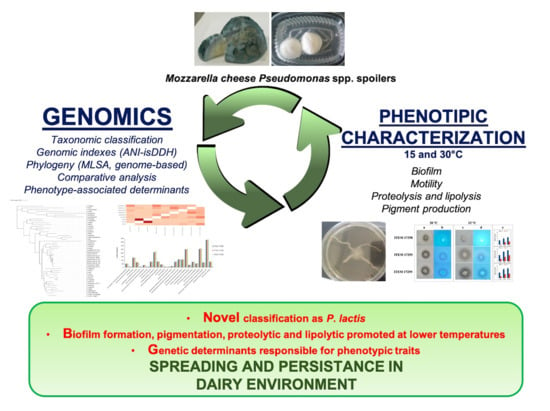
Genomic Analysis of Three Cheese-Borne Pseudomonas lactis with Biofilm and Spoilage-Associated Behavior
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Condition
2.2. Genome Sequencing and Assembly
2.3. Bioinformatic Methods
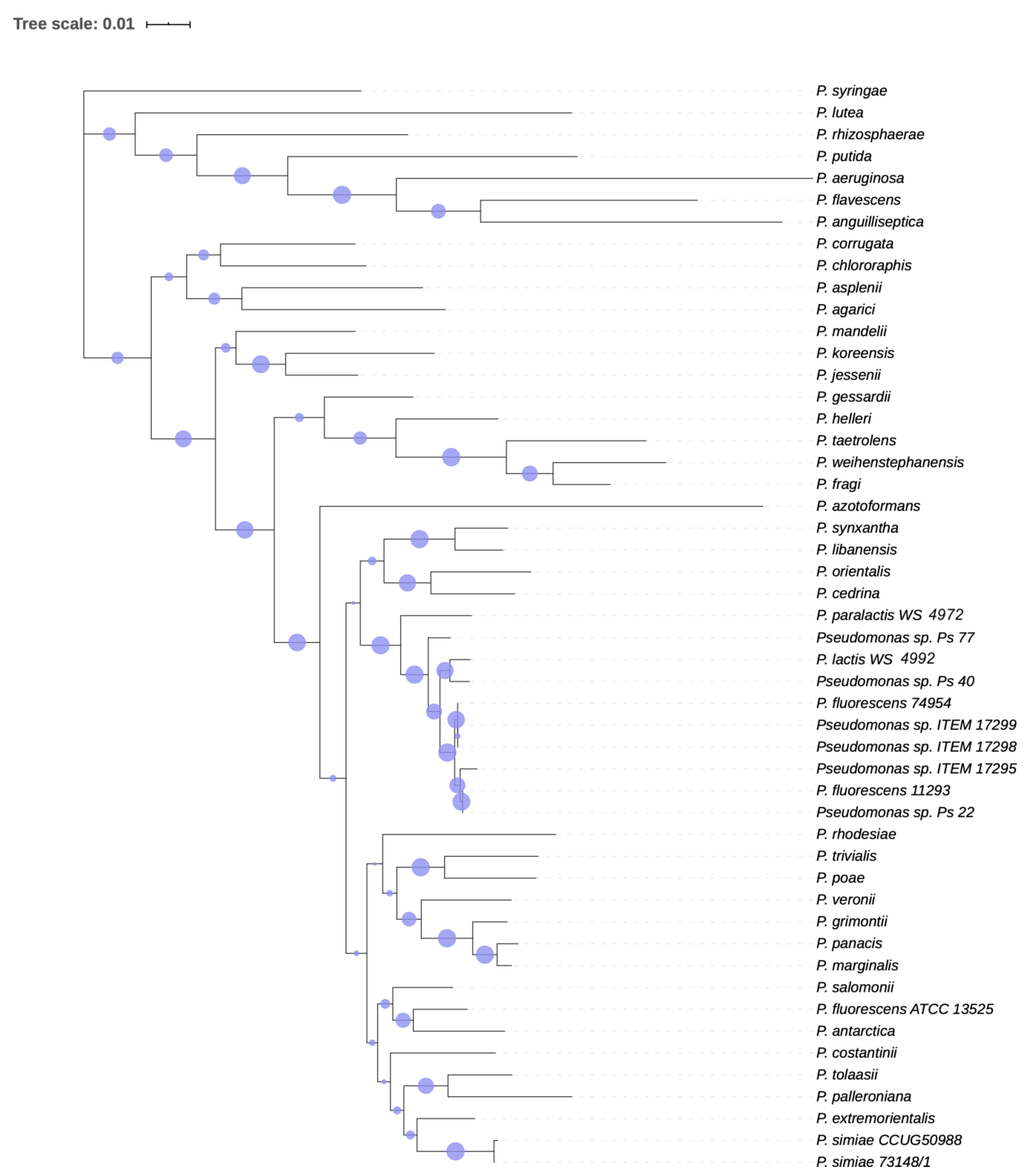
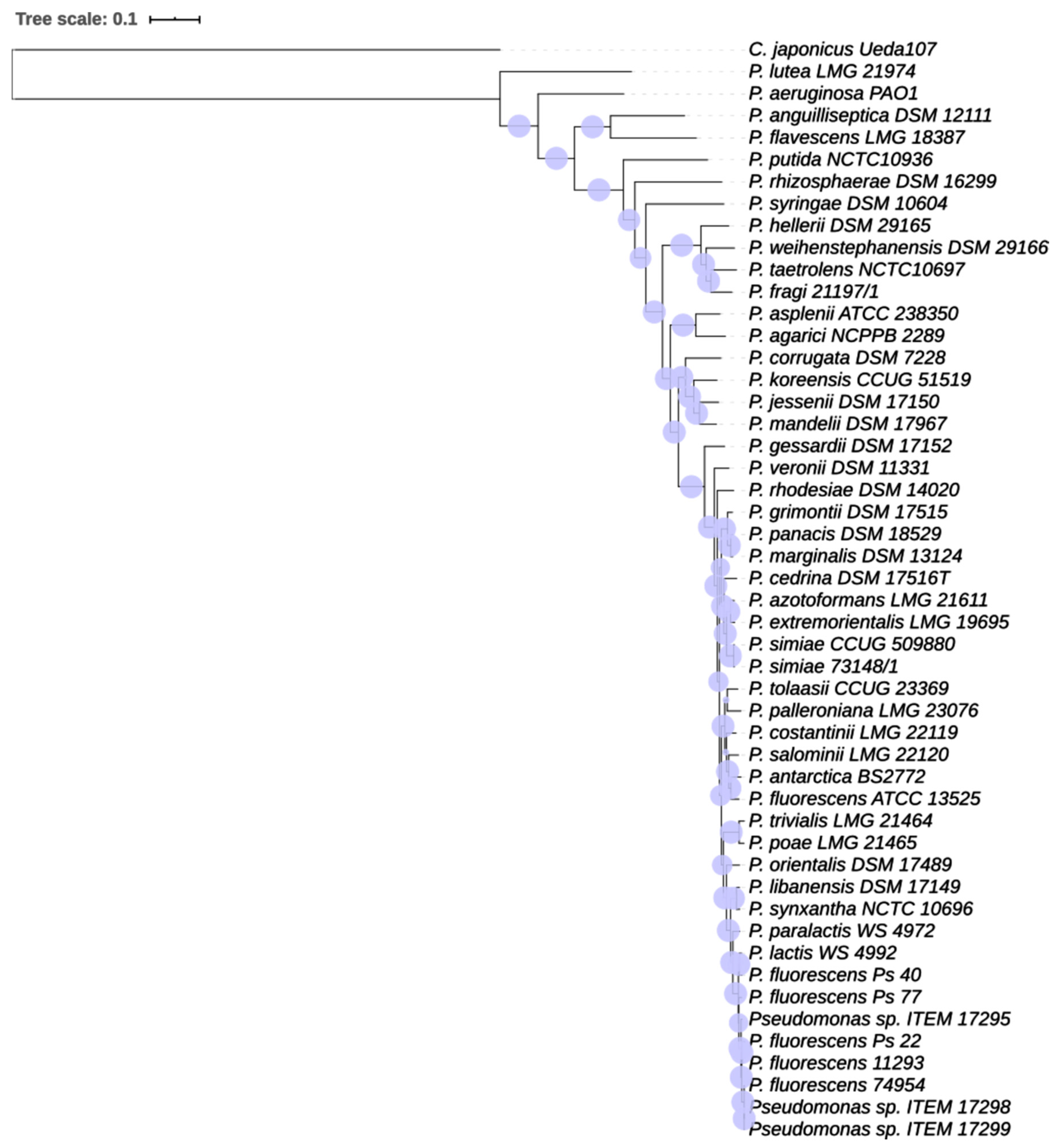
2.4. Phylogenetic Analysis
2.5. Phenotypic Analysis under Two Temperatures of Incubation
2.5.1. Static Biofilm Formation and Motility Assays
2.5.2. Exopolysaccharides (EPS) Production by Congo Red Binding Assay
2.5.3. Proteolytic and Lipolytic Activity
2.5.4. Pigment Production
2.6. Statistical Analyses
3. Results and Discussion
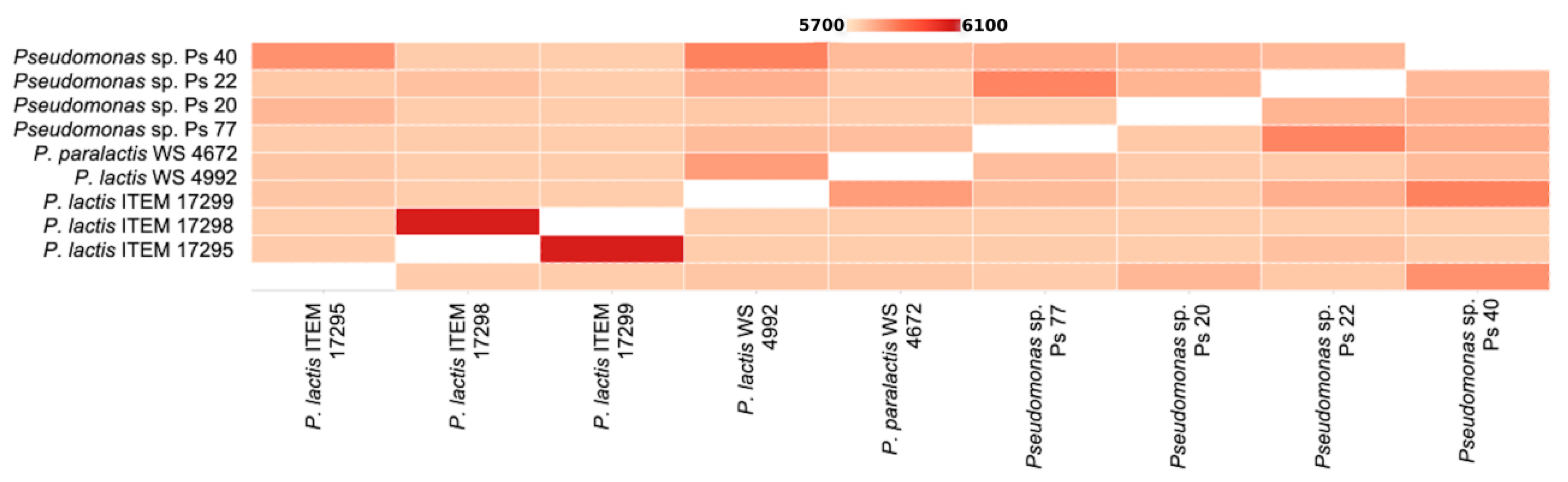
3.1. Genomic Analysis of Pseudomonas spp. Strains
3.1.1. General Features of Pseudomonas sp. Genomes
3.1.2. Phylogenetic Analysis
3.1.3. Protein Functional Classification
3.2. Phenotypic and Genomic Characterization
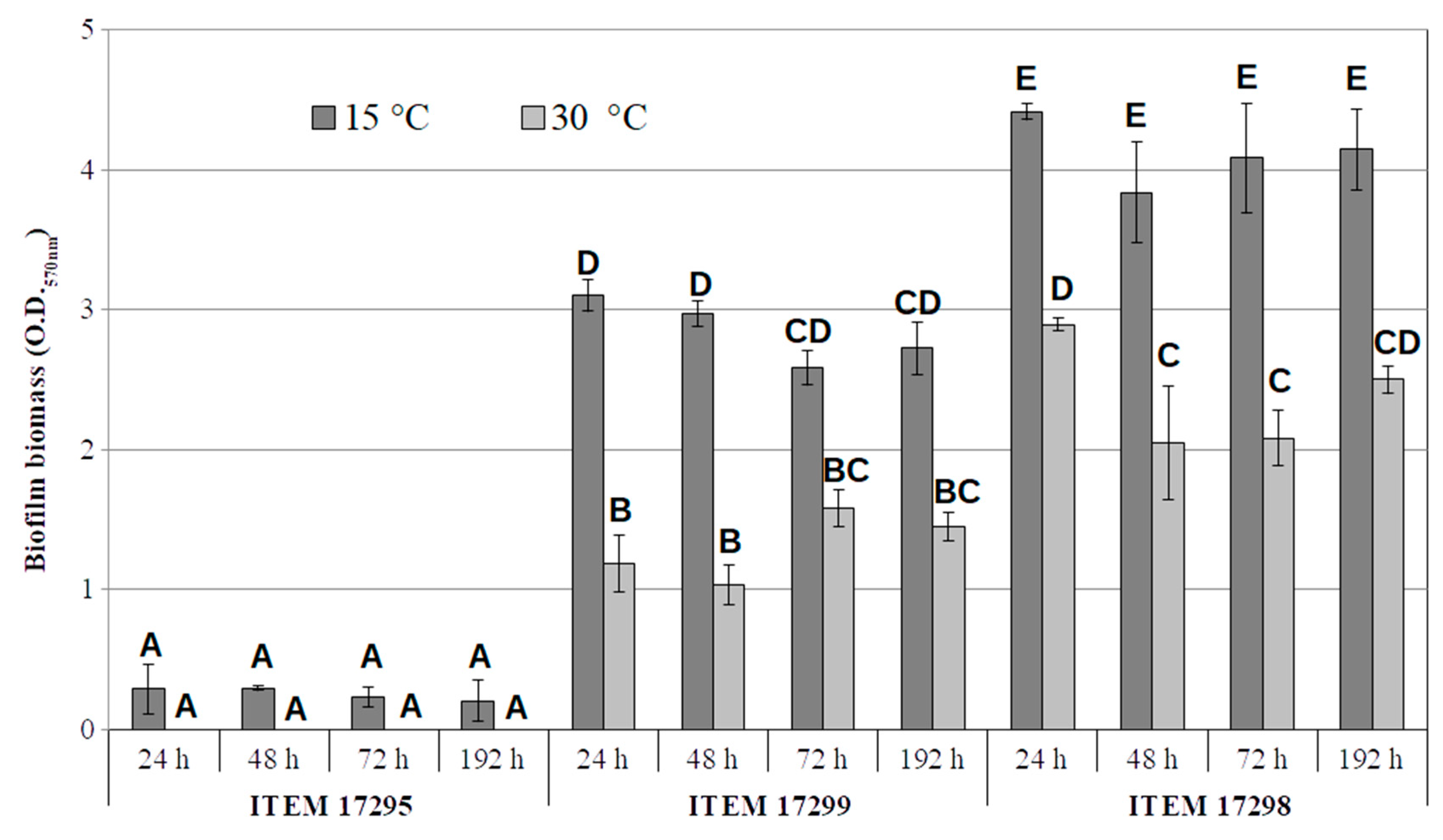
Biofilm and Motility
3.3. Pigment Production
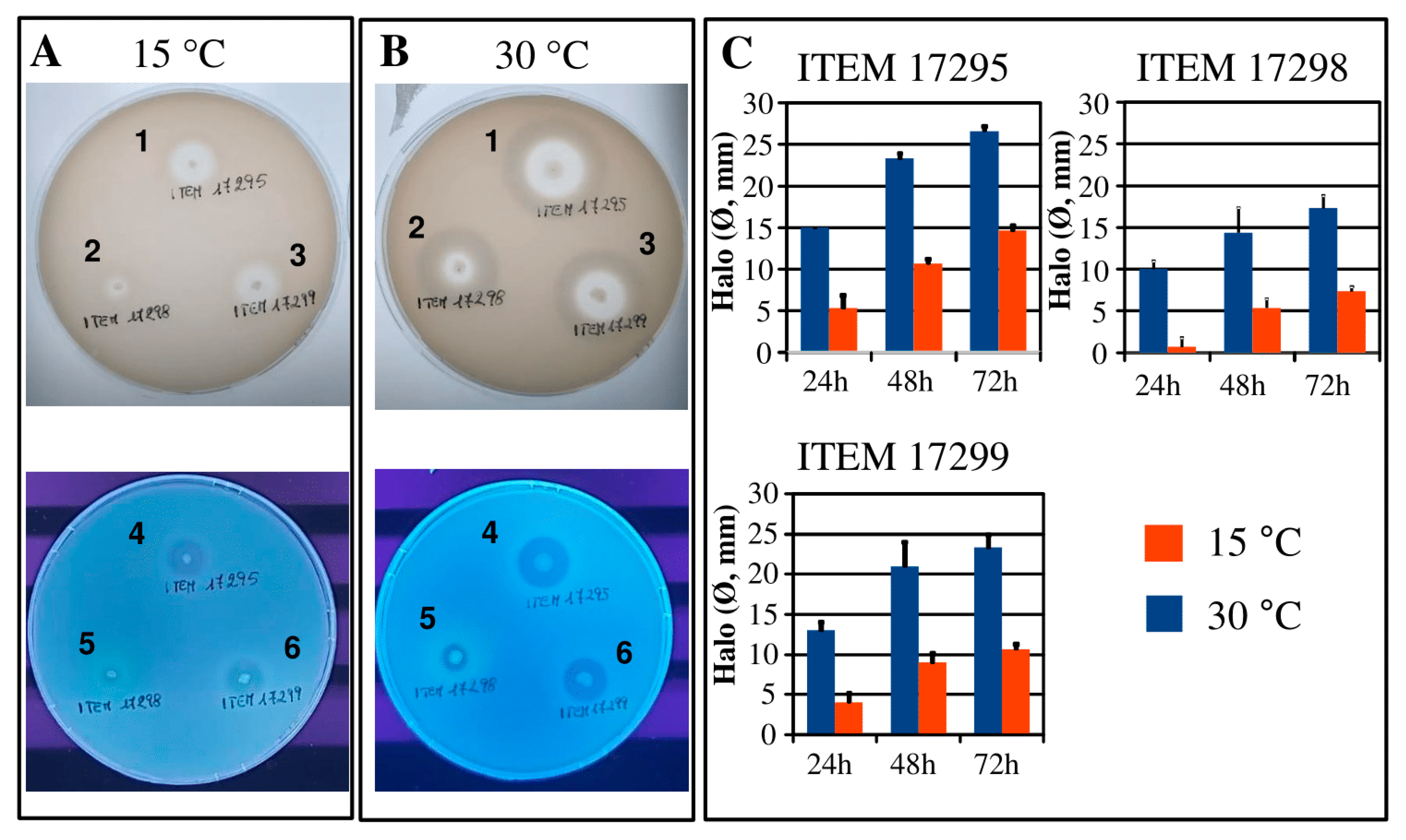
3.4. Protease and Lipase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MLST | Multilocus Sequence Typing |
| COG | Clusters of Orthologous Groups |
| CV | Crystal Violet |
| EPS | Extracellular Polymeric Substances |
| QS | Quorum Sensing |
| TA | Toxin–Antitoxin |
| RAxML | Randomized Axelerated Maximum Likelihood |
| GO | Gene Ontology |
References
- Gellatly, S.G.; Hancock, R.E.W. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Wang, L.; Wang, W.; Wu, G.; Tao, F.; Xu, P.; Deng, Z.; Tang, H. Regulatory mechanism of nicotine degradation in Pseudomonas putida. mBio 2019, 10, e602–e619. [Google Scholar] [CrossRef] [Green Version]
- Naz, T.; Khan, M.D.; Ahmed, I.; Rehman, S.U.; Rha, E.S.; Malook, I.; Jamil, M. Biosorption of heavy metals by Pseudomonas species isolated from sugar industry. Toxicol. Ind. Health. 2016, 32, 1619–1627. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E. The current status on the taxonomy of Pseudomonas revisited: An update. Infect. Genet. Evol. 2018, 57, 106–116. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 2009, 9, 1132–1147. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, A.; De Meyer, S.; Trujillo, M.E. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Chung, M.; Munro, J.B.; Tettelin, H.; Dunning Hotopp, J.C. Using core genome alignments to assign bacterial species. mSystems 2018, 3, e00236-18. [Google Scholar] [CrossRef] [Green Version]
- Gomila, M.; Peña, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 2015, 6, 214. [Google Scholar] [CrossRef] [Green Version]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [PubMed] [Green Version]
- von Neubeck, M.; Huptas, C.; Gluck, C.; Krewinkel, M.; Stoeckel, M.; Stressler, T.; Fischer, L.; Hinrichs, J.; Scherer, S.; Wenning, M. Pseudomonas lactis spp. nov. and Pseudomonas paralactis spp. nov.; isolated from bovine raw milk. Int. J. Syst. Evol. Microbiol. 2017, 67, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, C.; Yamada, K.; Takeuchi, H.; Inokuchi, Y.; Kashiwagi, A.; Toba, T. A lytic bacteriophage for controlling Pseudomonas lactis in raw cow’s milk. Appl. Environ. Microbiol. 2018, 84, e00111–e00118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baruzzi, F.; Lagonigro, R.; Quintieri, L.; Morea, M.; Caputo, L. Occurrence of non-lactic acid bacteria populations involved in protein hydrolysis of cold-stored high moisture mozzarella cheese. Food Microbiol. 2012, 30, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Arslan, S.; Eyi, A.; Özdemir, F. Spoilage potentials and antimicrobial resistance of Pseudomonas spp. isolated from cheeses. J. Dairy Sci. 2011, 94, 5851–5856. [Google Scholar] [CrossRef] [Green Version]
- Raposo, A.; Pérez, E.; de Faria, C.T.; Ferrús, M.A.; Carrascosa, C. Food spoilage by Pseudomonas spp.—An Overview. In Foodborne Pathogens and Antibiotic Resistance; Singh, O.V., Ed.; Wiley & Sons: New York, NY, USA, 2017; pp. 41–58. [Google Scholar]
- Caputo, L.; Quintieri, L.; Bianchi, D.M.; Decastelli, L.; Monaci, L.; Visconti, A.; Baruzzi, F. Pepsin-digested bovine lactoferrin prevents mozzarella cheese blue discoloration caused by Pseudomonas fluorescens. Food Microbiol. 2015, 46, 15–24. [Google Scholar] [CrossRef]
- Decimo, M.; Cabeza, M.C.; Ordóñez, J.A.; De Noni, I.; Brasca, M. Volatile organic compounds associated with milk spoilage by psychrotrophic bacteria. Int. J. Dairy Technol. 2018, 71, 593–600. [Google Scholar] [CrossRef]
- De Filippis, F.; La Storia, A.; Villani, F.; Ercolini, D. Strain-level diversity analysis of Pseudomonas fragi after in situ pangenome reconstruction shows distinctive spoilage-associated metabolic traits clearly selected by different storage conditions. Appl. Environ. Microbiol. 2109, 85, e02212-18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wei, J.; Yuan, Y.; Yue, T. Diversity and characterization of spoilage-associated psychrotrophs in food in cold chain. Int. J. Food Microbiol. 2019, 290, 86–95. [Google Scholar] [CrossRef]
- Teh, K.H.; Flint, S.; Palmer, J.; Andrewes, P.; Bremer, P.; Lindsay, D. Biofilm—An unrecognised source of spoilage enzymes in dairy products? Int. Dairy J. 2014, 34, 32–40. [Google Scholar] [CrossRef]
- Quintieri, L.; Zühlke, D.; Fanelli, F.; Caputo, L.; Liuzzi, V.C.; Logrieco, A.F.; Hirschfeld, C.; Becher, D.; Riedel, K. Proteomic analysis of the food spoiler Pseudomonas fluorescens ITEM 17298 reveals the antibiofilm activity of the pepsin-digested bovine lactoferrin. Food Microbiol. 2019, 82, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.H.; Murphy, S.C.; Ralyea, R.D.; Wiedmann, M.; Boor, K.J. When cheese gets the blues: Pseudomonas fluorescens as the causative agent of cheese spoilage. J. Dairy Sci. 2011, 94, 3176–3183. [Google Scholar] [CrossRef] [PubMed]
- Quintieri, L.; Fanelli, F.; Zühlke, D.; Caputo, L.; Logrieco, A.F.; Albrecht, D.; Riedel, K. Biofilm and pathogenesis-related proteins in the foodborne P. fluorescens ITEM 17298 with distinctive phenotypes during cold storage. Front. Microbiol. 2020, 11, 991. [Google Scholar] [CrossRef] [PubMed]
- Quintieri, L.; Fanelli, F.; Caputo, L. Antibiotic resistant Pseudomonas spp. spoilers in fresh dairy products: An underestimated risk and the control strategies. Foods 2019, 8, 372. [Google Scholar] [CrossRef] [Green Version]
- Quintieri, L.; Caputo, L.; Morea, M.; Baruzzi, F. Control of Mozzarella spoilage bacteria by using bovine lactoferrin pepsin-digested hydrolysate. In Worldwide Research Efforts in the Fighting against Microbial Pathogens; Mendez-Vilas, A., Ed.; Brown Walker Press: Boca-Raton, FL, USA, 2013; pp. 118–122. [Google Scholar]
- Guidone, A.; Zotta, T.; Matera, A.; Ricciardi, A.; De Filippis, F.; Ercolini, D.; Parente, E. The microbiota of high-moisture mozzarella cheese produced with different acidification methods. Int. J. Food Microbiol. 2016, 216, 9–17. [Google Scholar] [CrossRef]
- Caputo, L.; Quintieri, L.; Cavalluzzi, M.M.; Lentini, G.; Habtemariam, S. Antimicrobial and antibiofilm activities of citrus water-extracts obtained by microwave-assisted and conventional methods. Biomedicines 2018, 6, 70. [Google Scholar] [CrossRef] [Green Version]
- Ding, T.; Li, T.; Li, J. Identification of natural product compounds as quorum sensing inhibitors in Pseudomonas fluorescens P07 through virtual screening. Bioorg. Med. Chem. 2018, 26, 4088–4099. [Google Scholar] [CrossRef]
- Quintieri, L.; Carito, A.; Pinto, L.; Calabrese, N.; Baruzzi, F.; Caputo, L. Application of lactoferricin B to control microbial spoilage in cold stored fresh foods. In Multidisciplinary Approach for Studying and Combating Microbial Pathogens; Mendèz-Vilas, A., Ed.; Brown Walker Press: Boca-Raton, FL, USA, 2015; pp. 58–62. [Google Scholar]
- Fanelli, F.; Liuzzi, V.C.; Quintieri, L.; Mulè, G.; Baruzzi, F.; Logrieco, A.F.; Caputo, L. Draft genome sequence of Pseudomonas fluorescens strain ITEM 17298, associated with cheese spoilage. Genome Announc. 2017, 5, e01141-17. [Google Scholar] [CrossRef] [Green Version]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-R, L.M.; Gunturu, S.; Harvey, W.T.; Rosselló-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) webserver: Taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacteria Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2107, 45, D535–D542. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. Peer J. Prepr. 2016, 4, e1900v1. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M.; Klenk, H.-P. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter dish biofilm formation assay. JoVE 2011, 47, e2437. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Curty, L.K.; Barja, F.; Van Delden, C.; Pechère, J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouker, G.; Jaeger, K.E. Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microbiol. 1987, 53, 211–213. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Sadiq, F.A.; Burmølle, M.; Liu, T.; He, G. Insights into bacterial milk spoilage with particular emphasis on the roles of heat-stable enzymes, biofilms, and quorum sensing. J. Food Prot. 2018, 81, 1651–1660. [Google Scholar] [CrossRef]
- Machado, I.; Silva, L.R.; Giaouris, E.D.; Melo, L.F.; Simões, M. Quorum sensing in food spoilage and natural-based strategies for its inhibition. Food Res. Int. 2020, 127, 108754. [Google Scholar] [CrossRef]
- Wang, X.Y.; Xie, J. Quorum sensing system-regulated proteins affect the spoilage potential of co-cultured Acinetobacter johnsonii and Pseudomonas fluorescens from spoiled bigeye tuna (Thunnus obesus) as determined by proteomic analysis. Front. Microbiol. 2020, 11, 940. [Google Scholar] [CrossRef]
- Rossi, C.; Chaves-López, C.; Serio, A.; Goffredo, E.; Goga, B.T.; Paparella, A. Influence of incubation conditions on biofilm formation by Pseudomonas fluorescens isolated from dairy products and dairy manufacturing plants. Ital. J. Food Saf. 2016, 5, 5793. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.; Serio, A.; Chaves-López, C.; Anniballi, F.; Auricchio, B.; Goffredo, E.; Cenci-Goga, B.T.; Lista, F.; Fillo, S.; Paparella, A. Biofilm formation, pigment production and motility in Pseudomonas spp. isolated from the dairy industry. Food Cont. 2018, 86, 241–248. [Google Scholar] [CrossRef]
- Wang, H.; Qi, J.; Dong, Y.; Li, Y.; Xu, X.; Zhou, G. Characterization of attachment and biofilm formation by meat-borne Enterobacteriaceae strains associated with spoilage. LWT 2017, 86, 399–407. [Google Scholar] [CrossRef]
- Nakamura, S.; Tohru, M. Flagella-Driven Motility of Bacteria. Biomolecules 2019, 9, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chattopadhyay, M.K.; Raghu, G.; Sharma, Y.V.; Biju, A.R.; Rajasekharan, M.V.; Shivaji, S. Increase in oxidative stress at low temperature in an Antarctic Bacterium. Curr. Microbiol. 2011, 62, 544–546. [Google Scholar] [CrossRef]
- Friedman, L.; Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 2004, 186, 4457–4465. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Preston, J.F., 3rd; Romeo, T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 2004, 186, 2724–2734. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Guo, Y.; Tang, K.; Wen, Z.; Li, B.; Zeng, Z.; Wang, X. MqsR/MqsA toxin/antitoxin system regulates persistence and biofilm formation in Pseudomonas putida KT2440. Front. Microbiol. 2017, 8, 840. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, K. Type II Toxin-Antitoxins Loci: The relBE Family. In Prokaryotic Toxin-Antitoxins; Gerdes, K., Ed.; Springer: Newcastle upon Tyne, UK, 2013; pp. 69–92. [Google Scholar]
- Fulcher, N.B.; Holliday, P.M.; Klem, E.; Cann, M.J.; Wolfgang, M.C. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol. Microbiol. 2010, 76, 889–904. [Google Scholar] [CrossRef] [Green Version]
- Starnbach, M.N.; Lory, S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 1992, 6, 459–469. [Google Scholar] [CrossRef]
- Totten, P.A.; Lara, J.C.; Lory, S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 1990, 172, 389–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, A.; Schirm, M.; Arora, S.K.; Thibault, P.; Logan, S.M.; Ramphal, R. Glycosylation of b-Type flagellin of Pseudomonas aeruginosa: Structural and genetic basis. J. Bacteriol. 2006, 188, 4395–4403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Andreani, N.A.; Carraro, L.; Zhang, L.; Vos, M.; Cardazzo, B. Transposon mutagenesis in Pseudomonas fluorescens reveals genes involved in blue pigment production and antioxidant protection. Food Microbiol. 2019, 82, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Ringel, M.T.; Brüser, T. The biosynthesis of pyoverdines. Microb. Cell. 2019, 5, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Cunliffe, H.E.; Merriman, T.R.; Lamont, I.L. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J. Bacteriol. 1995, 177, 2744–2750. [Google Scholar] [CrossRef] [Green Version]
- Andreani, N.A.; Carraro, L.; Martino, M.E.; Fondi, M.; Fasolato, L.; Miotto, G.; Magro, M.; Vianello, F.; Cardazzo, B. A genomic and transcriptomic approach to investigate the blue pigment phenotype in Pseudomonas fluorescens. Int. J. Food Microbiol. 2015, 213, 88–98. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, C.N.; Woods, R.G.; Beacham, I.R. Regulation of the aprX-lipA operon of Pseudomonas fluorescens B52: Differential regulation of the proximal and distal genes, encoding protease and lipase, by ompR-envZ. FEMS Microbiol. Lett. 2004, 241, 243–248. [Google Scholar] [CrossRef] [Green Version]
- Dufour, D.; Nicodème, M.; Perrin, C.; Driou, A.; Brusseaux, E.; Humbert, G.; Gaillard, J.L. Molecular typing of industrial strains of Pseudomonas spp. isolated from milk and genetical and biochemical characterization of an extracellular protease produced by one of them. Int. J. Food Microbiol. 2008, 125, 188–196. [Google Scholar] [CrossRef]
- Woods, R.G.; Burger, M.; Beven, C.A.; Beacham, I.R. The aprX–lipA operon of Pseudomonas fluorescens B52: A molecular analysis of metalloprotease and lipase production. Microbiology 2001, 147, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Marchand, S.; Vandriesche, G.; Coorevits, A.; Coudijzer, K.; De Jonghe, V.; Dewettinck, K.; De Vos, P.; Devreese, B.; Heyndrickx, M.; De Block, J. Heterogeneity of heat-resistant proteases from milk Pseudomonas species. Int. J. Food Microbiol. 2009, 133, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Machado, S.G.; Baglinière, F.; Marchand, S.; Van Coillie, E.; Vanetti, M.C.; De Block, J.; Heyndrickx, M. The Biodiversity of the microbiota producing heat-resistant enzymes responsible for spoilage in processed bovine milk and dairy products. Front. Microbiol. 2017, 8, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, T.F.; Martin, L.W.; Lamont, I.L. Activation of a cell surface signaling pathway in Pseudomonas aeruginosa requires ClpP protease and new sigma factor synthesis. Front. Microbiol. 2017, 8, 2442. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.G.; Luke, R.K.J. Siderophore production and utilization by milk spoilage Pseudomonas species. J. Dairy Sci. 2010, 93, 1355–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilderman, P.J.; Vasil, A.I.; Johnson, Z.; Wilson, M.J.; Cunliffe, H.E.; Lamont, I.L.; Vasil, M.L. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 2001, 69, 5385–5394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirienko, D.R.; Kang, D.; Kirienko, N.V. Novel pyoverdine inhibitors mitigate Pseudomonas aeruginosa pathogenesis. Front. Microbiol. 2019, 9, 3317. [Google Scholar] [CrossRef] [Green Version]
| Features | ITEM 17295 | ITEM 17298 | ITEM 17299 |
|---|---|---|---|
| Genome size (bp) | 6,288,321 | 6,288,436 | 6,244,929 |
| GC (%) | 59.83 | 59.75 | 59.78 |
| Number of contigs | 89 | 117 | 137 |
| Completeness | 93.7% | 93.7% | 93.7% |
| Quality | 89.2 (excellent) | 89.2 (excellent) | 89.2 (excellent) |
| Contig N50 | 351,021 | 146,994 | 139,064 |
| Genes (total) | 5736 | 5910 | 5752 |
| Genes (coding) | 5595 | 5734 | 5617 |
| Coding density | 88.81% | 88.60% | 88.55% |
| rRNAs | 2, 1, 1 (5S, 16S, 23S) | 3, 1, 1 (5S, 16S, 23S) | 2, 1, 1 (5S, 16S, 23S) |
| tRNAs | 68 | 55 | 54 |
| Biofilm Formation Step | Genes | ITEM 17295 | ITEM 17298 | ITEM 17299 | |
|---|---|---|---|---|---|
| Attachment | Switching from planktonic to biofilm state | csrA/rsmA | GIB65_24765 | PROKKA_01891 | GIB64_12670 |
| c-di-GMP regulated gacS/gacA | GIB65_18615/ GIB65_01605 | PROKKA_02681/ PROKKA_01410 | GIB64_19800 GIB64_28045 | ||
| luxQ | GIB65_08270 | PROKKA_00312 | GIB64_13935 | ||
| luxR | GIB65_04815 | PROKKA_03671 | GIB64_14810 | ||
| rhlR/rhlI | GIB65_06785/ GIB65_06780 | PROKKA_01310/ PROKKA_01311 | GIB64_11890/ GIB64_11895 | ||
| sagS | GIB65_08650 | PROKKA_03896 | GIB64_26065 | ||
| hptB | GIB65_07985 | PROKKA_01759 | GIB64_17280 | ||
| hsbR-hsbA | GIB65_07985- GIB65_07990 | PROKKA_01760- PROKKA_01761 | GIB64_17280- GIB64_17285 | ||
| ladS/retS | GIB65_05330/ GIB65_20530 | PROKKA_03621/ PROKKA_03260 | GIB64_14560/ GIB64_03985 | ||
| c-di-GMP regulated bifA | GIB65_02210 | PROKKA_05637 | GIB64_07760 | ||
| cAMP Phosphodiesterase cdpA | GIB65_19990 | PROKKA_03152 | GIB64_21855 | ||
| rbdA | GIB65_07415 | PROKKA_01243 | GIB64_00920 | ||
| ydaM | GIB65_04035 | PROKKA_01515 | GIB64_25020 | ||
| yfiB | GIB65_04455 | PROKKA_01205 | GIB64_26920 | ||
| c-di-GMP regulated tpbB-yfiN | GIB65_04460 | PROKKA_01204 | GIB64_26915 | ||
| yfiR | GIB65_04465 | PROKKA_01203 | GIB64_26910 | ||
| mqsR/mqsA | pseudo */ GIB65_14795 | PROKKA_01685/ PROKKA_01686 | GIB64_02008/ GIB64_02009 | ||
| chpB/chpS | NA | PROKKA_00869/ PROKKA_00870 | GIB64_11655/ GIB64_11650 | ||
| Pilus biosynthesis and twitching motility | pilz domain-containing protein | GIB65_04785 | PROKKA_03677 | GIB64_14840 | |
| pilGHIJ | GIB65_22310 GIB65_22295 | PROKKA_00456- PROKKA_00459 | GIB64_13350 GIB64_13365 | ||
| pilA | GIB64_11150 | PROKKA_00968 | GIB65_15860 | ||
| Flagellar gene expression (swimming, swarming motility) | fleQ | GIB65_08040 | PROKKA_01770 | GIB64_17225 | |
| fliA | GIB65_07910 | PROKKA_01745 | GIB64_17355 | ||
| flgM | GIB65_27400 | PROKKA_05108 | GIB64_24150 | ||
| Microbial cell adhesion and/or swarming motility | rhlA | GIB65_22015 | PROKKA_02434 | GIB64_28365 | |
| pvdQ | GIB65_09880 | PROKKA_00251 | GIB64_05820 | ||
| pleD | GIB65_20585 | PROKKA_03271 | GIB64_04040 | ||
| cyclic-Di-GMP phosphodiesterase bifA | GIB65_02210 | PROKKA_05637 | GIB64_07760 | ||
| Adhesion | pgaABCD | GIB65_26770 GIB65_26785 | PROKKA_04557 PROKKA_04560 | GIB64_08340 GIB64_08355 | |
| Chemotaxis | cheA/cheW | GIB65_22290/ GIB65_22295 | PROKKA_00460/ PROKKA_00461 | GIB64_13370/ GIB64_13375 | |
| cheBAZY | GIB65_07890 GIB65_07905 | PROKKA_01741 PROKKA_01744 | GIB64_17375 GIB64_17360 | ||
| wspRFEDCBA | GIB65_13580 GIB65_13610 | PROKKA_04814 PROKKA_04820 | GIB64_04640- GIB64_04610 | ||
| SurfaceSensing | cAMP-vfr | GIB65_21375 | PROKKA_04713 | GIB64_01775 | |
| Microcolonies formation and maturation | QS-signals | trpE/trpG | GIB65_21405/ GIB65_21395 | PROKKA_04707/ PROKKA_04709 | GIB64_01805/ GIB64_01795 |
| fabG | GIB65_27840 | PROKKA_02061 | GIB64_16215 | ||
| ppkA | GIB65_23520 | PROKKA_03312 | GIB64_09480 | ||
| Cell-cell signals | type VI secretion system T6SS | GIB65_23600 GIB65_23520 | PROKKA_03296 PROKKA_03311 | GIB64_09560 GIB64_09480 | |
| type VI secretion system protein TssB | GIB65_19080 GIB65_19150 | PROKKA_04236 PROKKA_04222 | GIB64_25965 GIB64_25895 | ||
| bfmR/bfmS | GIB65_09040/ GIB65_09045 | PROKKA_00299/ PROKKA_00300 | GIB64_14000/ NA | ||
| Eps formation | pslABCDEFGHIJKL | GIB65_16955 GIB65_16900 | PROKKA_04436 PROKKA_04447 | GIB64_17790 GIB64_17845 | |
| pelABCDEFG | NA 1 | PROKKA_02829- PROKKA_02835 | GIB64_21390- GIB64_21420 | ||
| Alginatebiosynthesis | mucR | GIB65_00065 | PROKKA_05333 | GIB64_26185 | |
| algD operon | GIB65_14860- GIB65_14920 | PROKKA_01700- PROKKA_01712 | GIB64_10145- GIB64_10205 | ||
| Detachment | chemotaxis | Biofilm dispersion protein BdlA | GIB65_02045, GIB65_16355 | PROKKA_03805, PROKKA_05334 | GIB64_15510, GIB64_03410 |
| Phytochrome-like protein cph2 | GIB65_12695 | PROKKA_00099 | GIB64_12320 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quintieri, L.; Caputo, L.; De Angelis, M.; Fanelli, F. Genomic Analysis of Three Cheese-Borne Pseudomonas lactis with Biofilm and Spoilage-Associated Behavior. Microorganisms 2020, 8, 1208. https://doi.org/10.3390/microorganisms8081208
Quintieri L, Caputo L, De Angelis M, Fanelli F. Genomic Analysis of Three Cheese-Borne Pseudomonas lactis with Biofilm and Spoilage-Associated Behavior. Microorganisms. 2020; 8(8):1208. https://doi.org/10.3390/microorganisms8081208
Chicago/Turabian StyleQuintieri, Laura, Leonardo Caputo, Maria De Angelis, and Francesca Fanelli. 2020. "Genomic Analysis of Three Cheese-Borne Pseudomonas lactis with Biofilm and Spoilage-Associated Behavior" Microorganisms 8, no. 8: 1208. https://doi.org/10.3390/microorganisms8081208
APA StyleQuintieri, L., Caputo, L., De Angelis, M., & Fanelli, F. (2020). Genomic Analysis of Three Cheese-Borne Pseudomonas lactis with Biofilm and Spoilage-Associated Behavior. Microorganisms, 8(8), 1208. https://doi.org/10.3390/microorganisms8081208