Microbiome Diversity and Community-Level Change Points within Manure-based small Biogas Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characteristics of the Biogas Plants and Sampling of Digester Content
2.2. Chemical Analyses
2.3. Inventory of the Taxonomic Microbiome Diversity
2.4. Microbial Community Profiling Using TRFLP and System Ecological Data Evaluation
3. Results and Discussion
3.1. Performance of the Analyzed Manure-Based Small Biogas Plants
3.2. Inventory of the Taxonomic Microbiome Diversity
3.3. Dynamic Variation of the Microbial Community Over Time
3.4. Community-Level Change Points and Potential Indicator Taxa
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ahammad, S.Z.; Sreekrishnan, T.R. Biogas: An Evolutionary Perspective in the Indian Context. In Green Fuels Technology; Soccol, C.R., Brar, S.K., Faulds, C., Ramos, L.P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 431–443. ISBN 978-3-319-30203-4. [Google Scholar]
- Chen, B.; Hayat, T.; Alsaedi, A. Biogas Systems in China; Springer International Publishing: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Kemausuor, F.; Adaramola, M.S.; Morken, J. A review of commercial biogas systems and lessons for Africa. Energies 2018, 11, 2984. [Google Scholar] [CrossRef] [Green Version]
- Tabatabaei, M.; Ghanavati, H. Biogas—Fundamentals, Process, and Operation; Springer International Publishing: Cham, Switzerland, 2018; Volume 6, pp. 35–49, 437–472. [Google Scholar]
- Hahn, H.; Krautkremer, B.; Hartmann, K.; Wachendorf, M. Review of concepts for a demand-driven biogas supply for flexible power generation. Renew. Sustain. Energy Rev. 2014, 29, 383–393. [Google Scholar] [CrossRef]
- Lauer, M.; Thrän, D. Flexible biogas in future energy systems - Sleeping beauty for a cheaper power generation. Energies 2018, 11, 761. [Google Scholar] [CrossRef] [Green Version]
- Theuerl, S.; Herrmann, C.; Heiermann, M.; Grundmann, P.; Landwehr, N.; Kreidenweis, U.; Prochnow, A. The future agricultural biogas plant in Germany: A vision. Energies 2019, 12, 396. [Google Scholar] [CrossRef] [Green Version]
- Agostini, A.; Ferdinando Battini, F.; Giuntoli, J.; Tabaglio, V.; Padella, M.; Baxter, D.; Marelli, L.; Amaducci, S. Environmentally sustainable biogas? The key role of manure co-digestion with energy crops. Energies 2015, 8, 5234–5265. [Google Scholar] [CrossRef]
- Dalgaard, T.; Olesen, J.E.; Petersen, S.O.; Petersen, B.M.; Jørgensen, U.; Kristensen, T.; Hutchings, N.J.; Gyldenkærne, S.; Hermansen, J.E. Developments in greenhouse gas emissions and net energy use in Danish agriculture e How to achieve substantial CO2 reductions? Environ. Pollut. 2011, 159, 3193–3203. [Google Scholar] [CrossRef]
- Meyer-Aurich, A.; Schattauer, A.; Hellebrand, H.-J.; Klauss, H.; Plöchl, M.; Berg, W. Impacts of uncertainties on greenhouse gas mitigation potential of biogas production from agricultural resources. Renew. Energy 2012, 37, 277–284. [Google Scholar] [CrossRef]
- Weiske, A.; Vabitsch, A.; Olesen, J.E.; Schelde, K.; Michel, J.; Friedrich, R.; Kaltschmitt, M. Mitigation of greenhouse gas emissions in European conventional and organic dairy farming. Agric. Ecosyst. Environ. 2006, 112, 221–232. [Google Scholar] [CrossRef]
- Agostini, A.; Battini, F.; Padella, M.; Giuntoli, J.; Baxter, D.; Marelli, L.; Amaducci, S. Economics of GHG emissions mitigation via biogas production from sorghum, maize and dairy farm manure digestion in the Po valley. Biomass Bioenerg. 2016, 89, 58–66. [Google Scholar] [CrossRef]
- Kalt, G.; Lauk, C.; Mayer, A.; Theurl, M.C.; Kaltenegger, K.; Winiwarter, W.; Erb, K.-H.; Matej, S.; Haberl, H. Greenhouse gas implications of mobilizing agricultural biomass for energy: A reassessment of global potentials in 2050 under different food-system pathways. Environ. Res. Lett. 2020, 15, 034066. [Google Scholar] [CrossRef]
- Scholz, L.; Meyer-Aurich, A.; Kirschke, D. Greenhouse gas mitigation potential and mitigation costs of biogas production in Brandenburg, Germany. AgBioForum 2011, 14, 133–141. [Google Scholar]
- Kaparaju, P.; Rintala, J. Mitigation of greenhouse gas emissions by adopting anaerobic digestion technology on dairy, sow and pig farms in Finland. Renew. Energy 2011, 36, 31–41. [Google Scholar] [CrossRef]
- Massé, D.I.; Talbot, G.; Gilbert, Y. On farm biogas production: A method to reduce GHG emissions and develop more sustainable livestock operations. Anim. Feed Sci. Technol. 2011, 166-167, 436–445. [Google Scholar] [CrossRef]
- Arthurson, V. Closing the global energy and nutrient cycles through application of biogas residue to agricultural land–Potential benefits and drawbacks. Energies 2009, 2, 226–242. [Google Scholar] [CrossRef] [Green Version]
- Valentinuzzi, F.; Cavani, L.; Porfido, C.; Terzano, R.; Pii, Y.; Cesco, S.; Marzadori, C.; Mimmo, T. The fertilising potential of manure-based biogas fermentation residues: Pelleted vs. liquid digestate. Heliyon 2020, 6, e03325. [Google Scholar] [CrossRef] [PubMed]
- Baute, K.A.; Robinson, D.E.; van Eerd, L.L.; Edson, M.; Sikkema, P.H.; Gilroyed, B.H. Survival of seeds from perennial biomass species during commercial-scale anaerobic digestion. Weed Res. 2016, 56, 258–266. [Google Scholar] [CrossRef]
- Fröschle, B.; Heiermann, M.; Lebuhn, M.; Messelhäusser, U.; Plöchl, M. Hygiene and sanitation in biogas plants. Adv. Biochem. Eng. Biotechnol. 2015, 151, 63–69. [Google Scholar] [PubMed]
- Insam, H.; Gomez-Brandon, M.; Ascher, J. Manure-based biogas fermentation residues—Friend or foe of soil fertility? Soil Biol. Biochem. 2015, 84, 1–14. [Google Scholar] [CrossRef]
- Massé, D.I.; Saady, N.M.C.; Gilbert, Y. Potential of biological processes to eliminate antibiotics in livestock manure: An overview. Animals 2014, 4, 146–163. [Google Scholar] [CrossRef] [Green Version]
- Daniel-Gromke, J.; Rensberg, N.; Denysenko, V.; Stinner, W.; Schmalfuß, T.; Scheftelowitz, M.; Nelles, M.; Liebetrau, J. Current developments in production and utilization of biogas and biomethane in Germany. Chem. Ing. Tech. 2018, 90, 17–35. [Google Scholar] [CrossRef]
- German Environment Agency [GEA]—UNFCCC-Submission. Submission under the United Nations Framework Convention on Climate Change and the Kyoto Protocol 2019, National Inventory Report for the German Greenhouse Gas Inventory 1990–2017; German Environment Agency: Dessau-Roßlau, Germany, 2019; Available online: https://www.umweltbundesamt.de/publikationen (accessed on 30 June 2020).
- Budde, J.; Prochnow, A.; Plöchl, M.; Suárez, T.; Heiermann, M. Energy balance, greenhouse gas emissions, and profitability of thermobarical pretreatment of cattle waste in anaerobic digestion. Waste Manag. 2016, 49, 390–410. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, W.; Wang, X.; Gabauer, W.; Ortner, M.; Li, Z. Tackling ammonia inhibition for efficient biogas production from chicken manure: Status and technical trends in Europe and China. Renew. Sustain. Energy Rev. 2018, 97, 186–199. [Google Scholar] [CrossRef]
- Hadin, Å. From waste problem to renewable energy resource – exploring horse manure as feedstock for anaerobic digestions. Doctor of Philosophy, University of Gävle, Gävle, Sweden, 9 November 2018. [Google Scholar]
- Mönch-Tegeder, M.; Lemmer, A.; Oechsner, H.; Jungbluth, T. Investigation of the methane potential of horse manure. Agric. Eng. Int.: CIGR J. 2013, 15, 161–172. [Google Scholar]
- Morozova, I.; Nikulina, N.; Oechsner, H.; Krümpel, J.; Lemmer, A. Effects of increasing nitrogen content on process stability and reactor performance in anaerobic digestion. Energies 2020, 13, 1139. [Google Scholar] [CrossRef] [Green Version]
- Budde, J.; Heiermann, M.; Suárez Quiñones, T.; Plöchl, M. Effects of thermobarical pretreatment of cattle waste as feedstock for anaerobic digestion. Waste Manag. 2014, 34, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Cu, T.T.T.; Nguyen, T.X.; Triolo, J.M.; Pedersen, L.; Le, V.D.; Le, P.D.; Sommer, S.G. Biogas production from vietnamese animal manure, plant residues and organic waste: Influence of biomass composition on methane yield. Asian-Australas. J. Anim. Sci. 2015, 28, 280–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kafle, G.K.; Chen, L. Comparison on batch anaerobic digestion of five different livestock manures and prediction of biochemical methane potential (BMP) using different statistical models. Waste Manag. 2016, 48, 492–502. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, C.; Idler, C.; Heiermann, M. Biogas crops grown in energy crop rotations: Linking chemical composition and methane production characteristics. Bioresour. Technol. 2016, 206, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Scheftelowitz, M.; Thrän, D. Unlocking the energy potential of manure - An assessment of the biogas production potential at the farm level in Germany. Agriculture 2016, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Rajagopal, R.; Massé, D.I.; Singh, G. A critical review on inhibition of anaerobic digestion process by excess ammonia. Bioresour. Technol. 2013, 143, 632–641. [Google Scholar] [CrossRef]
- Westerholm, M.; Moestedt, J.; Schnürer, A. Biogas production through syntrophic acetate oxidation and deliberate operating strategies for improved digester performance. Appl. Energy 2016, 179, 124–135. [Google Scholar] [CrossRef] [Green Version]
- Theuerl, S.; Klang, J.; Prochnow, A. Process Disturbances in Agricultural Biogas Production—Causes, mechanisms and effects on the biogas microbiome: A review. Energies 2019, 12, 365. [Google Scholar] [CrossRef] [Green Version]
- Alsouleman, K.; Linke, B.; Klang, J.; Klocke, M.; Krakat, N.; Theuerl, S. Reorganisation of a mesophilic biogas microbiome as response to a stepwise increase of ammonium nitrogen induced by poultry manure supply. Bioresour. Technol. 2016, 208, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Klang, J.; Szewzyk, U.; Bock, D.; Theuerl, S. Nexus between the microbial diversity level and the stress tolerance within the biogas process. Anaerobe 2019, 56, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Isaksson, S.; Karlsson Lindsjö, O.; Schnürer, A. Microbial community adaptability to altered temperature conditions determines the potential for process optimisation in biogas production. Appl. Energy 2018, 226, 838–848. [Google Scholar] [CrossRef]
- Niu, Q.; Kubota, K.; Qiao, W.; Jing, Z.; Zhang, Y.; Yu-You, L. Effect of ammonia inhibition on microbial community dynamic and process functional resilience in mesophilic methane fermentation of chicken manure. J. Chem. Technol. Biotechnol. 2014, 90, 2161–2169. [Google Scholar] [CrossRef]
- Lv, Z.; Leite, A.F.; Harms, H.; Glaser, K.; Liebetrau, J.; Kleinsteuber, S.; Nikolausz, M. Microbial community shifts in biogas reactors upon complete or partial ammonia inhibition. Appl. Microbiol. Biotechnol. 2019, 103, 519–533. [Google Scholar] [CrossRef]
- Gutleben, J.; Chaib De Mares, M.; van Elsas, J.D.; Smidt, H.; Overmann, J.; Sipkema, D. The multi-omics promise in context: From sequence to microbial isolate. Crit. Rev. Microbiol. 2018, 44, 212–229. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 2018, 3, e00055-18. [Google Scholar] [CrossRef] [Green Version]
- Schnürer, A. Cristal ball—Know thy microorganism – why metagenomics is not enough! Microb. Biotechnol. 2013, 6, 3–16. [Google Scholar] [CrossRef]
- De Vrieze, J.; Ijaz, U.Z.; Saunders, A.M.; Theuerl, S. Terminal restriction fragment length polymorphism is an “old school” reliable technique for swift microbial community screening in anaerobic digestion. Sci. Rep. 2018, 8, 16818. [Google Scholar] [CrossRef]
- Hassa, J.; Maus, I.; Off, S.; Pühler, A.; Scherer, P.; Klocke, M.; Schlüter, A. Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants. Appl. Microbiol. Biotechnol. 2018, 102, 5045–5063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyer, R.; Schallert, K.; Büdel, A.; Zoun, R.; Dorl, S.; Behne, A.; Kohrs, F.; Püttker, S.; Siewert, C.; Muth, T.; et al. A Robust and universal metaproteomics workflow for research studies and routine diagnostics within 24 h using phenol extraction, FASP digest, and the MetaProteomeAnalyzer. Front. Microbiol. 2019, 10, 1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paliy, O.; Shankar, V. Application of multivariate statistical techniques in microbial ecology. Mol. Ecol. 2016, 25, 1032–1057. [Google Scholar] [CrossRef] [Green Version]
- King, R.; Baker, M. Use, misuse, and limitations of Threshold Indicator Taxa Analysis [TITAN] for natural resource management. In Application of Threshold Concepts in Natural Resource Decision Makin; Guntenspergen, G., Ed.; Springer: New York, NY, USA, 2014. [Google Scholar]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [Green Version]
- Karimi, B.; Maron, P.A.; Chemidlin-Prevost Boure, N.; Bernard, N.; Gilbert, D.; Ranjard, L. Microbial diversity and ecological networks as indicators of environmental quality. Environ. Chem. Lett. 2017, 15, 265–267. [Google Scholar] [CrossRef]
- Röttjers, L.; Faust, K. From hairballs to hypotheses – biological insights from microbial networks. FEMS Microbiol. Rev. 2018, 42, 761–780. [Google Scholar] [CrossRef] [Green Version]
- De Vrieze, J.; Saunders, A.M.; He, Y.; Fang, J.; Nielsen, P.H.; Verstraete, W.; Boon, N. Ammonia and temperature determine potential clustering in the anaerobic digestion microbiome. Water Res. 2015, 75, 312–323. [Google Scholar] [CrossRef]
- Mei, R.; Nobu, M.K.; Narihiro, T.; Kuroda, K.; Muñoz Sierra, J.; Wu, Z.; Ye, L.; Lee, P.K.H.; Lee, P.H.; van Lier, J.B.; et al. Operation-driven heterogeneity and overlooked feed-associated populations in global anaerobic digester microbiome. Water Res. 2017, 124, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Sundberg, C.; Al-Soud, W.A.; Larsson, M.; Alm, E.; Yekta, S.S.; Svensson, B.H.; Sørensen, S.J.; Karlsson, A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 2013, 85, 612–626. [Google Scholar] [CrossRef] [Green Version]
- Ziels, R.M.; Svensson, B.H.; Sundberg, C.; Larsson, M.; Karlsson, A.; Yekta, S.S. Microbial rRNA gene expression and co-occurrence profiles associate with biokinetics and elemental composition in full-scale anaerobic digesters. Microb. Biotechnol. 2018, 11, 694–709. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Lahti, L.; Gonze, D.; de Vos, W.M.; Raes, J. Metagenomics meets time series analysis: Unraveling microbial community dynamics. Curr. Opin. Microbiol. 2015, 25, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shade, A.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 2013, 7, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Calusinska, M.; Goux, X.; Fossépré, M.; Muller, E.E.L.; Wilmes, P.; Delfosse, P. A year of monitoring 20 mesophilic full-scale bioreactors reveals the existence of stable but different core microbiomes in bio-waste and wastewater anaerobic digestion systems. Biotechnol. Biofuels 2018, 11, 196. [Google Scholar] [CrossRef] [PubMed]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Kusel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F.; et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.J.; Knights, D.; Garcia, M.L.; Scalfone, N.B.; Smith, S.; Yarasheski, K.; Cummings, T.A.; Beers, A.R.; Knight, R.; Angenent, L.T. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc. Natl. Acad. Sci. USA 2011, 108, 4158–4163. [Google Scholar] [CrossRef] [Green Version]
- Bonk, F.; Popp, D.; Weinrich, S.; Sträuber, H.; Kleinsteuber, S.; Harms, H.; Centler, F. Intermittent fasting for microbes: How discontinuous feeding increases functional stability in anaerobic digestion. Biotechnol. Biofuels 2018, 11, 274. [Google Scholar] [CrossRef] [Green Version]
- Theuerl, S.; Klang, J.; Heiermann, M.; De Vrieze, J. Marker microbiome clusters are determined by operational parameters and specific key taxa combinations in anaerobic digestion. Bioresour. Technol. 2018, 263, 128–135. [Google Scholar] [CrossRef]
- Liebetrau, J.; Pfeiffer, D.; Thrän, D. Collection of Methods for Biogas—Methods to Determine Parameters for Analysis Purposes and Parameters that Describe Processes in the Biogas Sector; DBFZ Deutsches Biomasseforschungszentrum gemeinnützige GmbH: Fischer Druck, Leipzig, Germany, 2016. [Google Scholar]
- Hansen, K.H.; Angelidaki, I.; Ahring, B.K. Anaerobic digestion of swine manure: Inhibition by ammonia. Water Res. 1998, 32, 5–12. [Google Scholar] [CrossRef]
- VDI 4630—Fermentation of Organic Materials—Characterization of the Substrate, Sampling, Collection of Material Data, Fermentation Tests; Beuth Verlag GmbH: Berlin, Germany, 2016.
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 30 June 2020).
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Joshi, N.A.; Fass, J.N. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files [version 1.33]. 2011. Available online: https://github.com/najoshi/sickle (accessed on 30 June 2020).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Zhang, L.; Loh, K.-C.; Lim, J.W.; Zhang, J. Bioinformatics analysis of metagenomics data of biogas-producing microbial communities in anaerobic digesters: A review. Renew. Sustain. Energy Rev. 2019, 100, 110–126. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Klang, J.; Theuerl, S.; Szewzyk, U.; Huth, M.; Tölle, R.; Klocke, M. Dynamic variation of the microbial community structure during the long-time mono-fermentation of maize and sugar beet silage. Microb. Biotechnol. 2015, 8, 764–775. [Google Scholar] [CrossRef] [Green Version]
- Rademacher, A.; Nolte, C.; Schönberg, M.; Klocke, M. Temperature increases from 55 to 75 °C in a two-phase biogas reactor result in fundamental alterations within the bacterial and archaeal community structure. Appl. Microbiol. Biotechnol. 2012, 96, 565–576. [Google Scholar] [CrossRef]
- R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, 2018. Available online: https://www.r-project.org/ (accessed on 30 June 2020).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package 2018. Available online: https://github.com/vegandevs/vegan (accessed on 30 June 2020).
- Clarke, K.R. Non-parametic multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Baker, M.E.; King, R.S. A new method for detecting and interpreting biodiversity and ecological community thresholds. Methods Ecol. Evol. 2010, 1, 25–37. [Google Scholar] [CrossRef]
- Carney, R.L. Microbial community dynamics within impacted coastal ecosystems. Ph.D. Thesis, School of Life Sciences, University of Technology Sydney, Sydney, Australia, February 2019. [Google Scholar]
- Simonin, M.; Voss, K.A.; Hassett, B.A.; Rocca, J.D.; Wang, S.-Y.; Bier, R.L.; Violin, C.R.; Wright, J.P.; Bernhardt, E.S. In search of microbial indicator taxa: Shifts in stream bacterial communities along an urbanization gradient. Environ. Microbiol. 2019, 21, 3653–3668. [Google Scholar] [CrossRef] [PubMed]
- Aschonitis, V.; Karydas, C.G.; Iatrou, M.; Mourelatos, S.; Metaxa, I.; Tziachris, P.; Iatrou, G. An integrated approach to assessing the soil quality and nutritional status of large and long-term cultivated rice agro-ecosystems. Agriculture 2019, 9, 80. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Lee, C. Response of a continuous anaerobic digester to temperature transitions: A critical range for restructuring the microbial community structure and function. Water Res. 2016, 89, 241–251. [Google Scholar] [CrossRef]
- Luo, G.; De Francisci, D.; Kougias, P.G.; Treu, L.; Zhu, X.; Angelidaki, I. New steady-state microbial community compositions and process performances in biogas reactors induced by temperature disturbances. Biotechnol. Biofuels 2015, 8, 3. [Google Scholar] [CrossRef] [Green Version]
- Weiland, P. Biogas production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010, 85, 849–860. [Google Scholar] [CrossRef]
- Li, Y.Q.; Liu, C.M.; Wachemo, A.C.; Li, X.J. Effects of liquid fraction of digestate recirculation on system performance and microbial community structure during serial anaerobic digestion of completely stirred tank reactors for corn stover. Energy 2018, 160, 309–317. [Google Scholar] [CrossRef]
- Zamanzadeh, M.; Hagen, L.H.; Svensson, K.; Linjordet, R.; Horn, S.J. Anaerobic digestion of food waste e Effect of recirculation and temperature on performance and microbiology. Water Res. 2016, 96, 246–254. [Google Scholar] [CrossRef]
- Maus, I.; Kim, Y.S.; Wibberg, D.; Stolze, Y.; Off, S.; Antonczyk, S.; Pühler, A.; Scherer, P.; Schlüter, A. Biphasic study to characterize agricultural biogas plants by high-throughput 16S rRNA gene amplicon sequencing and microscopic analysis. J. Microbiol. Biotechnol. 2017, 27, 321–334. [Google Scholar] [CrossRef] [Green Version]
- Nobu, M.K.; Dodsworth, J.A.; Murugapiran, S.K.; Rinke, C.; Gies, E.A.; Webster, G.; Schwientek, P.; Kille, P.; Parkes, J.R.; Sass, H.; et al. Phylogeny and physiology of candidate phylum ‘Atribacteria’ (OP9/JS1) inferred from cultivation independent genomics. ISME J. 2016, 10, 273–286. [Google Scholar] [CrossRef]
- Leng, L.; Yang, P.; Singh, S.; Zhuang, H.; Xu, L.; Chen, W.-H.; Dolfing, J.; Li, D.; Zhang, Y.; Zeng, H.; et al. A review on the bioenergetics of anaerobic microbial metabolism close to the thermodynamic limits and its implications for digestion applications. Bioresour. Technol. 2018, 247, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.E.L.; Henneberger, R.; Huber, H.; Moissl-Eichinger, C. Microbial syntrophy: Interaction for the common good. FEMS Microbiol. Rev. 2013, 37, 384–406. [Google Scholar] [CrossRef] [PubMed]
- Schnürer, A.; Nordberg, A. Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Sci. Technol. 2008, 57, 735–740. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Hennebel, T.; Boon, N.; Verstraete, W. Methanosarcina: The rediscovered methanogen for heavy duty biomethanation. Bioresour. Technol. 2012, 112, 1–9. [Google Scholar] [CrossRef]
- Lewin, G.R.; Carlos, C.; Chevrette, M.G.; Horn, H.A.; McDonald, B.R.; Stankey, R.J.; Fox, B.G.; Currie, C.R. Evolution and ecology of Actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 2016, 70, 235–254. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Dong, D.; Wang, H.; Müller, K.; Qin, Y.; Wang, H.; Wu, W. Metagenomic analysis of microbial consortia enriched from compost: New insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol. Biofuels 2016, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Hülsemann, B.; Zhou, L.; Merkle, W.; Hassa, J.; Müller, J.; Oechsner, H. Biomethane potential test: Influence of inoculum and the digestion system. Appl. Sci. 2020, 10, 2589. [Google Scholar] [CrossRef] [Green Version]
- Allison, S.D.; Martiny, J.B. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [Green Version]
- Drosg, B. Process monitoring in biogas plants. In IEA Bioenergy Task 37—Energy from Biogas; Frost, P., Baxter, D., Eds.; IEA Bioenergy: Paris, France, 2013. [Google Scholar]
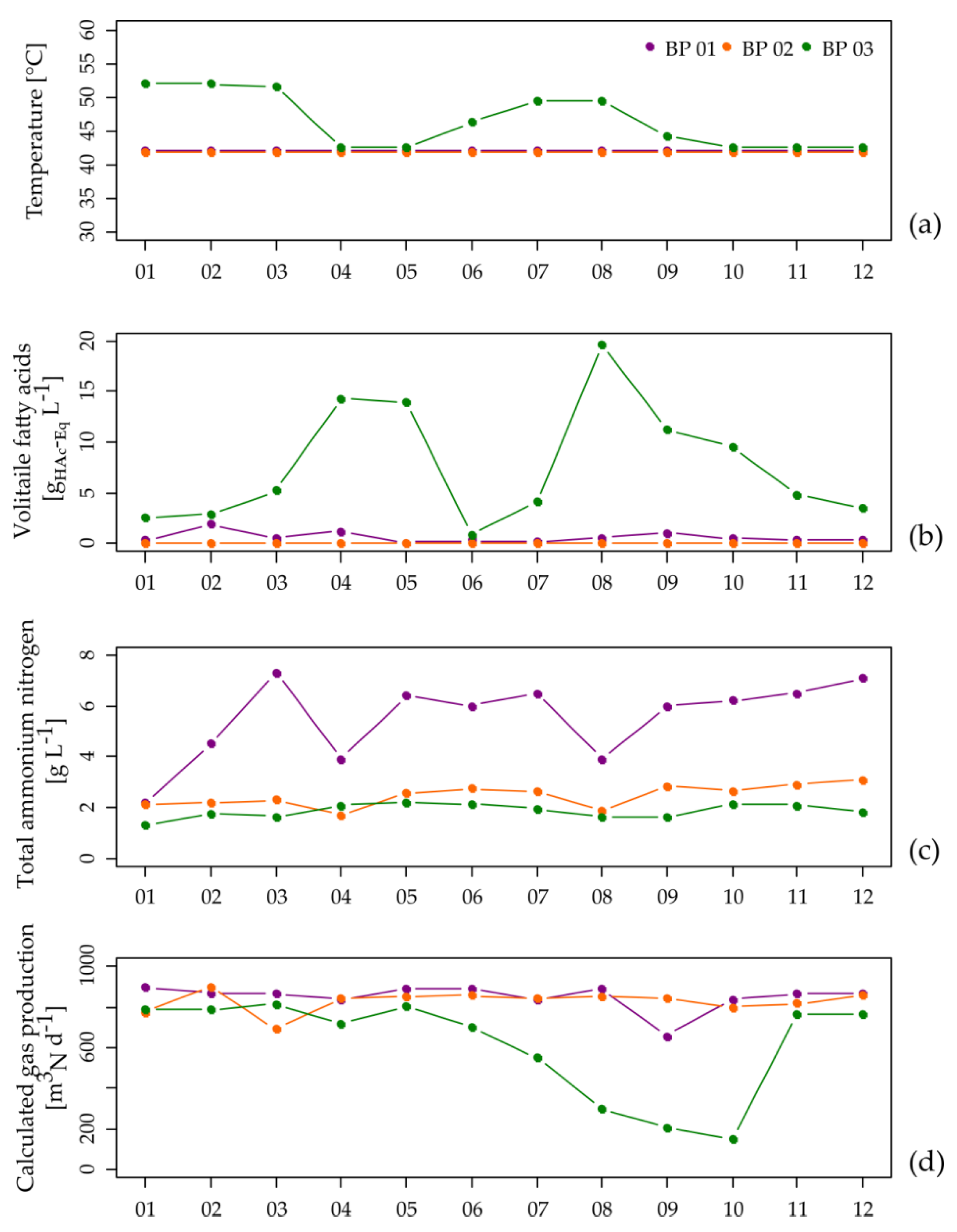

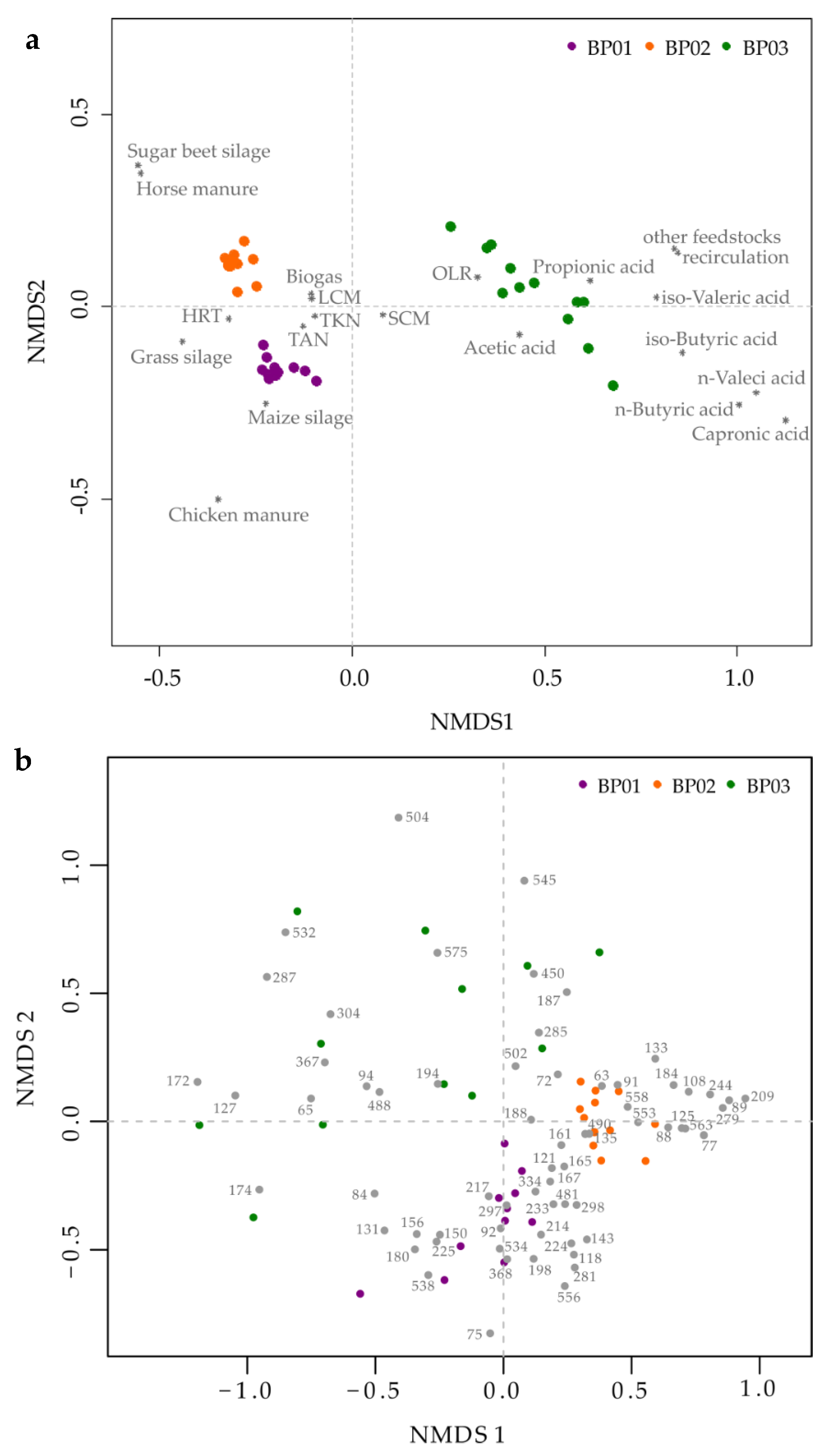
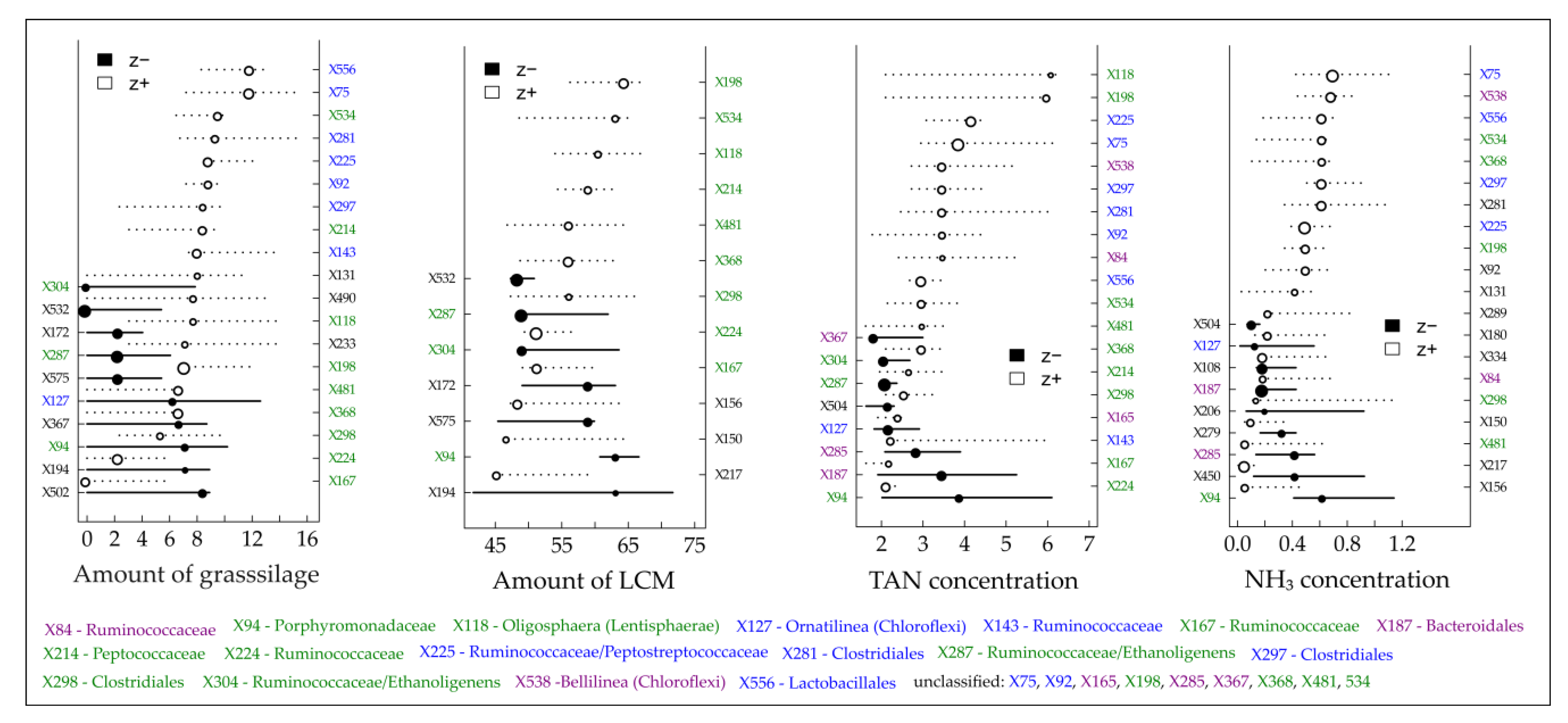
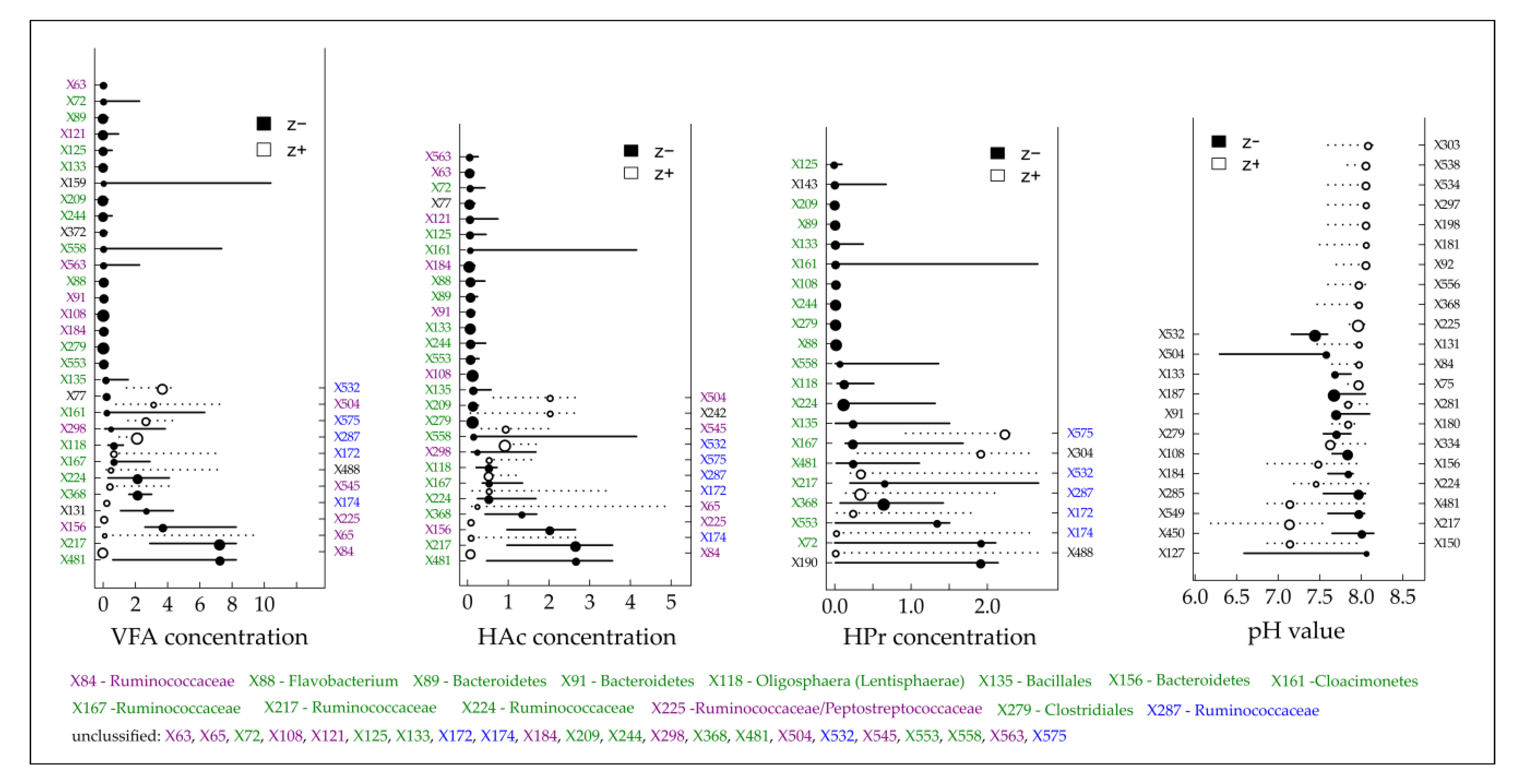
| BP (Fermenter Type) | Feedstock Supply [tFM d−1] | OLR/HRT [kgVS m−3 d−1]/[d] | Temperature [°C] |
|---|---|---|---|
| BP 01 (CSTR) | Maize silage: 0.6 ± 0.2 (5.1%) Grass silage: 1.6 ± 0.4 (14.6%) Liquid cattle manure: 8.3 ± 0.4 (65.8%) Solid cattle manure: 0.7 ± 0.5 (5.7%) Chicken manure: 1.1 ± 0.2 (9.7%) | OLR: 2.0 ± 0.4 HRT: 77.2 ± 5.8 | 41.9 ± 0.1 |
| BP 02 (CSTR) | Maize silage: 0.1 ± 0.1 (0.5%) Grass silage: 1.3 ± 0.2 (8.2%) Sugar beet silage: 0.1 ± 0.1 (0.5%) Liquid cattle manure: 11.0 ± 0.4 (64.8%) Solid cattle manure: 0.3 ± 0.1 (1.6%) Horse manure: 4.2 ± 1.3 (27.8%) | OLR: 1.9 ± 0.5 HRT: 58.7 ± 5.6 | 41.9 ± 0.0 |
| BP 03 (PF) | Liquid cattle manure: 13.5 ± 1.8 (49.5%) Solid cattle manure: 2.1 ± 0.6 (8.2%) Others 1: 1.3 ± 0.5 (3.8%) Recirculate: 13.2 ± 2.5 (56.6%) | OLR: 17.2 ± 2.1 HRT: 7.2 ± 1.0 | 46.5 ± 4.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theuerl, S.; Klang, J.; Hülsemann, B.; Mächtig, T.; Hassa, J. Microbiome Diversity and Community-Level Change Points within Manure-based small Biogas Plants. Microorganisms 2020, 8, 1169. https://doi.org/10.3390/microorganisms8081169
Theuerl S, Klang J, Hülsemann B, Mächtig T, Hassa J. Microbiome Diversity and Community-Level Change Points within Manure-based small Biogas Plants. Microorganisms. 2020; 8(8):1169. https://doi.org/10.3390/microorganisms8081169
Chicago/Turabian StyleTheuerl, Susanne, Johanna Klang, Benedikt Hülsemann, Torsten Mächtig, and Julia Hassa. 2020. "Microbiome Diversity and Community-Level Change Points within Manure-based small Biogas Plants" Microorganisms 8, no. 8: 1169. https://doi.org/10.3390/microorganisms8081169
APA StyleTheuerl, S., Klang, J., Hülsemann, B., Mächtig, T., & Hassa, J. (2020). Microbiome Diversity and Community-Level Change Points within Manure-based small Biogas Plants. Microorganisms, 8(8), 1169. https://doi.org/10.3390/microorganisms8081169






