The Physiological Responses of Escherichia coli Triggered by Phosphoribulokinase (PrkA) and Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco)
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Media and Cultivations
2.3. Analytical Methods
3. Results
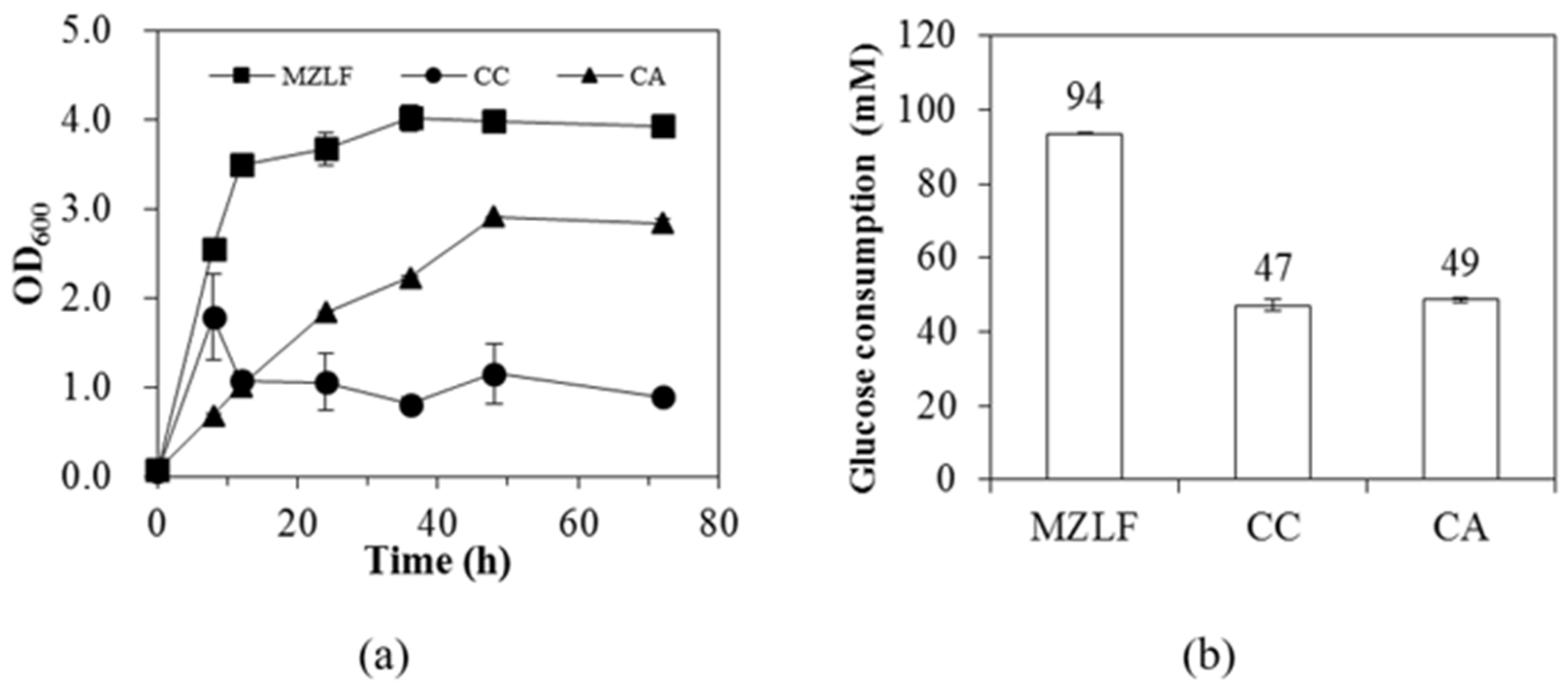
3.1. Phenotypes of E. coli Strains CC and CA
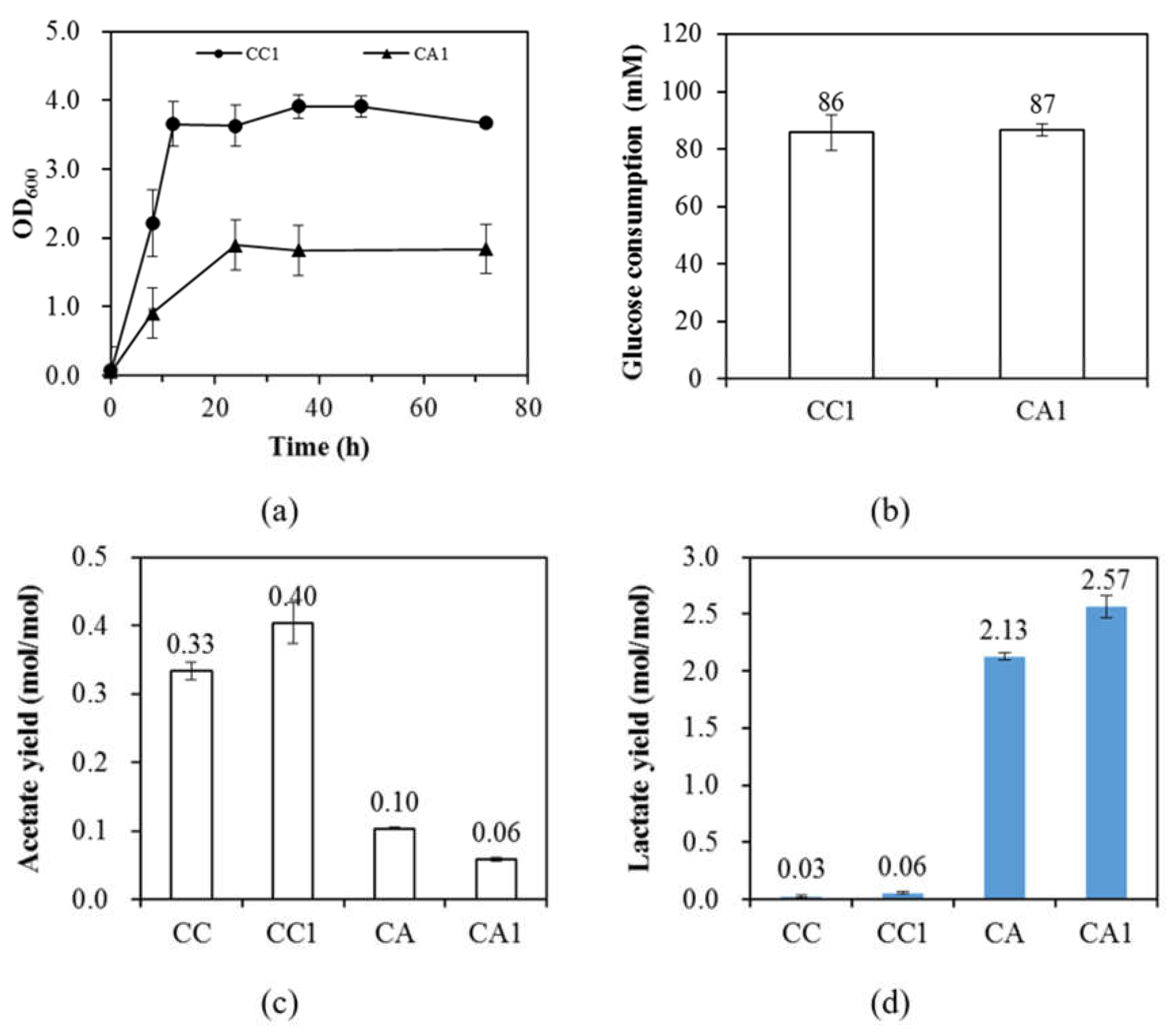
3.2. The Presence of PrkA Further Creates the Demand for ATP
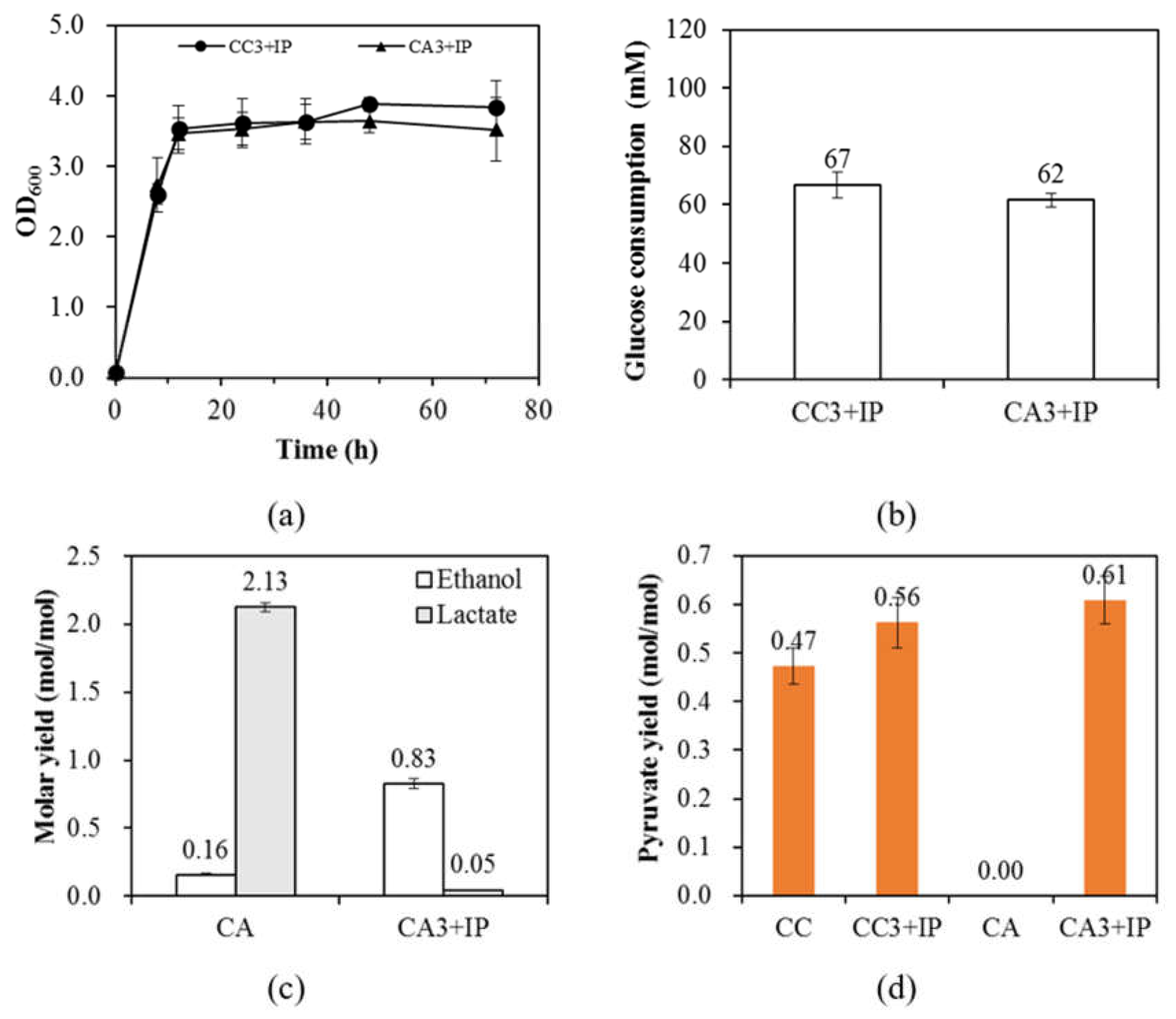
3.3. The Overexpression of Rubisco Strongly Directs Carbon into Biomass and Ethanol Production
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nowicka, B.; Kruk, J. Powered by light: Phototrophy and photosynthesis in prokaryotes and its evolution. Microbiol. Res. 2016, 186–187, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, A.A. Synthesis and assembly of bacterial and higher plant Rubisco subunits in Escherichia coli. Photosynth. Res. 1988, 17, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Van der Vies, S.M.; Bradley, D.; Gatenby, A.A. Assembly of cyanobacterial and higher plant ribulose bisphosphate carboxylase subunits into functional homologous and heterologous enzyme molecules in Escherichia coli. EMBO J. 1986, 5, 2439–2444. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, Y.; Yoshida, S.; Fujihashi, M.; Kitagawa, K.; Doi, T.; Atomi, H.; Imanaka, T.; Miki, K. Structure-based catalytic optimization of a type III rubisco from a hyperthermophile. J. Biol. Chem. 2010, 285, 39339–39347. [Google Scholar] [CrossRef]
- Mueller-cajar, O.; Whitney, S.M. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem. J. 2008, 414, 205–214. [Google Scholar] [CrossRef]
- Parikh, M.R.; Greene, D.N.; Woods, K.K.; Matsumura, I. Directed evolution of RuBisCO hypermorphs through genetic selection in engineered E. coli. Protein Eng. Des. Sel. 2006, 19, 113–119. [Google Scholar] [CrossRef]
- Zhuang, Z.-Y.; Li, S.-Y. Rubisco-based engineered Escherichia coli for in situ carbon dioxide recycling. Bioresour. Technol. 2013, 150, 79–88. [Google Scholar] [CrossRef]
- Li, Y.-H.; Ou-Yang, F.-Y.; Yang, C.-H.; Li, S.-Y. The coupling of glycolysis and the Rubisco-based pathway through the non-oxidative pentose phosphate pathway to achieve low carbon dioxide emission fermentation. Bioresour. Technol. 2015, 187, 189–197. [Google Scholar] [CrossRef]
- Yang, C.-H.; Liu, E.-J.; Chen, Y.-L.; Ou-Yang, F.-Y.; Li, S.-Y. The comprehensive profile of fermentation products during in situ CO2 recycling by Rubisco-based engineered Escherichia coli. Microb. Cell Factories 2016, 15, 1–10. [Google Scholar] [CrossRef]
- Tseng, I.T.; Chen, Y.-L.; Chen, C.-H.; Shen, Z.-X.; Yang, C.-H.; Li, S.-Y. Exceeding the theoretical fermentation yield in mixotrophic Rubisco-based engineered Escherichia coli. Metab. Eng. 2018, 47, 445–452. [Google Scholar] [CrossRef]
- Nikel, P.I.; Zhu, J.; San, K.-Y.; Méndez, B.S.; Bennett, G.N. Metabolic flux analysis of Escherichia coli creB and arcA mutants reveals shared control of carbon catabolism under microaerobic growth conditions. J. Bacteriol. 2009, 191, 5538–5548. [Google Scholar] [CrossRef] [PubMed]
- Fong, S.S.; Nanchen, A.; Palsson, B.O.; Sauer, U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J. Biol. Chem. 2006, 281, 8024–8033. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Eiteman, M.A.; DeWitt, K.; Altman, E. Homolactate fermentation by metabolically engineered Escherichia coli Strains. Appl. Environ. Microbiol. 2007, 73, 456–464. [Google Scholar] [CrossRef]
- Castaño-Cerezo, S.; Pastor, J.M.; Renilla, S.; Bernal, V.; Iborra, J.L.; Cánovas, M. An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli. Microb. Cell Fact. 2009, 8, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.P.; Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1995, 158, 9–14. [Google Scholar] [CrossRef]
- Chen, S.-K.; Chin, W.-C.; Tsuge, K.; Huang, C.-C.; Li, S.-Y. Fermentation approach for enhancing 1-butanol production using engineered butanologenic Escherichia coli. Bioresour. Technol. 2013, 145, 204–209. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Beard, K.F.M.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef]
- Sauer, U.; Eikmanns, B.J. The PEP—Pyruvate—Oxaloacetate node as the switch point for carbon flux distribution in bacteria: We dedicate this paper to Rudolf, K.; Thauer, Director of the Max-Planck-Institute for Terrestrial Microbiology in Marburg, Germany, on the occasion of his 65th birthday. FEMS Microbiol. Rev. 2005, 29, 765. [Google Scholar]
- Dittrich, C.R.; Bennett, G.N.; San, K.-Y. Characterization of the acetate-producing pathways in Escherichia coli. Biotechnol. Progr. 2005, 21, 1062–1067. [Google Scholar] [CrossRef]
- Chang, Y.-Y.; Wang, A.-Y.; Cronan, J.E. Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS (katF) gene. Mol. Microbiol. 1994, 11, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Koebmann, B.J.; Westerhoff, H.V.; Snoep, J.L.; Nilsson, D.; Jensen, P.R. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 2002, 184, 3909–3916. [Google Scholar] [CrossRef] [PubMed]
- Yomano, L.P.; York, S.W.; Shanmugam, K.T.; Ingram, L.O. Deletion of methylglyoxal synthase gene (mgsA) increased sugar co-metabolism in ethanol-producing Escherichia coli. Biotechnol. Lett. 2009, 31, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.W.; Saini, M.; Lin, L.-J.; Chiang, C.-J.; Chao, Y.-P. Systematic engineering of Escherichia coli for d-lactate production from crude glycerol. J. Agric. Food Chem. 2015, 63, 9583–9589. [Google Scholar] [CrossRef]
- Grabar, T.B.; Zhou, S.; Shanmugam, K.T.; Yomano, L.P.; Ingram, L.O. Methylglyoxal bypass identified as source of chiral contamination in l (+) and d (−)-lactate fermentations by recombinant Escherichia coli. Biotechnol. Lett. 2006, 28, 1527–1535. [Google Scholar] [CrossRef]
- Mazumdar, S.; Blankschien, M.D.; Clomburg, J.M.; Gonzalez, R. Efficient synthesis of L-lactic acid from glycerol by metabolically engineered Escherichia coli. Microb. Cell Fact. 2013, 12, 1–11. [Google Scholar] [CrossRef]
- Weber, J.; Kayser, A.; Rinas, U. Metabolic flux analysis of Escherichia coli in glucose-limited continuous culture. II. Dynamic response to famine and feast, activation of the methylglyoxal pathway and oscillatory behaviour. Microbiology 2005, 151, 707–716. [Google Scholar] [CrossRef]
- Knoop, H.; Gründel, M.; Zilliges, Y.; Lehmann, R.; Hoffmann, S.; Lockau, W.; Steuer, R. Flux Balance analysis of cyanobacterial metabolism: The metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol. 2013, 9, e1003081. [Google Scholar] [CrossRef]
- Shastri, A.A.; Morgan, J.A. Flux balance analysis of photoautotrophic metabolism. Biotechnol. Progr. 2005, 21, 1617–1626. [Google Scholar] [CrossRef]
- Partridge, J.D.; Sanguinetti, G.; Dibden, D.P.; Roberts, R.E.; Poole, R.K.; Green, J. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 2007, 282, 11230–11237. [Google Scholar] [CrossRef]

| Name | Descriptions | Reference |
|---|---|---|
| Bacterial strains | ||
| E. coli BL21 (DE3) | F-, dcm, ompT, gal, lon, hsdSB(rB-, mB-), λ(DE3[lacI, lacUV5-T7 gene 1, ind1, sam7,nin5]) | A gift from Prof. Huang, Chieh-Chen at NCHU, Taiwan |
| MZLF | E. coli BL21(DE3) Δzwf, Δldh, Δfrd | [9] |
| CC | E. coli BL21(DE3) Δzwf, Δldh, Δfrd, Δppc | This study |
| CC1 | E. coli BL21(DE3) Δzwf, Δldh, Δfrd, Δppc harboring PBAD-his6-prkA-pACYC184 | This study |
| CC3 | E. coli BL21(DE3) Δzwf, Δldh, Δfrd, Δppc harboring rbcLS-pET30a + (M259T) | This study |
| CA | E. coli BL21(DE3) Δzwf, Δldh, Δfrd, Δppc, Δpta | This study |
| CA1 | E. coli BL21(DE3) Δzwf, Δldh, Δfrd, Δppc, Δpta harboring PBAD-his6-prkA-pACYC184 | This study |
| CA3 | E. coli BL21(DE3) Δzwf, Δldh, Δfrd, Δppc, Δpta harboring rbcLS-pET30a + (M259T) | This study |
| Plasmids | ||
| pKD46 | araC, bla, oriR101, repA101(Ts), araC-ParaB-γ-β-exo (encode λ Red recombinases), temperature-conditional replicon | [15] |
| pkD13 | bla, FRT-kan-FRT | [15] |
| pCP20 | FLP+, λ cI857 +,λ PR Pepts, bla, catF | [16] |
| PBAD-his6-prkA-pACYC184 | Recombinant plasmid carries prkA gene (derived from Synechococcus PCC7492) for the overexpresion of phosphoribulokinase (PrkA) under the control of PBAD | [6] |
| rbcLS-pET30a + (M259T) | Recombinant plasmid carries engineered rbcLS gene (originated from Synechococcus PCC6301) for the overexpresion of engineered Rubisco (M259T) under the control of PT7 | [6] |
| Name | Sequence |
|---|---|
| PPChf-HP1 | CGTGAAGGATACAGGGCTATCAAACGATAAGATGGGGTGTCTGGGGTAAT GTGTAGGCTGGAGCTGCTTC |
| PPChr-HP2 | ATTTCAGAAAACCCTCGCGCAAAAGCACGAGGGTTTGCAGAAGAGGAAGAA TTCCGGGGATCCGTCGACC |
| PPCa-U | CGTGAAGGAT ACAGGGCTATC |
| KANk2- k2 | CGGTGCCCTGAATGAACTGC |
| pta-HP1 | ACACCGCCAGCTCAGCTGGCGGTGCTGTTTTGTAACCCGCCAAATCGGCGGTAACGAAAGAGGATAAACC GTGTAGGCTGGAGCTGCTTC |
| pta-HP2 | TAAAAAACCGGAAATAGTGATTATTTCCGGTTCAGATATCCGCAGCGCAAAGCTGCGGATGATGACGAGAATTCCGGGGATCCGTCGACC |
| acka-pta-U-new | GTGTCATCATGCGCTACGCT |
| Strain 1 | Fermentation Product Yield (mol/mol) 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Glucose Consumption (mM) | Formate | Acetate | Ethanol | Succinate | Pyruvate | Lactate | CO2 | Biomass | |
| MZLF | 94 ± 0 | 1.15 ± 0.00 | 0.37 ± 0.00 | 0.84 ± 0.00 | 0.009 ± 0.000 | 0.41 ± 0.00 | 0.13 ± 0.01 | 0.13 ± 0.02 | 0.62 ± 0.02 |
| CC | 47 ± 2 | 1.13 ± 0.06 | 0.33 ± 0.01 | 0.82 ± 0.06 | 0.006 ± 0.002 | 0.47 ± 0.04 | 0.03 ± 0.01 | 0.09 ± 0.05 | 0.25 ± 0.02 |
| CC1 | 86 ± 6 | 1.20 ± 0.12 | 0.40 ± 0.03 | 0.95 ± 0.18 | 0.006 ± 0.001 | 0.46 ± 0.10 | 0.06 ± 0.01 | 0.10 ±0.01 | 0.66 ± 0.06 |
| CC3 + IP | 67 ± 5 | 1.06 ± 0.09 | 0.29 ± 0.03 | 0.76 ± 0.10 | 0.008 ± 0.002 | 0.56 ± 0.05 | 0.05 ± 0.01 | 0.04 ± 0.01 | 0.79 ± 0.09 |
| CA | 49 ± 1 | 0.27 ± 0.01 | 0.10 ± 0.00 | 0.16 ± 0.01 | 0.017 ± 0.000 | 0.00 ± 0.00 | 2.13 ± 0.03 | 0.09 ± 0.00 | 0.66 ± 0.01 |
| CA1 | 87 ± 2 | 0.16 ± 0.00 | 0.06 ± 0.00 | 0.04 ± 0.00 | 0.003 ± 0.000 | 0.02 ± 0.00 | 2.57 ± 0.10 | 0.04 ± 0.00 | 0.30 ± 0.01 |
| CA3 + IP | 62 ± 2 | 1.16 ± 0.05 | 0.33 ± 0.02 | 0.83 ± 0.04 | 0.010 ± 0.002 | 0.61 ± 0.05 | 0.05 ± 0.00 | 0.04 ± 0.01 | 0.85 ± 0.07 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, E.-J.; Tseng, I.-T.; Chen, Y.-L.; Pang, J.-J.; Shen, Z.-X.; Li, S.-Y. The Physiological Responses of Escherichia coli Triggered by Phosphoribulokinase (PrkA) and Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco). Microorganisms 2020, 8, 1187. https://doi.org/10.3390/microorganisms8081187
Liu E-J, Tseng I-T, Chen Y-L, Pang J-J, Shen Z-X, Li S-Y. The Physiological Responses of Escherichia coli Triggered by Phosphoribulokinase (PrkA) and Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco). Microorganisms. 2020; 8(8):1187. https://doi.org/10.3390/microorganisms8081187
Chicago/Turabian StyleLiu, En-Jung, I-Ting Tseng, Yi-Ling Chen, Ju-Jiun Pang, Zhi-Xuan Shen, and Si-Yu Li. 2020. "The Physiological Responses of Escherichia coli Triggered by Phosphoribulokinase (PrkA) and Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco)" Microorganisms 8, no. 8: 1187. https://doi.org/10.3390/microorganisms8081187
APA StyleLiu, E.-J., Tseng, I.-T., Chen, Y.-L., Pang, J.-J., Shen, Z.-X., & Li, S.-Y. (2020). The Physiological Responses of Escherichia coli Triggered by Phosphoribulokinase (PrkA) and Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco). Microorganisms, 8(8), 1187. https://doi.org/10.3390/microorganisms8081187





