Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in an M-SHIME® Model
Abstract
1. Introduction
2. Materials and Methods
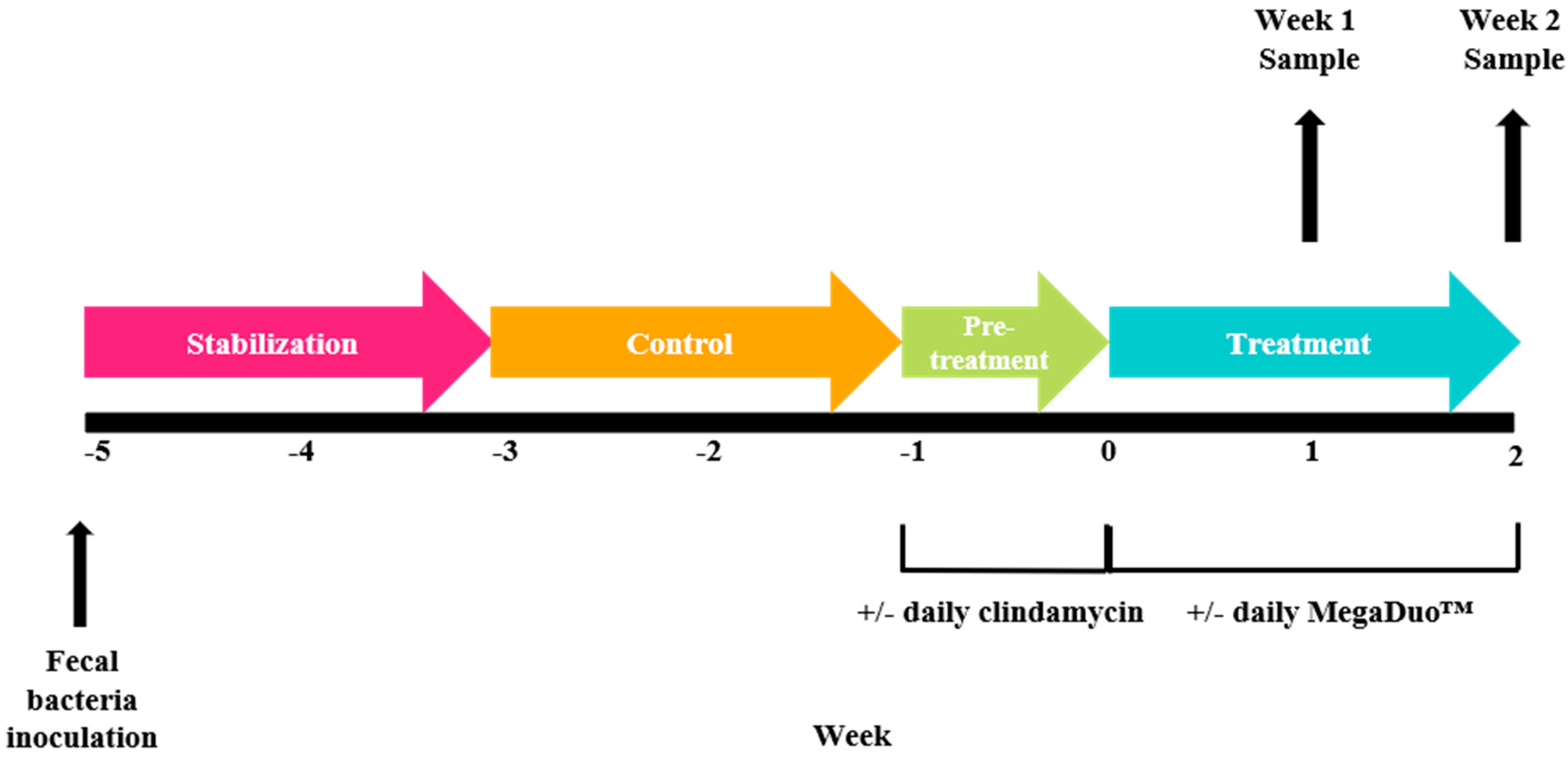
2.1. M-SHIME® Model.
2.2. Microbial Community Analysis
2.3. Caco-2/THP1-Blue™ Co-Culture Model.
2.4. Measurement of NFκB Activity
2.5. Cytokine and Chemokine Measurements
2.6. Statistical Methods
3. Results
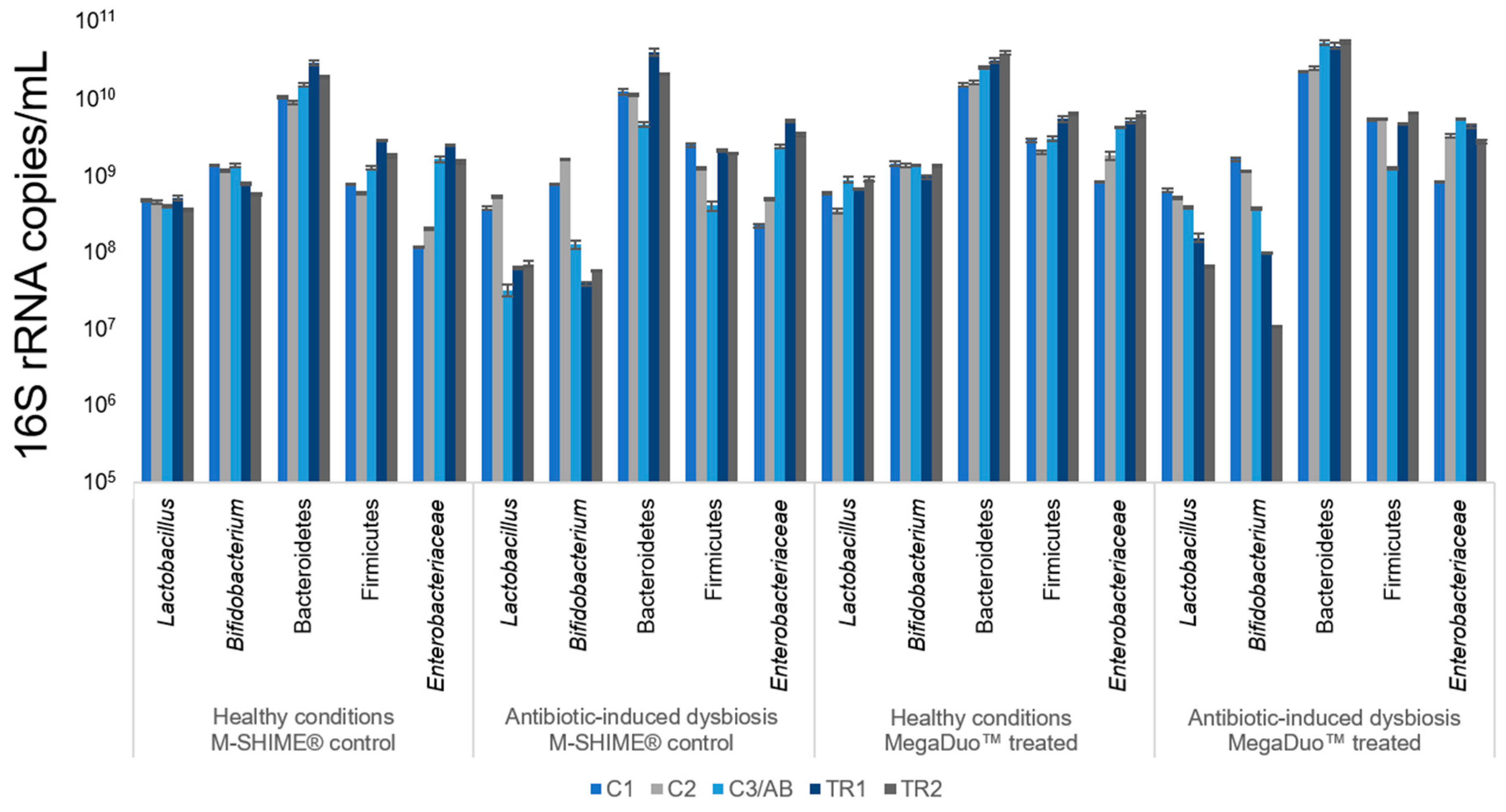
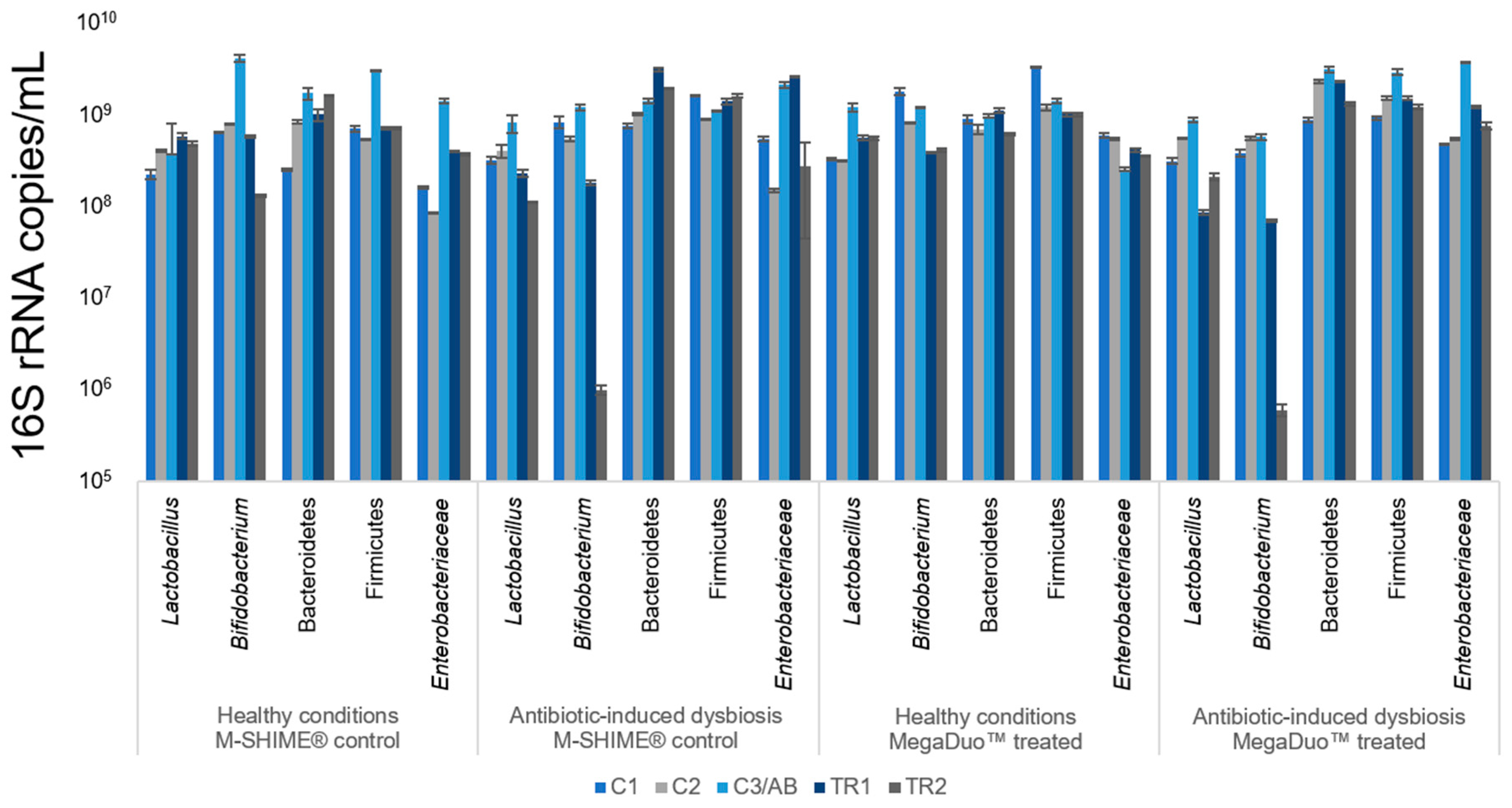
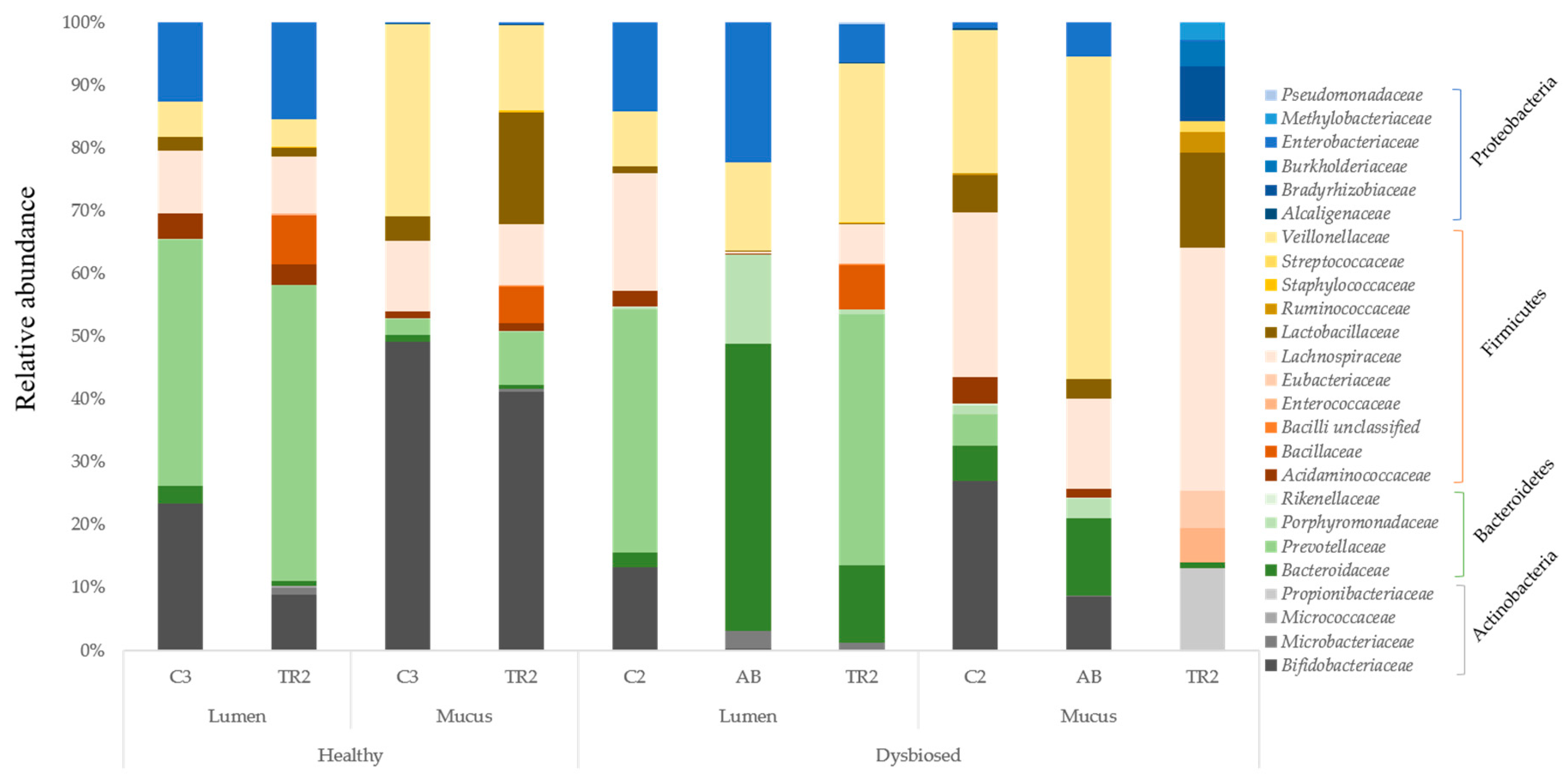
3.1. Microbial Community Analysis
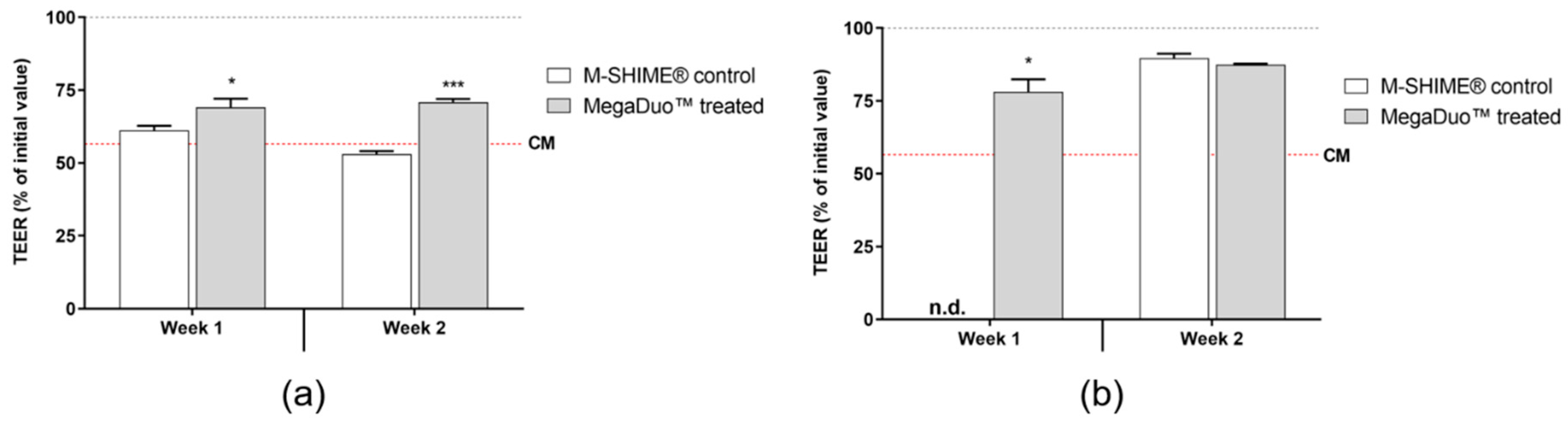
3.2. Transepithelial Electrical Resistance
3.3. Immune Markers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Virgin, H.W.; Todd, J.A. Metagenomics and personalized medicine. Cell 2011, 147, 44–56. [Google Scholar] [CrossRef]
- Priyadarshini, M.; Kotlo, K.U.; Dudeja, P.K.; Layden, B.T. Role of short chain fatty acid receptors in intestinal physiology and pathophysiology. Compr. Physiol. 2018, 8, 1091–1115. [Google Scholar] [PubMed]
- Peng, L.; He, Z.; Chen, W.; Holzman, I.R.; Lin, J. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr. Res. 2007, 61, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Francescone, R.; Hou, V.; Grivennikov, S.I. Cytokines, IBD, and colitis-associated cancer. Inflamm. Bowel. Dis. 2015, 21, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Neumann, C.; Scheffold, A.; Rutz, S. Functions and regulation of T cell-derived interleukin-10. Semin. Immunol. 2019, 44, 101344. [Google Scholar] [CrossRef] [PubMed]
- Satsu, H.; Ishimoto, Y.; Nakano, T.; Mochizuki, T.; Iwanaga, T.; Shimizu, M. Induction by activated macrophage-like THP-1 cells of apoptotic and necrotic cell death in intestinal epithelial Caco-2 monolayers via tumor necrosis factor-alpha. Exp. Cell Res. 2006, 312, 3909–3919. [Google Scholar] [CrossRef] [PubMed]
- Foey, A.D. Butyrate regulation of distinct macrophage subsets: Opposing effects on M1 and M2 macrophages. Int. J. Probiotics Prebiotics 2001, 6, 147–158. [Google Scholar]
- Larraufie, P.; Doré, J.; Lapaque, N.; Blottière, H.M. TLR ligands and butyrate increase Pyy expression through two distinct but inter-regulated pathways. Cell Microbiol. 2017, 19. [Google Scholar]
- Vinolo, M.A.R.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, M.; van der Poorten, D. The gut microbiome. Aust. Fam. Physician 2017, 46, 206–211. [Google Scholar] [PubMed]
- Dudek-Wicher, R.K.; Junka, A.; Bartoszewicz, M. The influence of antibiotics and dietary components on gut microbiota. Prz. Gastroenterol. 2018, 13, 85–92. [Google Scholar] [CrossRef]
- Knoop, K.A.; McDonald, K.G.; Kulkarni, D.H.; Newberry, R.D. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 2016, 65, 1100–1109. [Google Scholar] [CrossRef]
- Tulstrup, M.V.; Christensen, E.G.; Carvalho, V.; Linninge, C.; Ahrné, S.; Højberg, O.; Licht, T.R.; Bahl, M.I. Antibiotic treatment affects intestinal permeability and gut microbial composition in Wistar rats dependent on antibiotic class. PLoS ONE 2015, 10, e0144854. [Google Scholar] [CrossRef]
- Guidelines for the evaluation of probiotics in food. Available online: https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf (accessed on 1 June 2020).
- Pyne, D.B.; West, N.P.; Cox, A.J.; Cripps, A.W. Probiotics supplementation for athletes—clinical and physiological effects. Eur. J. Sport Sci. 2015, 15, 63–72. [Google Scholar] [CrossRef]
- Cutting, S.M. Bacillus probiotics. Food Microbiol. 2011, 28, 214–220. [Google Scholar] [CrossRef]
- Eishaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as potential probiotics: Status, concerns, and future perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef]
- Kristjansson, J.K. Thermiphilic Bacteria; CRC Press: Boca Raton, FL, USA, 1991; pp. 26–29. [Google Scholar]
- Konuray, G.; Erginkaya, Z. Potential use of Bacillus coagulans in the food industry. Foods 2018, 7, 92. [Google Scholar] [CrossRef]
- Hyronimus, B.; Le Marrec, C.; Sassi, A.H.; Deschamps, A. Acid and bile tolerance of spore-forming lactic acid bacteria. Int. J. Food Microbiol. 2000, 61, 193–197. [Google Scholar] [CrossRef]
- EFSA Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA J. 2013, 11, 3449.
- Hong, H.A.; Khaneja, R.; Tam, N.M.K.; Cazzato, A.; Tan, S.; Urdaci, M.; Brisson, A.; Gasbarrini, A.; Barnes, I.; Cutting, S.M. Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 2009, 160, 134–143. [Google Scholar] [CrossRef]
- Hong, H.A.; To, E.; Baccigalupi, L.; Ricca, E.; Cutting, S.M. Defining the natural habitat of Bacillus spore formers. Res. Microbiol. 2009, 160, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Tam, N.K.; Uyen, N.Q.; Hong, H.A.; Duc le, H.; Hoa, T.T.; Serra, C.R.; Henriques, A.O.; Cutting, S.M. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 2006, 188, 2692–2700. [Google Scholar] [CrossRef] [PubMed]
- McFarlin, B.K.; Henning, A.L.; Bowman, E.M.; Gary, M.A.; Carbajal, K.M. Oral spore-based probiotic supplementation was associated with reduced incidence of post-prandial dietary endotoxin, triglycerides, and disease risk biomarkers. World J. Gastrointest. Pathophysiol. 2017, 8, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Roos, S.; Eeckhaut, V.; MacKenzie, D.A.; Derde, M.; Verstraete, W.; Marzorati, M.; Possemiers, S.; Vanhoecke, B.; Van Immerseel, F.; et al. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb. Biotechnol. 2012, 5, 106–115. [Google Scholar] [CrossRef]
- Molly, K.; Vande Woestyne, M.; Verstraete, W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, T.; Kerckhof, F.-M.; Van de Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Possemiers, S.; Verthé, K.; Uyttendaele, S.; Verstraete, W. PCR-DDGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Micrbiol. Ecol. 2004, 49, 495–507. [Google Scholar] [CrossRef]
- Vilchez-Vargas, R.; Geffers, R.; Suarez-Diez, M.; Conte, I.; Waliczek, A.; Kaser, V.S.; Kralova, M.; Junca, H.; Pieper, D.H. Analysis of the microbial gene landscape and transcriptome for aromatic pollutants and alkane degradation using a novel internally calibrated microarray system. Environ. Microbiol. 2013, 15, 1016–1039. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef]
- Rinttilä, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef]
- Furet, J.P.; Firmesse, O.; Guormelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Doré, J.; Corthier, G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef]
- Nakano, S.; Kobayashi, T.; Funabiki, K.; Matsumura, A.; Nagao, Y.; Yamada, T. Development of a PCR assay for detection of Enterobacteriaceae in foods. J. Food Prot. 2003, 66, 1798–1804. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Kozich, J.J.; Wescott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Wescott, S.L. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2011, 77, 3219–3226. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedge, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Daguet, D.; Pinheiro, I.; Verheist, A.; Possemiers, S.; Marzorati, M. Arabinogalactan and fructooligosaccharides improve the gut barrier function in distinct areas of the colon in the Simulator of the Human Intestinal Microbial Ecosystem. J. Funct. Foods 2016, 20, 369–379. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Suva, M.A.; Sureja, V.P.; Kheni, D.B. Novel insight on probiotic Bacillus subtilis: Mechanism of action and clinical applications. J. Curr. Res. Sci. Med. 2016, 2, 65–72. [Google Scholar] [CrossRef]
- Horosheva, T.V.; Vodyanoy, V.; Sorokulova, I. Efficacy of Bacillus probiotics in prevention of antibiotic-associated diarrhea: A randomized, double-blind, placebo-controlled clinical trial. JMM Case Rep. 2014, 1, 1–6. [Google Scholar] [CrossRef]
- Pastorelli, L.; De Salvo, C.; Mercado, J.R.; Vecchi, M.; Pizarro, T.T. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: Lessons learned from animal models and human genetics. Front. Immunol. 2013, 4, 280. [Google Scholar] [CrossRef]
- Hun, L. Bacillus coagulans significantly improved abdominal pain and bloating in patients with IBS. Postrad. Med. 2009, 121, 119–124. [Google Scholar] [CrossRef]
- Kalman, D.S.; Schwartz, H.I.; Alvarez, P.; Feldman, S.; Pezzullo, J.C.; Krieger, D.R. A prospective, randomized, double-blind, placebo-controlled parallel-group dual site trial to evaluate the effects of a Bacillus coagulans-based product on functional intestinal gas symptoms. BMC Gastroenterol. 2009, 9, 85. [Google Scholar] [CrossRef]
- Dolin, B.J. Effects of a proprietary Bacillus coagulans preparation on symptoms of diarrhea-predominant irritable bowel syndrome. Methods Find. Exp. Clin. Pharm. 2009, 31, 655–659. [Google Scholar] [CrossRef]

| Cytokine/Chemokine | Dysbiosis (Clindamycin Pre-treatment) | |||||
|---|---|---|---|---|---|---|
| Week 1 | Week 2 | |||||
| M-SHIME® Control | MegaDuo™ Treated | p Value | M-SHIME® Control | MegaDuo™ Treated | p Value | |
| TNFα a | 6585.5 (194.7) | 6970.0 (305.4) | 0.8227 | 9172.6 (622.3) | 6604.7 (343.9) | 0.0067 |
| IL-8 b | 336.2 (50.2) | 343.7 (51.2) | 0.9906 | 372.3 (21.5) | 391.9 (29.6) | 0.9502 |
| MCP1 a | 7068.4 (1129.6) | 4071.1 (696.0) | 0.0699 | 8388.8 (601.0) | 8032.9 (835.3) | 0.9480 |
| CXCL10 a | 781.5 (78.6) | 641.5 (124.3) | 0.4049 | 563.8 (29.4) | 555.21 (23.5) | 0.9962 |
| IL-10 a | 10.3 (0.4) | 10.7 (0.9) | 0.8938 | 9.9 (0.8) | 12.1 (0.4) | 0.0777 |
| IL-6 a | 510.6 (14.7) | 370.3 (26.4) | 0.0070 | 483.5 (24.0) | 483.9 (29.5) | >0.999 |
| Cytokine/Chemokine | Healthy | |||||
|---|---|---|---|---|---|---|
| Week 1 | Week 2 | |||||
| M-SHIME® Control | MegaDuo™ Treated | p Value 1 | M-SHIME® Control | MegaDuo™ Treated | p Value | |
| TNFα 2 | n.d. | 5827.8 (202.2) | 0.0047 | 7251.4 (275.6) | 7124.6 (137.4) | 0.9140 |
| IL-8 3 | n.d. | 249.6 (61.6) | 0.0238 | 427.3 (22.0) | 372.9 (35.8) | 0.5785 |
| MCP1 2 | n.d. | 5778.6 (331.5) | 0.0359 | 8595.7 (339.3) | 11,516.0 (1212.1) | 0.0304 |
| CXCL10 2 | n.d. | 645.3 (23.8) | 0.9326 | 655.9 (22.5) | 703.0 (18.5) | 0.3057 |
| IL-10 2 | n.d. | 14.9 (1.3) | 0.3786 | 17.0 (1.0) | 19.8 (1.0) | 0.2049 |
| IL-6 2 | n.d. | 746.4 (34.4) | 0.9165 | 764.7 (31.9) | 865.7 (36.5) | 0.1294 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzorati, M.; Van den Abbeele, P.; Bubeck, S.S.; Bayne, T.; Krishnan, K.; Young, A.; Mehta, D.; DeSouza, A. Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in an M-SHIME® Model. Microorganisms 2020, 8, 1028. https://doi.org/10.3390/microorganisms8071028
Marzorati M, Van den Abbeele P, Bubeck SS, Bayne T, Krishnan K, Young A, Mehta D, DeSouza A. Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in an M-SHIME® Model. Microorganisms. 2020; 8(7):1028. https://doi.org/10.3390/microorganisms8071028
Chicago/Turabian StyleMarzorati, Massimo, Pieter Van den Abbeele, Sarah S. Bubeck, Thomas Bayne, Kiran Krishnan, Aicacia Young, Dilip Mehta, and Anselm DeSouza. 2020. "Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in an M-SHIME® Model" Microorganisms 8, no. 7: 1028. https://doi.org/10.3390/microorganisms8071028
APA StyleMarzorati, M., Van den Abbeele, P., Bubeck, S. S., Bayne, T., Krishnan, K., Young, A., Mehta, D., & DeSouza, A. (2020). Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in an M-SHIME® Model. Microorganisms, 8(7), 1028. https://doi.org/10.3390/microorganisms8071028





