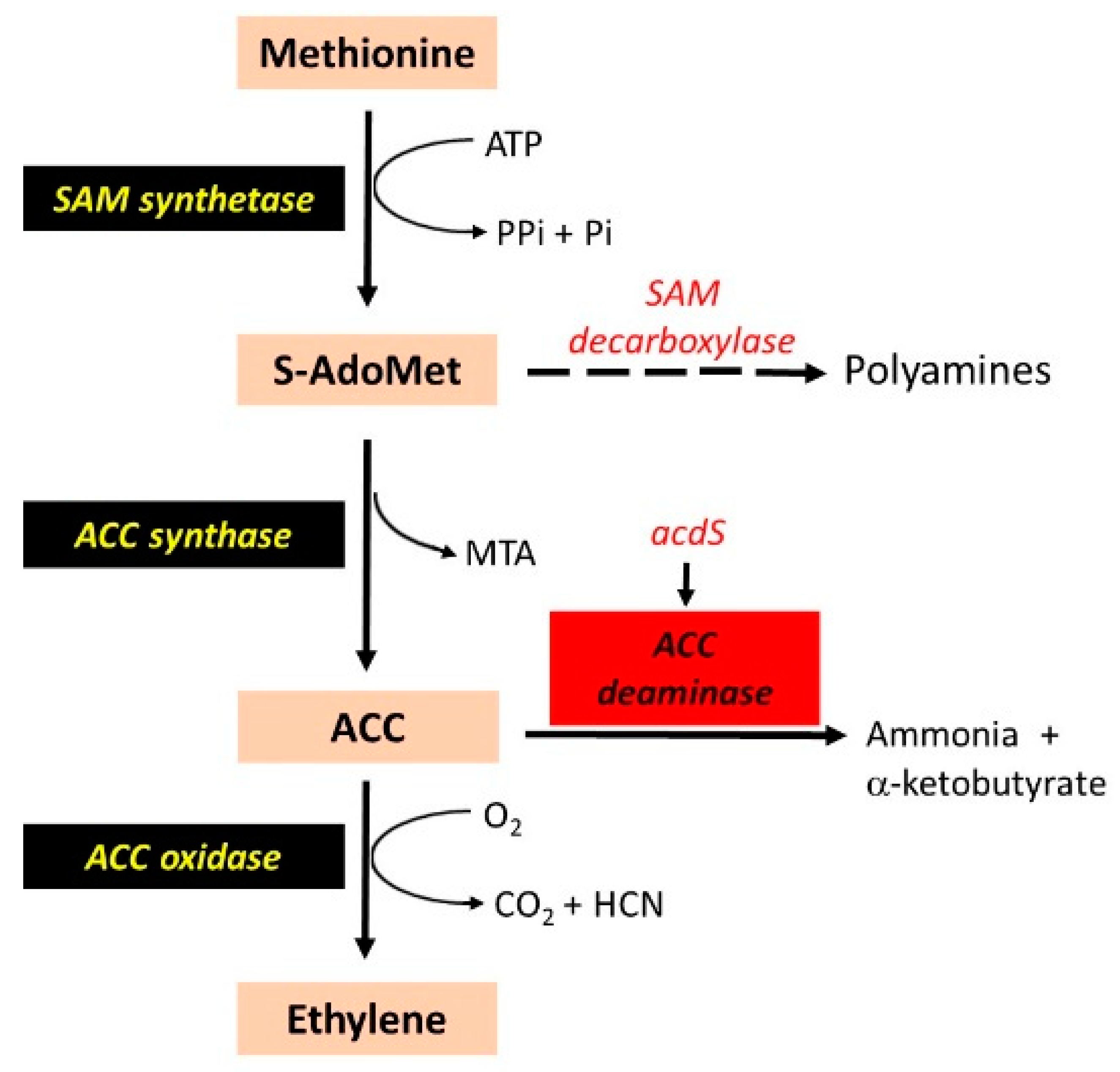
3. Reduction in Ethylene Synthesis and Nodulation
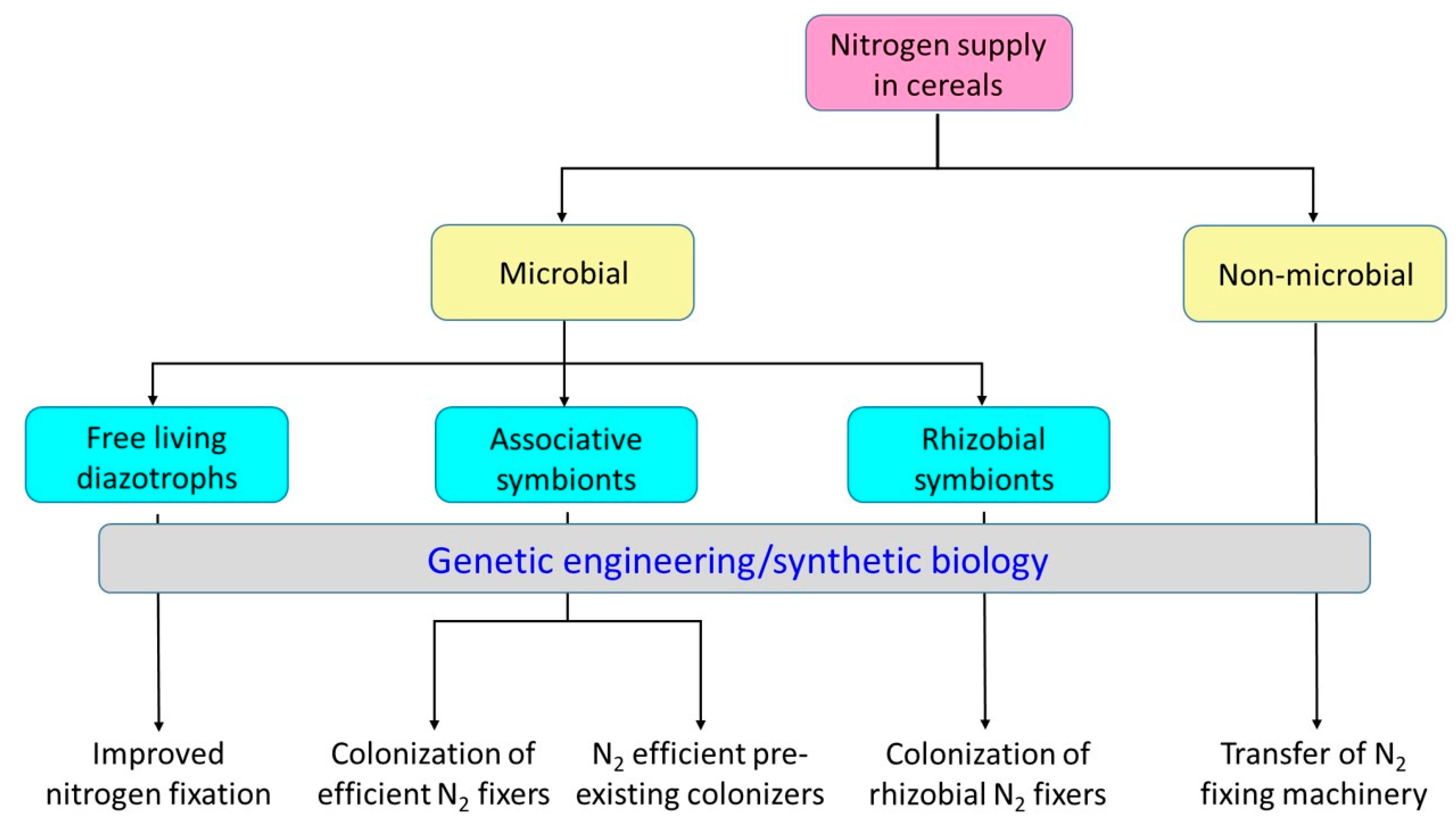
4. Nitrogen Fixation in Cereals
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smil, V. Enriching the Earth: Fritz Haber, Carl Bosch, and the Transformation of World Food Production; The MIT Press: Cambridge, MA, USA, 2001; 338p, ISBN 0-262-19449-X. [Google Scholar]
- Strebel, O.; Duynisveld, W.H.M.; Böttcher, J. Nitrate pollution of groundwater in western Europe. Agric. Ecosyst. Environ. 1989, 26, 189–214. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Herridge, D.F.; Peoples, M.B.; Boddey, R.M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 2008, 311, 1–18. [Google Scholar] [CrossRef]
- Lindström, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef]
- Sands, D.C.; Morris, C.E.; Dratz, E.A.; Pilgeram, A.L. Elevating optimal human nutrition to a central goal of plant breeding and production of plant-based foods. Plant Sci. 2009, 177, 377–389. [Google Scholar] [CrossRef]
- Voisin, A.S.; Guéguen, J.; Huyghe, C.; Jeuffroy, M.H.; Magrini, M.B.; Meynard, J.M.; Mougel, C.; Pellerin, S.; Pelzer, E. Legumes for feed, food, biomaterials and bioenergy in Europe: A review. Agron. Sustain. Dev. 2014, 34, 361–380. [Google Scholar] [CrossRef]
- Via, V.D.; Zanetti, M.E.; Blanco, F. How legumes recognize rhizobia. Plant Signal. Behav. 2016, 11. [Google Scholar] [CrossRef]
- Mavingui, P.; Flores, M.; Romero, D.; Martinez-Romero, E.; Palacios, R. Generation of Rhizobium strains with improved symbiotic properties by random DNA amplification (RDA). Nat. Biotechnol. 1997, 15, 564–569. [Google Scholar] [CrossRef]
- Prithiviraj, B.; Zhou, X.; Souleimanov, A.; Kahn, W.; Smith, D. A host-specific bacteria-to-plant signal molecule (Nod factor) enhances germination and early growth of diverse crop plants. Planta 2003, 216, 437–445. [Google Scholar] [CrossRef]
- Gautam, K.; Schwinghamer, T.D.; Smith, D.L. The response of soybean to nod factors and a bacteriocin. Plant Signal. Behav. 2016, 11, e1241934. [Google Scholar] [CrossRef]
- Hogg, B.; Davies, A.E.; Wilson, K.E.; Bisseling, T.; Downie, J.A. Competitive nodulation blocking of cv. Afghanistan pea is related to high levels of nodulation factors made by some strains of Rhizobium leguminosarum bv. viciae. Mol. Plant Microbe Interact. 2002, 15, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Sanjuan, J.; Olivares, J. Multicopy plasmids carring the Klebsiella pneumoniae nifA gene enhance Rhizobium meliloti nodulation competitiveness on alfalfa. Mol. Plant Microbe Interact. 1991, 4, 5. [Google Scholar] [CrossRef]
- Wang, T.; Zhao, X.; Shi, H.; Sun, L.; Li, Y.; Li, Q.; Zhang, H.; Chen, S.; Li, J. Positive and negative regulation of transferred nif genes mediated by indigenous GlnR in Gram-positive Paenibacillus polymyxa. PLoS Genet. 2018, 14, e1007629. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yu, G.; Shen, S.; Zhu, J. Functional difference between Sinorhizobium meliloti NifA and Enterobacter cloacae NifA. Sci. China Ser. C Life Sci. 2004, 47, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Bosworth, A.H.; Williams, M.K.; Albrecht, K.A.; Kwiatkowski, R.; Beynon, J.; Hankinson, T.R.; Ronson, C.W.; Cannon, F.; Wacek, T.J.; Triplett, E.W. Alfalfa yield response to inoculation with recombinant strains of Rhizobium meliloti with an extra copy of dctABD and/or modified nifA expression. Appl. Environ. Microbiol. 1994, 60, 3815–3832. [Google Scholar] [CrossRef]
- Peralta, H.; Mora, Y.; Salazar, E.; Encarnación, S.; Palacios, R.; Mora, J. Engineering the nifH promoter region and abolishing poly-β -hydroxybutyrate accumulation in Rhizobium etli enhance nitrogen fixation in symbiosis with Phaseolus vulgaris. Appl. Environ. Microbiol. 2004, 70, 3272–3281. [Google Scholar] [CrossRef] [PubMed]
- da-Silva, J.R.; Menéndez, E.; Eliziário, F.; Mateos, P.F.; Alexandre, A.; Oliveira, S. Heterologous expression of nifA or nodD genes improves chickpea-Mesorhizobium symbiotic performance. Plant Soil 2019, 436, 607–621. [Google Scholar] [CrossRef]
- da-Silva, J.R.; Paço, A.; Alexandre, A.; Brígido, C.; Menéndez, E. Genetic engineering as a strategy to improve rhizobial symbiotic performance. In Agricultural Research Updates; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2018; Volume 24, pp. 45–84. [Google Scholar]
- Mongiardini, E.J.; Pérez-Giménez, J.; Althabegoiti, M.J.; Covelli, J.; Quelas, J.I.; López-García, S.L.; Lodeiro, A.R. Overproduction of the rhizobial adhesin RapA1 increases competitiveness for nodulation. Soil Biol. Biochem. 2009, 41, 2017–2020. [Google Scholar] [CrossRef]
- Robleto, E.A.; Kmiecik, K.; Oplinger, E.S.; Nienhuis, J.; Triplett, E.W. Trifolitoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Appl. Environ. Microbiol. 1998, 64, 2630–2633. [Google Scholar] [CrossRef]
- Chien, H.-L.; Huang, W.-Z.; Tsai, M.-Y.; Cheng, C.-H.; Liu, C.-T. Overexpression of the chromosome partitioning gene parA in Azorhizobium caulinodans ORS571 alters the bacteroid morphotype in Sesbania rostrata stem nodules. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Janczarek, M.; Jaroszuk-Ściseł, J.; Skorupska, A. Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Antonie Leeuwenhoek 2009, 96, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.M. Increased production of the exopolysaccharide succinoglycan enhances Sinorhizobium meliloti 1021 symbiosis with the host plant Medicago truncatula. J. Bacteriol. 2012, 194, 4322–4331. [Google Scholar] [CrossRef] [PubMed]
- Paço, A.; Brígido, C.; Alexandre, A.; Mateos, P.F.; Oliveira, S. The symbiotic performance of chickpea rhizobia can be improved by additional copies of the clpB chaperone gene. PLoS ONE 2016, 11, e148221. [Google Scholar] [CrossRef] [PubMed]
- da-Silva, J.R.; Alexandre, A.; Brigido, C.; Oliveira, S. Can stress response genes be used to improve the symbiotic performance of rhizobia? AIMS Microbiol. 2017, 3, 365–382. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.R.; Brito, B.; Imperial, J.; Palacios, J.M.; Ciampitti, I.A.; Ruiz-Argüeso, T.; Hungria, M. Hydrogen-uptake genes improve symbiotic efficiency in common beans (Phaseolus vulgaris L.). Antonie Leeuwenhoek 2020, 113, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Ike, A.; Sriprang, R.; Ono, H.; Murooka, Y.; Yamashita, M. Bioremediation of cadmium contaminated soil using symbiosis between leguminous plant and recombinant rhizobia with the MTL4 and the PCS genes. Chemosphere 2007, 66, 1670–1676. [Google Scholar] [CrossRef]
- Ike, A.; Sriprang, R.; Ono, H.; Murooka, Y.; Yamashita, M. Promotion of metal accumulation in nodule of Astragalus sinicus by the expression of the iron-regulated transporter gene in Mesorhizobium huakuii subsp. rengei B3. J. Biosci. Bioeng. 2008, 105, 642–648. [Google Scholar] [CrossRef]
- Drewniak, L.; Dziewit, L.; Ciezkowska, M.; Gawor, J.; Gromadka, R.; Sklodowska, A. Structural and functional genomics of plasmid pSinA of Sinorhizobium sp. M14 encoding genes for the arsenite oxidation and arsenic resistance. J. Biotechnol. 2013, 164, 479–488. [Google Scholar] [CrossRef]
- Delgadillo, J.; Lafuente, A.; Doukkali, B.; Redondo-Gómez, S.; Mateos-Naranjo, E.; Caviedes, M.A.; Pajuelo, E.; Rodríguez-Llorente, I.D. Improving legume nodulation and Cu rhizostabilization using a genetically modified rhizobia. Environ. Technol. 2015, 36, 1237–1245. [Google Scholar] [CrossRef]
- Pérez-Palacios, P.; Romero-Aguilar, A.; Delgadillo, J.; Doukkali, B.; Caviedes, M.A.; Rodríguez-Llorente, I.D.; Pajuelo, E. Double genetically modified symbiotic system for improved Cu phytostabilization in legume roots. Environ. Sci. Pollut. Res. 2017, 24, 14910–14923. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Y.; Cao, T.; Chen, J.; Rosen, B.P.; Zhao, F.J. Arsenic methylation by a genetically engineered Rhizobium-legume symbiont. Plant Soil 2017, 416, 259–269. [Google Scholar] [CrossRef] [PubMed]
- González-Sánchez, A.; Cubillas, C.A.; Miranda, F.; Dávalos, A.; García-de los Santos, A. The ropAe gene encodes a porin-like protein involved in copper transit in Rhizobium etli CFN42. MicrobiologyOpen 2018, 7. [Google Scholar] [CrossRef]
- Tsyganov, V.E.; Tsyganova, A.V.; Gorshkov, A.P.; Seliverstova, E.V.; Kim, V.E.; Chizhevskaya, E.P.; Belimov, A.A.; Serova, T.A.; Ivanova, K.A.; Kulaeva, O.A.; et al. Efficacy of a plant-microbe system: Pisum sativum (L.) cadmium-tolerant mutant and Rhizobium leguminosarum strains, expressing pea metallothionein genes PsMT1 and PsMT2, for cadmium phytoremediation. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Rastogi, V.; Labes, M.; Finan, T.; Watson, R. Overexpression of the dctA gene in Rhizobium meliloti: Effect on transport of C4 dicarboxylates and symbiotic nitrogen fixation. Can. J. Microbiol. 1992, 38, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Van Dillewijn, P.; Martínez-Abarca, F.; Toro, N. Multicopy vectors carrying the Klebsiella pneumoniae nifA gene do not enhance the nodulation competitiveness of Sinorhizobium meliloti on alfalfa. Mol. Plant Microbe Interact. 1998, 11, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 1994, 58, 352–386. [Google Scholar] [CrossRef]
- Zhao, J.; Dai, X.; Xu, L.; Zhu, J.; Shen, S.; Yu, G. Extra-copy nifA enhances the nodulation efficiency of Sinorhizobium fredii. Chin. Sci. Bull. 2002, 47, 565–567. [Google Scholar] [CrossRef]
- Castillo, M.; Flores, M.; Mavingui, P.; Martínez-Romero, E.; Palacios, R.; Hernández, G. Increase in alfalfa nodulation, nitrogen fixation, and plant growth by specific DNA amplification in Sinorhizobium meliloti. Appl. Environ. Microbiol. 1999, 65, 2716–2722. [Google Scholar] [CrossRef]
- Spaink, H.P.; Okker, R.J.; Wijffelman, C.A.; Tak, T.; Goosen-de Roo, L.; Pees, E.; van Brussel, A.A.; Lugtenberg, B.J. Symbiotic properties of rhizobia containing a flavonoid-independent hybrid nodD product. J. Bacteriol. 1989, 171, 4045–4053. [Google Scholar] [CrossRef]
- Machado, D.; Krishnan, H.B. nodD alleles of Sinorhizobium fredii USDA191 differentially influence soybean nodulation, nodC expression, and production of exopolysaccharides. Curr. Microbiol. 2003, 47, 134–137. [Google Scholar] [CrossRef]
- Vinardell, J.M.; Ollero, F.J.; Hidalgo, Á.; López-Baena, F.J.; Medina, C.; Ivanov-Vangelov, K.; Parada, M.; Madinabeitia, N.; Del Rosario Espuny, M.; Bellogín, R.A.; et al. NolR regulates diverse symbiotic signals of Sinorhizobium fredii HH103. Mol. Plant Microbe Interact. 2004, 17, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Redondo, F.J.; De La Peña, T.C.; Morcillo, C.N.; Lucas, M.M.; Pueyo, J.J. Overexpression of flavodoxin in bacteroids induces changes in antioxidant metabolism leading to delayed senescence and starch accumulation in alfalfa root nodules. Plant Physiol. 2009, 149, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Shvaleva, A.; de la Peña, T.C.; Rincón, A.; Morcillo, C.N.; de la Torre, V.S.G.; Lucas, M.M.; Pueyo, J.J. Flavodoxin overexpression reduces cadmium-induced damage in alfalfa root nodules. Plant Soil 2010, 326, 109–121. [Google Scholar] [CrossRef]
- Redondo, F.J.; de la Peña, T.C.; Lucas, M.M.; Pueyo, J.J. Alfalfa nodules elicited by a flavodoxin-overexpressing Ensifer meliloti strain display nitrogen-fixing activity with enhanced tolerance to salinity stress. Planta 2012, 236, 1687–1700. [Google Scholar] [CrossRef] [PubMed]
- Coba de la Pena, T.; Redondo, F.J.; Fillat, M.F.; Lucas, M.M.; Pueyo, J.J. Flavodoxin overexpression confers tolerance to oxidative stress in beneficial soil bacteria and improves survival in the presence of the herbicides paraquat and atrazine. J. Appl. Microbiol. 2013, 115, 236–246. [Google Scholar] [CrossRef]
- Jamet, A.; Mandon, K.; Puppo, A.; Hérouart, D. H2O2 is required for optimal establishment of the Medicago sativa/Sinorhizobium meliloti symbiosis. J. Bacteriol. 2007, 189, 8741–8745. [Google Scholar] [CrossRef]
- Yurgel, S.N.; Soberon, M.; Sharypova, L.A.; Miranda, J.; Morera, C.; Simarov, B.V. Isolation of Sinorhizobium meliloti Tn5 mutants with altered cytochrome terminal oxidase expression and improved symbiotic performance. FEMS Microbiol. Lett. 1998, 165, 167–173. [Google Scholar] [CrossRef][Green Version]
- Soberón, M.; López, O.; Morera, C.; Girard, M.D.L.; Tabche, M.L.; Miranda, J. Enhanced nitrogen fixation in a Rhizobium etli ntrC mutant that overproduces the Bradyrhizobium japonicum symbiotic terminal oxidase cbb3. Appl. Environ. Microbiol. 1999, 65, 2015–2019. [Google Scholar] [CrossRef]
- Talbi, C.; Sánchez, C.; Hidalgo-Garcia, A.; González, E.M.; Arrese-Igor, C.; Girard, L.; Bedmar, E.J.; Delgado, M.J. Enhanced expression of Rhizobium etli cbb3 oxidase improves drought tolerance of common bean symbiotic nitrogen fixation. J. Exp. Bot. 2012, 63, 5035–5043. [Google Scholar] [CrossRef]
- Orikasa, Y.; Nodasaka, Y.; Ohyama, T.; Okuyama, H.; Ichise, N.; Yumoto, I.; Morita, N.; Wei, M.; Ohwada, T. Enhancement of the nitrogen fixation efficiency of genetically-engineered Rhizobium with high catalase activity. J. Biosci. Bioeng. 2010, 110, 397–402. [Google Scholar] [CrossRef]
- Shrivastava, A.K.; Pandey, S.; Dietz, K.J.; Singh, P.K.; Singh, S.; Rai, R.; Rai, L.C. Overexpression of AhpC enhances stress tolerance and N2–fixation in Anabaena by upregulating stress responsive genes. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 2576–2588. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.J.; Vig, S.; Nareshkumar, G.; Archana, G. Effect of transgenic rhizobacteria overexpressing Citrobacter braakii appA on Phytate-P availability to mung bean plants. J. Microbiol. Biotechnol. 2010, 20, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Kumar, A.; Archana, G.; Gattupalli, N.K. Ensifer meliloti overexpressing Escherichia coli phytase gene (AppA) improves phosphorus (P) acquisition in maize plants. Sci. Nat. 2016, 103, s00114–s00116. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Guinel, F.C.; Glick, B.R. Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl. Environ. Microbiol. 2003, 69, 4396–4402. [Google Scholar] [CrossRef]
- Ma, W.; Charles, T.C.; Glick, B.R. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl. Environ. Microbiol. 2004, 70, 5891–5897. [Google Scholar] [CrossRef]
- Pii, Y.; Crimi, M.; Cremonese, G.; Spena, A.; Pandolfini, T. Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol. 2007, 7. [Google Scholar] [CrossRef]
- Bianco, C.; Defez, R. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 2009, 60, 3097–3107. [Google Scholar] [CrossRef]
- Imperlini, E.; Bianco, C.; Lonardo, E.; Camerini, S.; Cermola, M.; Moschetti, G.; Defez, R. Effects of indole-3-acetic acid on Sinorhizobium meliloti survival and on symbiotic nitrogen fixation and stem dry weight production. Appl. Microbiol. Biotechnol. 2009, 83, 727–738. [Google Scholar] [CrossRef]
- Bianco, C.; Defez, R. Improvement of phosphate solubilization and Medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appl. Environ. Microbiol. 2010, 76, 4626–4632. [Google Scholar] [CrossRef]
- Defez, R.; Esposito, R.; Angelini, C.; Bianco, C. Overproduction of indole-3-acetic acid in free-living rhizobia induces transcriptional changes resembling those occurring in nodule bacteroids. Mol. Plant Microbe Interact. 2016, 29, 484–495. [Google Scholar] [CrossRef]
- Defez, R.; Andreozzi, A.; Dickinson, M.; Charlton, A.; Tadini, L.; Pesaresi, P.; Bianco, C. Improved drought stress response in alfalfa plants nodulated by an IAA over-producing Rhizobium strain. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Camerini, S.; Senatore, B.; Lonardo, E.; Imperlini, E.; Bianco, C.; Moschetti, G.; Rotino, G.L.; Campion, B.; Defez, R. Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch. Microbiol. 2008, 190, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Conforte, V.P.; Echeverria, M.; Sánchez, C.; Ugalde, R.A.; Menéndez, A.B.; Lepek, V.C. Engineered ACC deaminase-expressing free-living cells of Mesorhizobium loti show increased nodulation efficiency and competitiveness on Lotus spp. J. Gen. Appl. Microbiol. 2010, 56, 331–338. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nascimento, F.; Brígido, C.; Alho, L.; Glick, B.R.; Oliveira, S. Enhanced chickpea growth-promotion ability of a Mesorhizobium strain expressing an exogenous ACC deaminase gene. Plant Soil 2012, 353, 221–230. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Brígido, C.; Glick, B.R.; Oliveira, S.; Alho, L. Mesorhizobium ciceri LMS-1 expressing an exogenous 1-aminocyclopropane-1-carboxylate (ACC) deaminase increases its nodulation abilities and chickpea plant resistance to soil constraints. Lett. Appl. Microbiol. 2012, 55, 15–21. [Google Scholar] [CrossRef]
- Brígido, C.; Nascimento, F.X.; Duan, J.; Glick, B.R.; Oliveira, S. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Mesorhizobium spp. reduces the negative effects of salt stress in chickpea. FEMS Microbiol. Lett. 2013, 349, 46–53. [Google Scholar] [CrossRef]
- Kong, Z.; Glick, B.R.; Duan, J.; Ding, S.; Tian, J.; McConkey, B.J.; Wei, G. Effects of 1-aminocyclopropane-1-carboxylate (ACC) deaminase-overproducing Sinorhizobium meliloti on plant growth and copper tolerance of Medicago lupulina. Plant Soil 2015, 391, 383–398. [Google Scholar] [CrossRef]
- Xu, J.; Li, X.L.; Luo, L. Effects of engineered Sinorhizobium meliloti on cytokinin synthesis and tolerance of alfalfa to extreme drought stress. Appl. Environ. Microbiol. 2012, 78, 8056–8061. [Google Scholar] [CrossRef]
- Podleáková, K.; Fardoux, J.; Patrel, D.; Bonaldi, K.; Novák, O.; Strnad, M.; Giraud, E.; Spíchal, L.; Nouwen, N. Rhizobial synthesized cytokinins contribute to but are not essential for the symbiotic interaction between photosynthetic bradyrhizobia and aeschynomene legumes. Mol. Plant Microbe Interact. 2013, 26, 1232–1238. [Google Scholar] [CrossRef]
- Robledo, M.; Rivera, L.; Jiménez-Zurdo, J.I.; Rivas, R.; Dazzo, F.; Velázquez, E.; Martínez-Molina, E.; Hirsch, A.M.; Mateos, P.F. Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb. Cell Fact. 2012, 11. [Google Scholar] [CrossRef]
- Robledo, M.; Menéndez, E.; Jiménez-Zurdo, J.I.; Rivas, R.; Velázquez, E.; Martínez-Molina, E.; Oldroyd, G.; Mateos, P.F. Heterologous expression of rhizobial CelC2 cellulase impairs symbiotic signaling and nodulation in Medicago truncatula. Mol. Plant Microbe Interact. 2018, 31, 568–575. [Google Scholar] [CrossRef]
- Van Dillewijn, P.; Soto, M.J.; Villadas, P.J.; Toro, N. Construction and environmental release of a Sinorhizobium meliloti strain genetically modified to be more competitive for alfalfa nodulation. Appl. Environ. Microbiol. 2001, 67, 3860–3865. [Google Scholar] [CrossRef]
- Boscari, A.; Van De Sype, G.; Le Rudulier, D.; Mandon, K. Overexpression of BetS, a Sinorhizobium meliloti high-affinity betaine transporter, in bacteroids from Medicago sativa nodules sustains nitrogen fixation during early salt stress adaptation. Mol. Plant Microbe Interact. 2006, 19, 896–903. [Google Scholar] [CrossRef][Green Version]
- Suárez, R.; Wong, A.; Ramírez, M.; Barraza, A.; Orozco, M.D.C.; Cevallos, M.A.; Lara, M.; Hernández, G.; Iturriaga, G. Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Mol. Plant Microbe Interact. 2008, 21, 958–966. [Google Scholar] [CrossRef]
- Moussaid, S.; Domínguez-Ferreras, A.; Muñoz, S.; Aurag, J.; Berraho, E.B.; Sanjuán, J. Increased trehalose biosynthesis improves Mesorhizobium ciceri growth and symbiosis establishment in saline conditions. Symbiosis 2015, 67, 103–111. [Google Scholar] [CrossRef]
- Joshi, F.; Chaudhari, A.; Joglekar, P.; Archana, G.; Desai, A. Effect of expression of Bradyrhizobium japonicum 61A152 fegA gene in Mesorhizobium sp., on its competitive survival and nodule occupancy on Arachis hypogea. Appl. Soil Ecol. 2008, 40, 338–347. [Google Scholar] [CrossRef]
- Joshi, F.R.; Desai, D.K.; Archana, G.; Desai, A.J. Enhanced survival and nodule occupancy of pigeon pea nodulating Rhizobium sp. ST1 expressing fegA gene of Bradyrhizobium japonicum 61A152. OnLine J. Biol. Sci. 2009, 9, 40–51. [Google Scholar] [CrossRef]
- Geetha, R.; Desai, A.J.; Archana, G. Effect of the expression of Escherichia coli fhuA gene in Rhizobium sp. IC3123 and ST1 in planta: Its role in increased nodule occupancy and function in pigeon pea. Appl. Soil Ecol. 2009, 43, 185–190. [Google Scholar] [CrossRef]
- Yurgel, S.N.; Kahn, M.L. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 2004, 28, 489–501. [Google Scholar] [CrossRef]
- Seefeldt, L.C.; Mortenson, L.E. Increasing nitrogenase catalytic efficiency for MgATP by changing serine 16 of its Fe protein to threonine: Use of Mn2+ to show interaction of serine 16 with Mg2+. Protein Sci. 1993, 2, 93–102. [Google Scholar] [CrossRef]
- Milton, R.D.; Minteer, S.D. Nitrogenase bioelectrochemistry for synthesis applications. Acc. Chem. Res. 2019, 52, 3351–3360. [Google Scholar] [CrossRef] [PubMed]
- Solomon, J.B.; Lee, C.C.; Jasniewski, A.J.; Rasekh, M.F.; Ribbe, M.W.; Hu, Y. Heterologous expression and engineering of the nitrogenase cofactor biosynthesis scaffold nifEN. Angew. Chem. Int. Ed. 2020, 59, 6887–6893. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, B.M.; Lukoyanov, D.; Yang, Z.Y.; Dean, D.R.; Seefeldt, L.C. Mechanism of nitrogen fixation by nitrogenase: The next stage. Chem. Rev. 2014, 114, 4041–4062. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.J.; Triplett, E.W. Toward more productive, efficient, and competitive nitrogen-fixing symbiotic bacteria. Crit. Rev. Plant Sci. 1996, 15, 191–234. [Google Scholar] [CrossRef]
- Cunningham, S.D.; Kapulnik, Y.; Brewin, N.J.; Phillips, D.A. Uptake hydrogenase activity determined by plasmid pRL6JI in Rhizobium leguminosarum does not increase symbiotic nitrogen fixation. Appl. Environ. Microbiol. 1985, 50, 791–794. [Google Scholar] [CrossRef]
- Brito, B.; Monza, J.; Imperial, J.; Ruiz-Argüeso, T.; Palacios, J.M. Nickel availability and hupSL activation by heterologous regulators limit symbiotic expression of the Rhizobium leguminosarum bv. viciae hydrogenase system in Hup(-) rhizobia. Appl. Environ. Microbiol. 2000, 66, 937–942. [Google Scholar] [CrossRef]
- Emerich, D.W.; Ruiz-Argüeso, T.; Ching, T.M.; Evans, H.J. Hydrogen-dependent nitrogenase activity and ATP formation in Rhizobium japonicum bacteroids. J. Bacteriol. 1979, 137, 153–160. [Google Scholar] [CrossRef]
- Pitcher, R.S.; Watmough, N.J. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta Bioenerg. 2004, 1655, 388–399. [Google Scholar] [CrossRef]
- Ribeiro, C.W.; Alloing, G.; Mandon, K.; Frendo, P. Redox regulation of differentiation in symbiotic nitrogen fixation. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1469–1478. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Sol, S.; Valkov, V.T.; Rogato, A.; Noguero, M.; Gargiulo, L.; Mele, G.; Lacombe, B.; Chiurazzi, M. Disruption of the Lotus japonicus transporter LjNPF2.9 increases shoot biomass and nitrate content without affecting symbiotic performances. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Contador, C.A.; Fan, K.; Lam, H.-M. Interaction and regulation of carbon, nitrogen, and phosphorus metabolisms in root nodules of legumes. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Graham, P.H.; Allan, D.L. Biological Nitrogen Fixation: Phosphorus—A critical future need? In Nitrogen Fixation: From Molecules to Crop Productivity; Pedrosa, F.O., Hungria, M., Yates, G., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 509–514. [Google Scholar] [CrossRef]
- Annan, H.; Golding, A.-L.; Zhao, Y.; Dong, Z. Choice of hydrogen uptake (Hup) status in legume-rhizobia symbioses. Ecol. Evol. 2012, 2, 2285–2290. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.T.; Ronson, C.W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 1998, 95, 5145–5149. [Google Scholar] [CrossRef] [PubMed]
- Triplett, E.W.; Sadowsky, M.J. Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 1992, 46, 399–428. [Google Scholar] [CrossRef] [PubMed]
- Ames, P.; Bergman, K. Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti. J. Bacteriol. 1981, 148, 728–729. [Google Scholar] [CrossRef]
- Yost, C.K.; Rochepeau, P.; Hynes, M.F. Rhizobium leguminosarum contains a group of genes that appear to code for methyl-accepting chemotaxis proteins. Microbiology 1998, 144, 1945–1956. [Google Scholar] [CrossRef]
- Miller, L.D.; Yost, C.K.; Hynes, M.F.; Alexandre, G. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol. Microbiol. 2007, 63, 348–362. [Google Scholar] [CrossRef]
- Oresnik, I.J.; Pacarynuk, L.A.; O’Brien, S.A.P.; Yost, C.K.; Hynes, M.F. Plasmid-encoded catabolic genes in Rhizobium leguminosarum bv. trifolii: Evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol. Plant Microbe Interact. 1998, 11, 1175–1185. [Google Scholar] [CrossRef]
- Richardson, J.S.; Hynes, M.F.; Oresnik, I.J. A genetic locus necessary for rhamnose uptake and catabolism in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 2004, 186, 8433–8442. [Google Scholar] [CrossRef]
- Jimenez-Zurdo, J.I.; Van Dillewijn, P.; Soto, M.J.; De Felipe, M.R.; Olivares, J.; Toro, N. Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol. Plant Microbe Interact. 1995, 8, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Yost, C.K.; Rath, A.M.; Noel, T.C.; Hynes, M.F. Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiology 2006, 152, 2061–2074. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Yip, C.B.; Geddes, B.A.; Oresnik, I.J.; Hynes, M.F. Glycerol utilization by Rhizobium leguminosarum requires an ABC transporter and affects competition for nodulation. Microbiology 2012, 158, 1369–1378. [Google Scholar] [CrossRef] [PubMed]
- van Egeraat, A.W.S.M. The possible role of homoserine in the development of Rhizobium leguminosarum in the rhizosphere of pea seedlings. Plant Soil 1975, 42, 381–386. [Google Scholar] [CrossRef]
- Murphy, P.J.; Heycke, N.; Banfalvi, Z. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid. Proc. Natl. Acad. Sci. USA 1987, 84, 493–497. [Google Scholar] [CrossRef]
- Hynes, M.F.; O’Connell, M.P. Host plant effect on competition among strains of Rhizobium leguminosarum. Can. J. Microbiol. 1990, 36, 864–869. [Google Scholar] [CrossRef]
- Vanderlinde, E.M.; Hynes, M.F.; Yost, C.K. Homoserine catabolism by Rhizobium leguminosarum bv. viciae 3841 requires a plasmid-borne gene cluster that also affects competitiveness for nodulation. Environ. Microbiol. 2014, 16, 205–217. [Google Scholar] [CrossRef]
- Murphy, P.J.; Heycke, N.; Trenz, S.P.; Ratet, P.; de Bruijn, F.J.; Schell, J. Synthesis of an opine-like compound, a rhizopine, in alfalfa nodules is symbiotically regulated. Proc. Natl. Acad. Sci. USA 1988, 85, 9133–9137. [Google Scholar] [CrossRef]
- Murphy, P.J.; Wexler, W.; Grzemski, W.; Rao, J.P.; Gordon, D. Rhizopines-Their role in symbiosis and competition. Soil Biol. Biochem. 1995, 27, 525–529. [Google Scholar] [CrossRef]
- Geddes, B.A.; Paramasivan, P.; Joffrin, A.; Thompson, A.L.; Christensen, K.; Jorrin, B.; Brett, P.; Conway, S.J.; Oldroyd, G.E.D.; Poole, P.S. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Hirsch, P.R. Plasmid-determined bacteriocin production by Rhizobium leguminosarum. J. Gen. Microbiol. 1979, 113, 219–228. [Google Scholar] [CrossRef]
- Oresnik, I.J.; Twelker, S.; Hynes, M.F. Cloning and characterization of a Rhizobium leguminosarum gene encoding a bacteriocin with similarities to RTX toxins. Appl. Environ. Microbiol. 1999, 65, 2833–2840. [Google Scholar] [CrossRef] [PubMed]
- Venter, A.P.; Twelker, S.; Oresnik, I.J.; Hynes, M.F. Analysis of the genetic region encoding a novel rhizobiocin from Rhizobium leguminosarum bv. viciae strain 306. Can. J. Microbiol. 2001, 47, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Breil, B.T.; Ludden, P.W.; Triplett, E.W. DNA sequence and mutational analysis of genes involved in the production and resistance of the antibiotic peptide trifolitoxin. J. Bacteriol. 1993, 175, 3693–3702. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Triplett, E.W.; Barta, T.M. Trifolitoxin production and nodulation are necessary for the expression of superior nodulation competitiveness by Rhizobium leguminosarum bv. trifolii strain T24 on clover. Plant Physiol. 1987, 85, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Robleto, E.A.; Borneman, J.; Triplett, E.W. Effects of bacterial antibiotic production on rhizosphere microbial communities from a culture-independent perspective. Appl. Environ. Microbiol. 1998, 64, 5020–5022. [Google Scholar] [CrossRef] [PubMed]
- Hashem, F.M.; Angle, J.S. Rhizobiophage effects on Bradyrhizobium japonicum, nodulation and soybean growth. Soil Biol. Biochem. 1988, 20, 69–73. [Google Scholar] [CrossRef]
- Halmillawewa, A. Isolation, Characterization, and Applications of Rhizobiophages. Ph.D. Thesis, University of Calgary, Calgary, AB, Canada, 2014. [Google Scholar] [CrossRef]
- Ouma, E.W.; Asango, A.M.; Maingi, J.; Njeru, E.M. Elucidating the potential of native rhizobial isolates to improve biological nitrogen fixation and growth of common bean and soybean in smallholder farming systems of Kenya. Int. J. Agron. 2016, 2016, 4569241. [Google Scholar] [CrossRef]
- Koskey, G.; Mburu, S.W.; Njeru, E.M.; Kimiti, J.M.; Ombori, O.; Maingi, J.M. Potential of native rhizobia in enhancing nitrogen fixation and yields of climbing beans (Phaseolus vulgaris L.) in contrasting environments of eastern Kenya. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Donati, A.J.; Lee, H.I.; Leveau, J.H.J.; Chang, W.S. Effects of Indole-3-Acetic acid on the transcriptional activities and stress tolerance of Bradyrhizobium japonicum. PLoS ONE 2013, 8, e76559. [Google Scholar] [CrossRef]
- Prasad, T.K. Mechanisms of chilling-induced oxidative stress injury and tolerance in developing maize seedlings: Changes in antioxidant system, oxidation of proteins and lipids, and protease activities. Plant J. 1996, 10, 1017–1026. [Google Scholar] [CrossRef]
- Cruz De Carvalho, M.H. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant. Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef]
- Slimen, I.B.; Najar, T.; Ghram, A.; Dabbebi, H.; Ben Mrad, M.; Abdrabbah, M. Reactive oxygen species, heat stress and oxidative-induced mitochondrial damage. A review. Int. J. Hyperth. 2014, 30, 513–523. [Google Scholar] [CrossRef]
- Hungria, M.; Vargas, M.A.T. Environmental factors affecting N2 fixation in grain legumes in the tropics, with an emphasis on Brazil. Field Crops Res. 2000, 65, 151–164. [Google Scholar] [CrossRef]
- Shiro, S.; Matsuura, S.; Saiki, R.; Sigua, G.C.; Yamamoto, A.; Umehara, Y.; Hayashi, M.; Saeki, Y. Genetic diversity and geographical distribution of indigenous soybean-nodulating bradyrhizobia in the United States. Appl. Environ. Microbiol. 2013, 79, 3610–3618. [Google Scholar] [CrossRef]
- Voth, W.; Jakob, U. Stress-activated chaperones: A first line of defense. Trends Biochem. Sci. 2017, 42, 899–913. [Google Scholar] [CrossRef]
- Brígido, C.; Alexandre, A.; Oliveira, S. Transcriptional analysis of major chaperone genes in salt-tolerant and salt-sensitive mesorhizobia. Microbiol. Res. 2012, 167, 623–629. [Google Scholar] [CrossRef]
- Anyanwu, B.O.; Ezejiofor, A.N.; Igweze, Z.N.; Orisakwe, O.E. Heavy metal mixture exposure and effects in developing nations: An update. Toxics 2018, 6, 65. [Google Scholar] [CrossRef]
- Fagorzi, C.; Checcucci, A.; Dicenzo, G.C.; Debiec-Andrzejewska, K.; Dziewit, L.; Pini, F.; Mengoni, A. Harnessing rhizobia to improve heavy-metal phytoremediation by legumes. Genes 2018, 9, 542. [Google Scholar] [CrossRef]
- Gupta, A.; Joia, J. Microbes as potential tool for remediation of heavy metals: A Review. J. Microb. Biochem. Technol. 2016, 8. [Google Scholar] [CrossRef]
- Pérez-Palacios, P.; Funes-Pinter, I.; Agostini, E.; Talano, M.A.; Ibáñez, S.G.; Humphry, M.; Edwards, K.; Rodríguez-Llorente, I.D.; Caviedes, M.A.; Pajuelo, E. Targeting Acr3 from: Ensifer medicae to the plasma membrane or to the tonoplast of tobacco hairy roots allows arsenic extrusion or improved accumulation. Effect of acr3 expression on the root transcriptome. Metallomics 2019, 11, 1864–1886. [Google Scholar] [CrossRef]
- Alves, L.R.; Rodrigues dos Reis, A.; Prado, E.R.; Lavres, J.; Pompeu, G.B.; Azevedo, R.A.; Gratão, P.L. New insights into cadmium stressful-conditions: Role of ethylene on selenium-mediated antioxidant enzymes. Ecotoxicol. Environ. Saf. 2019, 186. [Google Scholar] [CrossRef]
- Guinel, F.C. Ethylene, a hormone at the center-stage of nodulation. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Larrainzar, E.; Riely, B.K.; Kim, S.C.; Carrasquilla-Garcia, N.; Yu, H.J.; Hwang, H.J.; Oh, M.; Kim, G.B.; Surendrarao, A.K.; Chasman, D.; et al. Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol. 2015, 169, 233–265. [Google Scholar] [CrossRef]
- Peters, N.K.; Crist-Estes, D.K. Nodule formation is stimulated by the ethylene inhibitor aminoethoxyvinylglycine. Plant Physiol. 1989, 91, 690–693. [Google Scholar] [CrossRef]
- Argueso, C.T.; Hansen, M.; Kieber, J.J. Regulation of ethylene biosynthesis. J. Plant Growth Regul. 2007, 26, 92–105. [Google Scholar] [CrossRef]
- López, S.M.Y.; Sánchez, M.D.M.; Pastorino, G.N.; Franco, M.E.E.; García, N.T.; Balatti, P.A. Nodulation and delayed nodule senescence: Strategies of two Bradyrhizobium japonicum isolates with high capacity to fix nitrogen. Curr. Microbiol. 2018, 75, 997–1005. [Google Scholar] [CrossRef]
- Tavares, M.J.; Nascimento, F.X.; Glick, B.R.; Rossi, M.J. The expression of an exogenous ACC deaminase by the endophyte Serratia grimesii BXF1 promotes the early nodulation and growth of common bean. Lett. Appl. Microbiol. 2018, 66, 252–259. [Google Scholar] [CrossRef]
- Ahmad, M.; Zahir, Z.A.; Asghar, H.N.; Asghar, M. Inducing salt tolerance in mung bean through coinoculation with rhizobia and plant-growthpromoting rhizobacteria containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 2011, 57, 578–589. [Google Scholar] [CrossRef]
- Pankievicz, V.C.S.; Irving, T.B.; Maia, L.G.S.; Ané, J.M. Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biol. 2019, 17. [Google Scholar] [CrossRef]
- Rogers, C.; Oldroyd, G.E.D. Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J. Exp. Bot. 2014, 65, 1939–1946. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Ormeño-Orrillo, E.; López-López, A.; Rogel, M.A.; Reyes-Hernández, B.J.; Martínez-Romero, J.C.; Reddy, P.M.; Martínez-Romero, E. Nitrogen fixation in cereals. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Inomura, K.; Bragg, J.; Follows, M.J. A quantitative analysis of the direct and indirect costs of nitrogen fixation: A model based on Azotobacter vinelandii. ISME J. 2017, 11, 166–175. [Google Scholar] [CrossRef]
- Mortenson, L.E.; Thorneley, R.N. Structure and function of nitrogenase. Annu. Rev. Biochem. 1979, 48, 387–418. [Google Scholar] [CrossRef]
- Halbleib, C.M.; Ludden, P.W. Regulation of biological nitrogen fixation. J. Nutr. 2000, 130, 1081–1084. [Google Scholar] [CrossRef]
- Peters, J.W.; Szilagyi, R.K. Exploring new frontiers of nitrogenase structure and mechanism. Curr. Opin. Chem. Biol. 2006, 10, 101–108. [Google Scholar] [CrossRef]
- Rutledge, H.L.; Tezcan, F.A. Electron transfer in nitrogenase. Chem. Rev. 2020. [Google Scholar] [CrossRef]
- Burén, S.; Rubio, L.M. State of the art in eukaryotic nitrogenase engineering. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Curatti, L.; Rubio, L.M. Challenges to develop nitrogen-fixing cereals by direct nif-gene transfer. Plant Sci. 2014, 225, 130–137. [Google Scholar] [CrossRef]
- Smanski, M.J.; Bhatia, S.; Zhao, D.; Park, Y.; Woodruff, L.B.A.; Giannoukos, G.; Ciulla, D.; Busby, M.; Calderon, J.; Nicol, R.; et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 2014, 32, 1241–1249. [Google Scholar] [CrossRef]
- Marchal, K.; Vanderleyden, J.; Janssens, F.A. The ‘oxygen paradox’ of dinitrogen-fixing bacteria. Biol. Fertil. Soils 2000, 30, 363–373. [Google Scholar] [CrossRef]
- Mus, F.; Colman, D.R.; Peters, J.W.; Boyd, E.S. Geobiological feedbacks, oxygen, and the evolution of nitrogenase. Free Radic. Biol. Med. 2019, 140, 250–259. [Google Scholar] [CrossRef]
- Ott, T.; van Dongen, J.T.; Günther, C.; Krusell, L.; Desbrosses, G.; Vigeolas, H.; Bock, V.; Czechowski, T.; Geigenberger, P.; Udvardi, M.K. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 2005, 15, 531–535. [Google Scholar] [CrossRef]
- Oelze, J. Respiratory protection of nitrogenase in Azotobacter species: Is a widely held hypothesis unequivocally supported by experimental evidence? FEMS Microbiol. Rev. 2000, 24, 321–333. [Google Scholar] [CrossRef]
- Mackerras, A.H.; Smith, G.D. Evidence for direct repression of nitrogenase by ammonia in the cyanobacterium Anabaena cylindrica. Biochem. Biophys. Res. Commun. 1986, 134, 835–844. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Alías-Villegas, C.; Bellogín, R.A.; Del Cerro, P.; Espuny, M.R.; Jiménez-Guerrero, I.; López-Baena, F.J.; Ollero, F.J.; Cubo, T. Plant growth promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014, 169, 325–336. [Google Scholar] [CrossRef]
- Chen, J.S.; Toth, J.; Kasap, M. Nitrogen-fixation genes and nitrogenase activity in Clostridium acetobutylicum and Clostridium beijerinckii. J. Ind. Microbiol. Biotechnol. 2001, 27, 281–286. [Google Scholar] [CrossRef]
- Bageshwar, U.K.; Srivastava, M.; Pardha-Saradhi, P.; Paul, S.; Gothandapani, S.; Jaat, R.S.; Shankar, P.; Yadav, R.; Biswas, D.R.; Kumar, P.A.; et al. An environmentally friendly engineered Azotobacter strain that replaces a substantial amount of urea fertilizer while sustaining the same wheat yield. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Bali, A.; Blanco, G.; Hill, S.; Kennedy, C. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl. Environ. Microbiol. 1992, 58, 1711–1718. [Google Scholar] [CrossRef]
- Ambrosio, R.; Ortiz-Marquez, J.C.F.; Curatti, L. Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae. Metab. Eng. 2017, 40, 59–68. [Google Scholar] [CrossRef]
- Orr, C.H.; James, A.; Leifert, C.; Cooper, J.M.; Cummings, S.P. Diversity and activity of free-living nitrogen-fixing bacteria and total bacteria in organic and conventionally managed soils. Appl. Environ. Microbiol. 2011, 77, 911–919. [Google Scholar] [CrossRef]
- Baldani, V.L.D.; Döbereiner, J. Host-plant specificity in the infection of cereals with Azospirillum spp. Soil Biol. Biochem. 1980, 12, 433–439. [Google Scholar] [CrossRef]
- Fox, J.E.; Gulledge, J.; Engelhaupt, E.; Burow, M.E.; McLachlan, J.A. Pesticides reduce symbiotic efficiency of nitrogen-fixing rhizobia and host plants. Proc. Natl. Acad. Sci. USA 2007, 104, 10282–10287. [Google Scholar] [CrossRef]
- Iniguez, A.L.; Dong, Y.; Triplett, E.W. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol. Plant Microbe Interact. 2004, 17, 1078–1085. [Google Scholar] [CrossRef]
- Geddes, B.A.; Ryu, M.H.; Mus, F.; Garcia Costas, A.; Peters, J.W.; Voigt, C.A.; Poole, P. Use of plant colonizing bacteria as chassis for transfer of N2-fixation to cereals. Curr. Opin. Biotechnol. 2015, 32, 216–222. [Google Scholar] [CrossRef]
- Fox, A.R.; Soto, G.; Valverde, C.; Russo, D.; Lagares, A., Jr.; Zorreguieta, Á.; Alleva, K.; Pascuan, C.; Frare, R.; Mercado-Blanco, J.; et al. Major cereal crops benefit from biological nitrogen fixation when inoculated with the nitrogen-fixing bacterium Pseudomonas protegens Pf-5 X940. Environ. Microbiol. 2016, 18, 3522–3534. [Google Scholar] [CrossRef]
- Ryu, M.H.; Zhang, J.; Toth, T.; Khokhani, D.; Geddes, B.A.; Mus, F.; Garcia-Costas, A.; Peters, J.W.; Poole, P.S.; Ané, J.M.; et al. Control of nitrogen fixation in bacteria that associate with cereals. Nat. Microbiol. 2020, 5, 314–330. [Google Scholar] [CrossRef]
- Downie, J.A. Legume haemoglobins: Symbiotic nitrogen fixation needs bloody nodules. Curr. Biol. 2005, 15, R196–R198. [Google Scholar] [CrossRef]
- Minchin, F.R.; James, E.K.; Becana, M. Oxygen diffusion, production of reactive oxygen and nitrogen species, and antioxidants in legume nodules. In Nitrogen-Fixing Leguminous Symbioses; Dilworth, M.J., James, E.K., Sprent, J.I., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 321–362. [Google Scholar] [CrossRef]
- Delaux, P.M.; Radhakrishnan, G.; Oldroyd, G. Tracing the evolutionary path to nitrogen-fixing crops. Curr. Opin. Plant Biol. 2015, 26, 95–99. [Google Scholar] [CrossRef]
- Zhang, X.X.; Tang, X.; Sheirdil, R.A.; Sun, L.; Ma, X.T. Rhizobium rhizoryzae sp. nov., isolated from rice roots. Int. J. Syst. Evol. Microbiol. 2014, 64, 1373–1377. [Google Scholar] [CrossRef]
- Mus, F.; Crook, M.B.; Garcia, K.; Garcia Costas, A.; Geddes, B.A.; Kouri, E.D.; Paramasivan, P.; Ryu, M.-H.; Oldroyd, G.E.D.; Poole, P.S.; et al. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 2016, 82, 3698–3710. [Google Scholar] [CrossRef] [PubMed]
- Brewin, N.J. An evolving story in biology and biotechnology: Legume root nodule symbiosis. Biochemist 2013, 35, 14–18. [Google Scholar] [CrossRef]
- Cocking, E.C. Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil 2003, 252, 169–175. [Google Scholar] [CrossRef]
- Dent, D.; Cocking, E. Establishing symbiotic nitrogen fixation in cereals and other non-legume crops: The greener nitrogen revolution. Agric. Food Secur. 2017, 6. [Google Scholar] [CrossRef]
- Van Deynze, A.; Zamora, P.; Delaux, P.M.; Heitmann, C.; Jayaraman, D.; Rajasekar, S.; Graham, D.; Maeda, J.; Gibson, D.; Schwartz, K.D.; et al. Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol. 2018, 16. [Google Scholar] [CrossRef]
- Bennett, A.B.; Pankievicz, V.C.S.; Ané, J.-M. A model for nitrogen fixation in cereal crops. Trends Plant Sci. 2020, 25, 226–235. [Google Scholar] [CrossRef]
- Bloch, S.E.; Clark, R.; Gottlieb, S.S.; Wood, L.K.; Shah, N.; Mak, S.-M.; Lorigan, J.G.; Johnson, J.; Davis-Richardson, A.G.; Williams, L.; et al. Biological nitrogen fixation in maize: Optimizing nitrogenase expression in a root-associated diazotroph. J. Exp. Bot. 2020. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; André, O.; Puech-Pagés, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A.; et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Altúzar-Molina, A.; Lozano, L.; Ortíz-Berrocal, M.; Ramírez, M.; Martínez, L.; de Lourdes Velázquez-Hernández, M.; Dhar-Ray, S.; Silvente, S.; Mariano, N.; Shishkova, S.; et al. Expression of the legume-specific nod factor receptor proteins alters developmental and immune responses in rice. Plant Mol. Biol. Rep. 2020, 38, 262–281. [Google Scholar] [CrossRef]
- Merrick, M.; Dixon, R. Why don’t plants fix nitrogen? Trends Biotechnol. 1984, 2, 162–166. [Google Scholar] [CrossRef]
- Ivleva, N.B.; Groat, J.; Staub, J.M.; Stephens, M. Expression of active subunit of nitrogenase via integration into plant organelle genome. PLoS ONE 2016, 11, e160951. [Google Scholar] [CrossRef] [PubMed]
- Eseverri, Á.; López-Torrejón, G.; Jiang, X.; Burén, S.; Rubio, L.M.; Caro, E. Use of synthetic biology tools to optimize the production of active nitrogenase Fe protein in chloroplasts of tobacco leaf cells. Plant Biotechnol. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xie, X.; Xiang, N.; Tian, Z.X.; Dixon, R.; Wang, Y.P. Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology. Proc. Natl. Acad. Sci. USA 2018, 115, E8509–E8517. [Google Scholar] [CrossRef]
- Liu, X.; Wang, M.; Song, Y.; Li, Y.; Liu, P.; Shi, H.; Li, Y.; Hao, T.; Zhang, H.; Jiang, W.; et al. Combined assembly and targeted integration of multigene for nitrogenase biosynthetic pathway in Saccharomyces cerevisiae. ACS Synth. Biol. 2019, 8, 1766–1775. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.S.; Tilbrook, K.; Warden, A.C.; Campbell, P.C.; Rolland, V.; Singh, S.P.; Wood, C.C. Expression of 16 nitrogenase proteins within the plant mitochondrial matrix. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Amarger, N. Genetically modified bacteria in agriculture. Biochimie 2002, 84, 1061–1072. [Google Scholar] [CrossRef]
- Hirsch, P.R. Release of transgenic bacterial inoculants—Rhizobia as a case study. Plant Soil 2005, 266, 1–10. [Google Scholar] [CrossRef]
- Vijayabharathi, R.; Sathya, A.; Gopalakrishnan, S. Formulation and commercialization of rhizobia: Asian scenario. In Agriculturally Important Microorganisms: Commercialization and Regulatory Requirements in Asia; Singh, H.B., Sarma, B.K., Keswani, C., Eds.; Springer: Singapore, 2016; pp. 47–67. [Google Scholar] [CrossRef]
- Owen, D.; Williams, A.P.; Griffith, G.W.; Withers, P.J.A. Use of commercial bio-inoculants to increase agricultural production through improved phosphrous acquisition. Appl. Soil Ecol. 2015, 86, 41–54. [Google Scholar] [CrossRef]
- Wozniak, C.A.; McClung, G.; Gagliardi, J.; Segal, M.; Matthews, K. Regulation of genetically engineered microorganisms under FIFRA, FFDCA and TSCA. In Regulation of Agricultural Biotechnology: The United States and Canada; Wozniak, C.A., McHughen, A., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 57–94. [Google Scholar] [CrossRef]
- Darch, H.; Shahsavarani, A. The Regulation of Organisms Used in Agriculture Under the Canadian Environmental Protection Act, 1999. In Regulation of Agricultural Biotechnology: The United States and Canada; Wozniak, C.A., McHughen, A., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 137–145. [Google Scholar] [CrossRef]
- Pandey, V.; Chandra, K. Agriculturally Important Microorganisms as Biofertilizers: Commercialization and Regulatory Requirements in Asia. In Agriculturally Important Microorganisms: Commercialization and Regulatory Requirements in Asia; Singh, H.B., Sarma, B.K., Keswani, C., Eds.; Springer: Singapore, 2016; pp. 133–145. [Google Scholar] [CrossRef]
| Group | Gene | Genotype | Phenotype | Reference |
|---|---|---|---|---|
| Adhesin biosynthesis | rapA1 | Overexpression in Rhizobium leguminosarum | Increased competitiveness and nodule occupation in red clover. | [20] |
| Antagonism related | TFX (peptide antibiotic trifolitoxin) | Production in Rhizobium etli | Higher rhizosphere competitiveness and nodulation. | [21] |
| Cellular replication | parA | Overexpression in Azorhizobium caulinodans | Single swollen bacteroid in one symbiosome, relatively narrow symbiosome space, and polyploid cells were observed when in symbiosis with Sesbania rostrate. | [22] |
| EPS biosynthesis | pssA and rosR | Overexpression in R. leguminosarum | Increased competitiveness and induced more nodules in clover plants. | [23] |
| exoY | Overexpression in Sinorhizobium meliloti | Higher shoot fresh weight and shoot length in Medicago truncatula. | [24] | |
| Heat stress | clpB | Extra copies in Mesorhizobium mediterraneum | Improvement in symbiosis under normal and acidic conditions. Overexpression of nodA and nodC. | [25] |
| groEL | Overexpression in Mesorhizobium | Improved symbiotic effectiveness in chickpea. | [26] | |
| Hydrogen uptake | hup | Gene from R. leguminosarum expressed in Rhizobium tropici and Rhizobium freirei | Increase in nodule efficiency and seed N content in Phaseolus vulgaris | [27] |
| Metal toxicity | MTL4 and AtPCS | Genes from Arabidopsis thaliana expressed in Mesorhizobium hauakuii | Increased Cd in nodules working on phytoremediation. | [28] |
| MTL4, AtPCS and AtIRT1 | Higher sensitivity and higher accumulation of Cd. Advantage in accumulation of Cu and As. | [29] | ||
| pSinA | Plasmid from Sinirhizobium inserted in several Alphaproteobacteria | Arsenic resistance and oxidation and heavy metal resistance. | [30] | |
| copAB | Gene from Pseudomonas fluorescens expressed in Sinorhizobium medicae | Improved root Cu accumulation without altering metal loading to shoots in M. truncatula. | [31] | |
| Improved root Cu tolerance in M. truncatula. | [32] | |||
| S-adenosyl-methionine methyltransferase | Gene from Chlamydomonas reinhardtii in R. leguminosarum bv. trifolii | Methylation of arsenite. | [33] | |
| ropAe | Deletion in R. etli | Cu tolerance enhanced. | [34] | |
| PsMT1 and PsMT2 | Metallothionein genes from pea expressed in R. leguminosarum | Improved tolerance to Cd depicting normal development of nodules. | [35] | |
| Molecular transport | dctA | Overexpression Rhizobium meliloti | Higher rate of nitrogen fixation in Medicago sativa. | [36] |
| nif genes | nifA | Gene from Klebsiella pneumonie overexpressed in S. meliloti | Increased nodulation competitiveness in alfalfa. | [13] |
| Did not affect S. meliloti competitiveness. | [37] | |||
| Extra copy in S. meliloti | Increased alfalfa biomass. | [16] | ||
| Overexpression in S. meliloti | Increased nodule formation efficiency and rhizopine synthesis. | [38] | ||
| Overexpression in Bradyrhizobium japonicum | Overexpression of groESL3. | |||
| Gene from K. pneumonie overexpressed in Sinorhizobium fredii | Accelerated nodulation and increased competitiveness in soybean. | [39] | ||
| Overexpression in S. meliloti | Improved nitrogen fixing efficiency in M. sativa. | [15] | ||
| nifHDK | Overexpression in R. etli | Increased nitrogenase activity and increased weight and yield in P. vulgaris. | [17] | |
| nod genes | Random DNA fragment | Random DNA duplication in R. tropici | More competitive strains for nodule formation in Macroptilium atropurprreum. | [9] |
| nodD1, nodABC and nifN | Overexpression in S. meliloti | Increase in nodulation, nitrogen fixation (acetylene reduction activity) and growth of alfalfa. | [40] | |
| nodD | Overexpression in R. leguminosarum | Increased nitrogen fixation in Vicia sativa and Trifolium repens. | [41] | |
| nodD1 | Delayed nodulation and reduced number of nodules on Vicia plants. | [42] | ||
| nodD2 | ||||
| nolR | Overexpression in S. fredii | Increased EPS production and less number of nodules on Glycine max. Increased number of nodules on Vigna unguiculata. | [43] | |
| Oxidative stress | fld | Gene from Anaboena variabilis Overexpressed in S. meliloti | Nodule senescence delayed in M. sativa. | [44] |
| Reduced structural alterations in alfalfa nodules. | [45] | |||
| Less decline in nitrogenase activity under salinity conditions. | [46] | |||
| Improves tolerance to oxidative stress and the survival in the presence of the herbicides paraquat and atrazine. | [47] | |||
| katB | Overexpression in S. meliloti | Aberration infection thread formation and delayed nodulation on M. sativa. | [48] | |
| cbb3 | Overexpression in B. japonicum | Increase in the symbiotic effectiveness and in O2 consumption rate (free-living cultures). | [49] | |
| Enhancement in symbiotic nitrogen fixation. | [50] | |||
| Overexpression in R. etli | Reduced sensitivity of symbiosis with P. vulgaris in drought conditions. | [51] | ||
| vktA (catalase) | Gene from Vibrio rumoiensis expressed in R. leguminosarum | Increased N fixation activity into nodules, reduced H2O2 production. | [52] | |
| ahpC | Overexpression in Anabaena | Lowered the peroxide, superoxide and malondialdehyde contents in Anabaena strains. | [53] | |
| Phosphate solubilization | appA | Gene from Citrobacter braakii overexpressed in rhizobia | Increased P content and shoot dry weight of Vigna radiataradiate. | [54] |
| Gene from Escherichia coli overexpressed in S. meliloti | Improvement of maize growth in low P soil. | [55] | ||
| Phytohormone modulation | acdS and lrpL (ACC deaminase) | Mutation in R. leguminosarum | Decreased nodulation in pea. | [56] |
| Genes from R. leguminosarum overexpressed in S. meliloti | Improved competitiveness, nodulation and shoot dry weight in alfalfa. | [57] | ||
| iaaM and tms2 | Overexpression in S. meliloti | Increased number of nodules in M. truncatula. | [58] | |
| Increased tolerance to UV, high salt, low pH and phosphate starvation. | [59] | |||
| Improved nitrogenase activity in nodules and increased stem dry weight. | [60] | |||
| Lower expression of ethylene signaling genes, released larger amounts of P-solubilizing organic acid and lower reduction in shoot dry-weight under P starvation on M. truncatula. | [61] | |||
| Induction of many of the transcriptional changes in free-living cells like those occur in nitrogen-fixing root nodule. Increased expression of nitrogen fixation genes and stress response-related genes. | [62] | |||
| Higher tolerance of alfalfa in drought conditions. Higher concentration of Rubisco and lower accumulation of ethylene in drought conditions. | [63] | |||
| Introduction of iaaM gene from Pseudomonas savastanoi and tms2 from Agrobacterium tumefaciens in R. leguminosarum | Fewer number of nodules (but heavier) and increased nitrogenase activity in vetch. | [64] | ||
| acdS (ACC deaminase) | Overexpression in Mesorhizobium loti | Higher nodulation in Lotus japonicus and Lotus tenuis, and improved competitiveness of the strain. | [65] | |
| Gene of Pseudomonas putida overexpressed in Mesorhizobium ciceri | Stimulated growth and increased nodulation on chickpea under normal and waterlogging stress conditions. | [66] | ||
| Increased nodulation, plant growth and biocontrol potential in chickpeas. | [67] | |||
| Improved growth of chickpea under saline conditions. | [68] | |||
| Gene of P. putida overexpressed in S. meliloti | Higher biomass of Medicago lupulina under copper stress. Enhancement of antioxidant defense system. | [69] | ||
| ipt (cytokinin) | ipt gene from Agrobacterium overexpressed in S. meliloti | Increased survival of nodules and increased production of antioxidants under drought conditions in alfalfa. | [70] | |
| miaA (cytokinin) | Mutation in Bradyrhizobium | Faster nodule formation and alteration of size and number of nodules in Aeschynomene. | [71] | |
| Polysaccharide biosynthesis | celC | Overexpression in R. leguminosarum | Reduction in biofilm formation, aberrant infection behavior, delay in nodulation and decreased root attachment in T. repens.. | [72] |
| Gene from R. leguminosarum overexpressed in S. meliloti | Delay in nodulation in M. truncatula. | [73] | ||
| Salinity and drought stress | putA | Overexpression in S. meliloti | Increased competitiveness in alfalfa plants under drought stress. | [74] |
| betS | Rapid acquisition of betaines and better maintenance of nitrogen fixation in salinized alfalfa. | [75] | ||
| otsA | Overexpression in R. etli | Improved number of nodules, nitrogenase activity and biomass in P. vulgaris. Plants recovered from drought stress. | [76] | |
| Gene from S. meliloti overexpressed in M. ciceri | Increased growth in saline media. Improved nodules formation and shoot biomass accumulation in chickpea growing in presence of NaCl. | [77] | ||
| Siderophore production | fegA | Gene from B. japonicum expressed in Mesorhizobium sp. | Increased growth and nodule occupancy in peanut plants. | [78] |
| fhuA | Gene from B. japonicum expressed in Rhizobium sp. | Increased growth and nodule occupancy in pigeon pea. | [79] | |
| Gene from E. coli overexpressed in Rhizobium ssp. | Increased nodulation and growth in pigeon pea. | [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goyal, R.K.; Schmidt, M.A.; Hynes, M.F. Molecular Biology in the Improvement of Biological Nitrogen Fixation by Rhizobia and Extending the Scope to Cereals. Microorganisms 2021, 9, 125. https://doi.org/10.3390/microorganisms9010125
Goyal RK, Schmidt MA, Hynes MF. Molecular Biology in the Improvement of Biological Nitrogen Fixation by Rhizobia and Extending the Scope to Cereals. Microorganisms. 2021; 9(1):125. https://doi.org/10.3390/microorganisms9010125
Chicago/Turabian StyleGoyal, Ravinder K., Maria Augusta Schmidt, and Michael F. Hynes. 2021. "Molecular Biology in the Improvement of Biological Nitrogen Fixation by Rhizobia and Extending the Scope to Cereals" Microorganisms 9, no. 1: 125. https://doi.org/10.3390/microorganisms9010125
APA StyleGoyal, R. K., Schmidt, M. A., & Hynes, M. F. (2021). Molecular Biology in the Improvement of Biological Nitrogen Fixation by Rhizobia and Extending the Scope to Cereals. Microorganisms, 9(1), 125. https://doi.org/10.3390/microorganisms9010125







