Asexual Evolution and Forest Conditions Drive Genetic Parallelism in Phytophthora ramorum
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolates and Phenotyping
2.2. DNA Extraction and Libraries
2.3. Microsatellite Analyses
2.4. SNP Calling
2.5. Structural Variant Calling
2.6. Test of Asexual Evolution
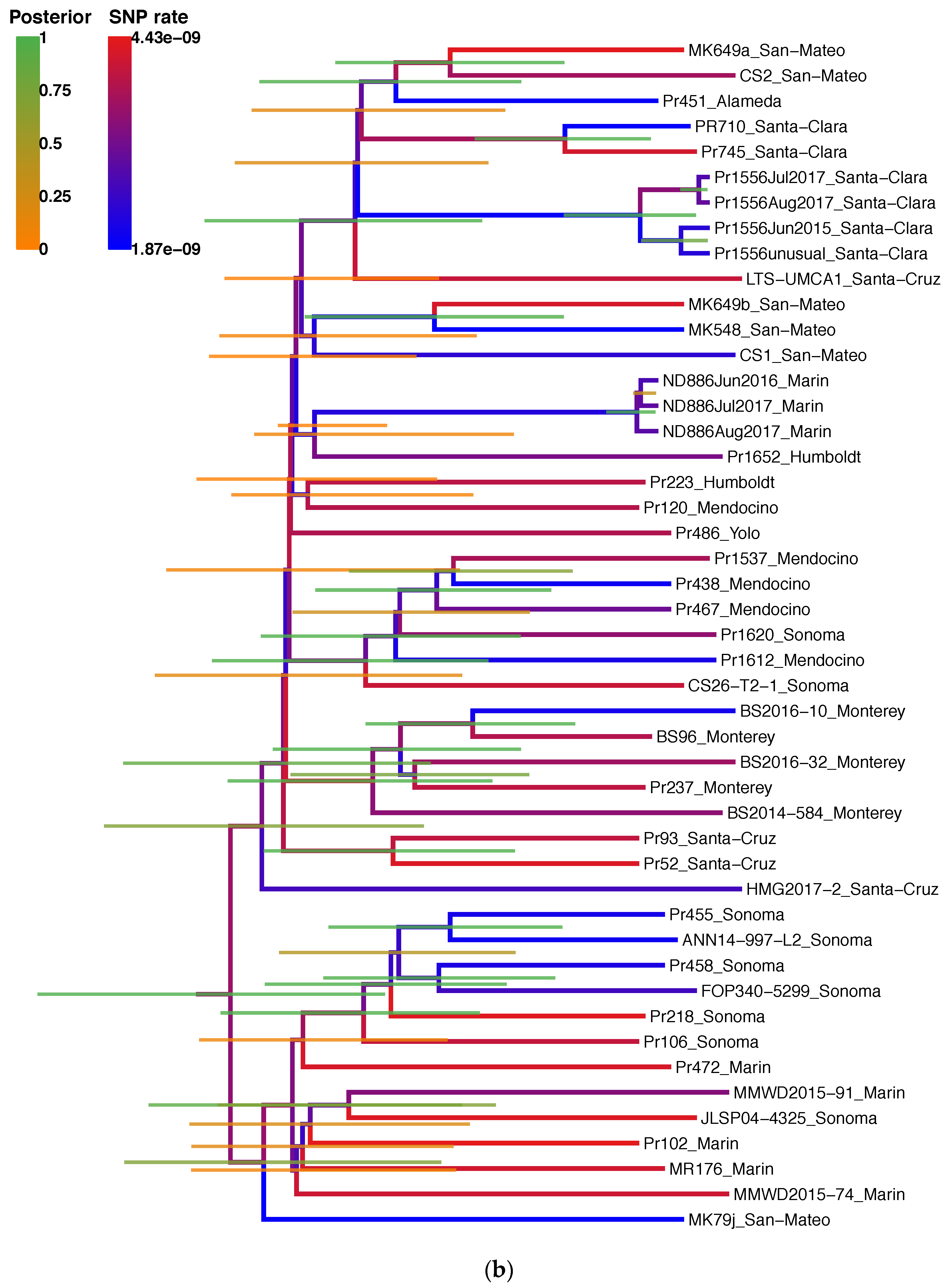
2.7. Phylogenetic Reconstruction and Ancestral State Reconstruction
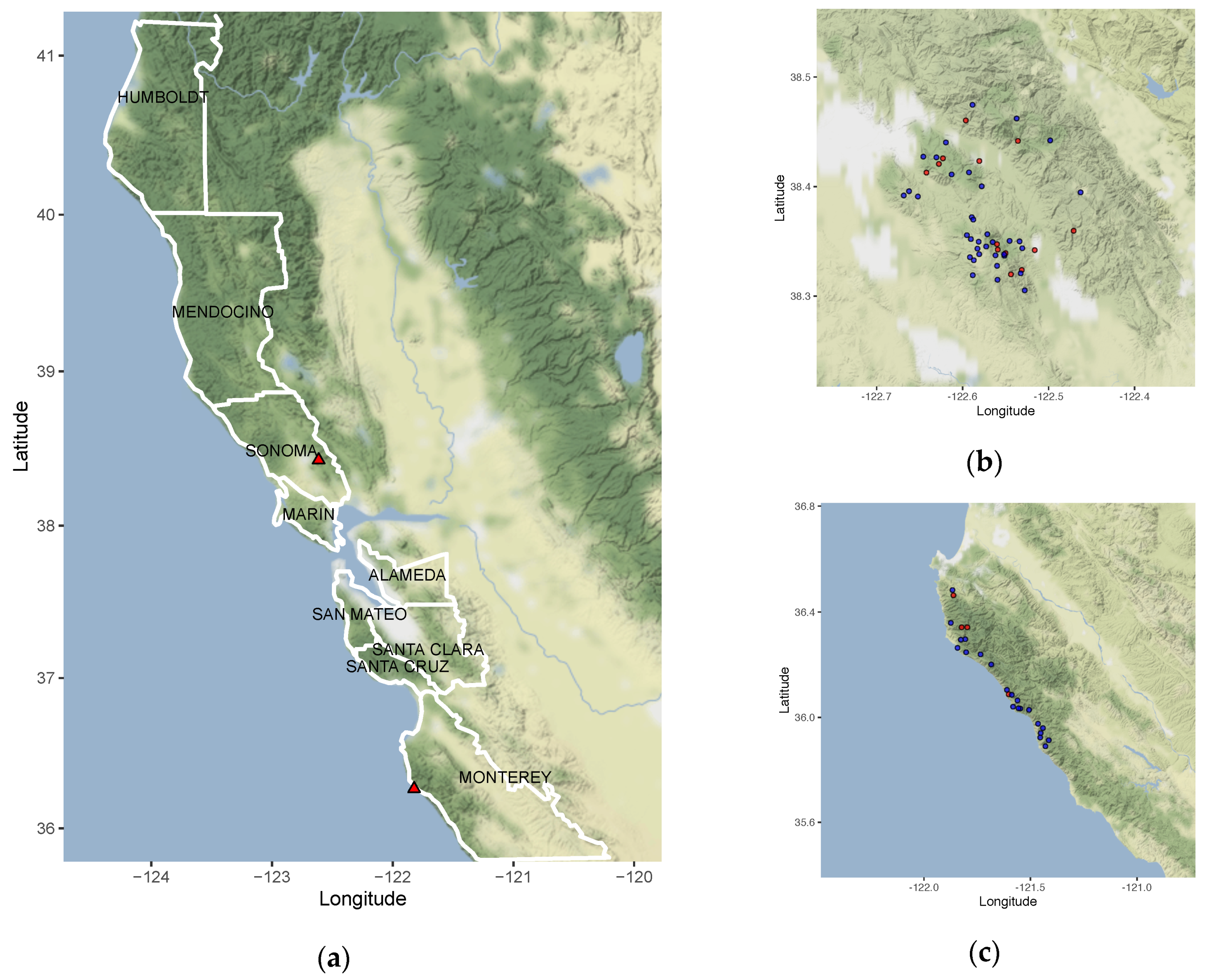
2.8. Environmental Sampling and Modelling
2.9. Association with Genomic Features
3. Results
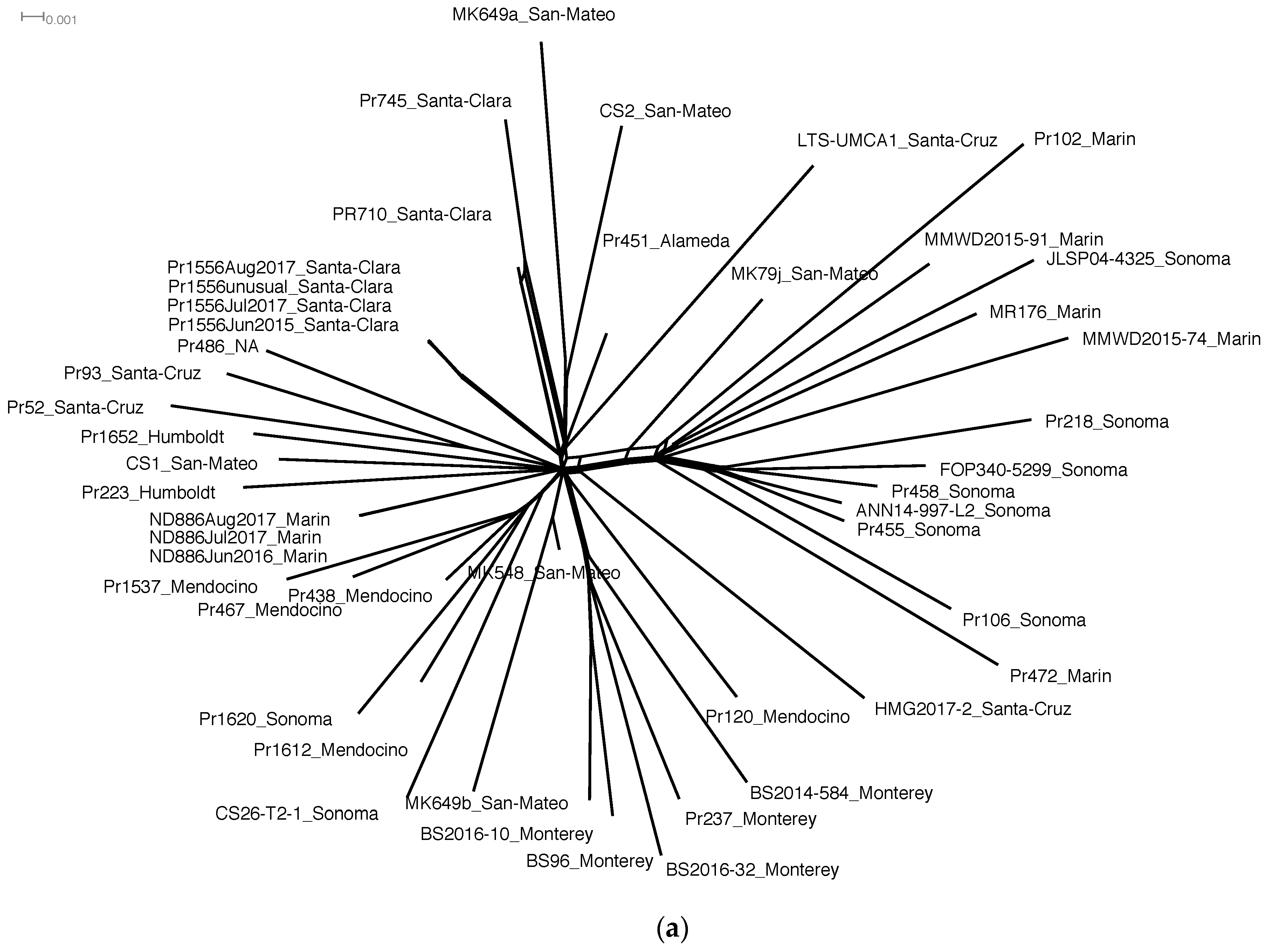
3.1. SNP Markers Confidently Reconstruct the Phylogeny of P. ramorum NA1
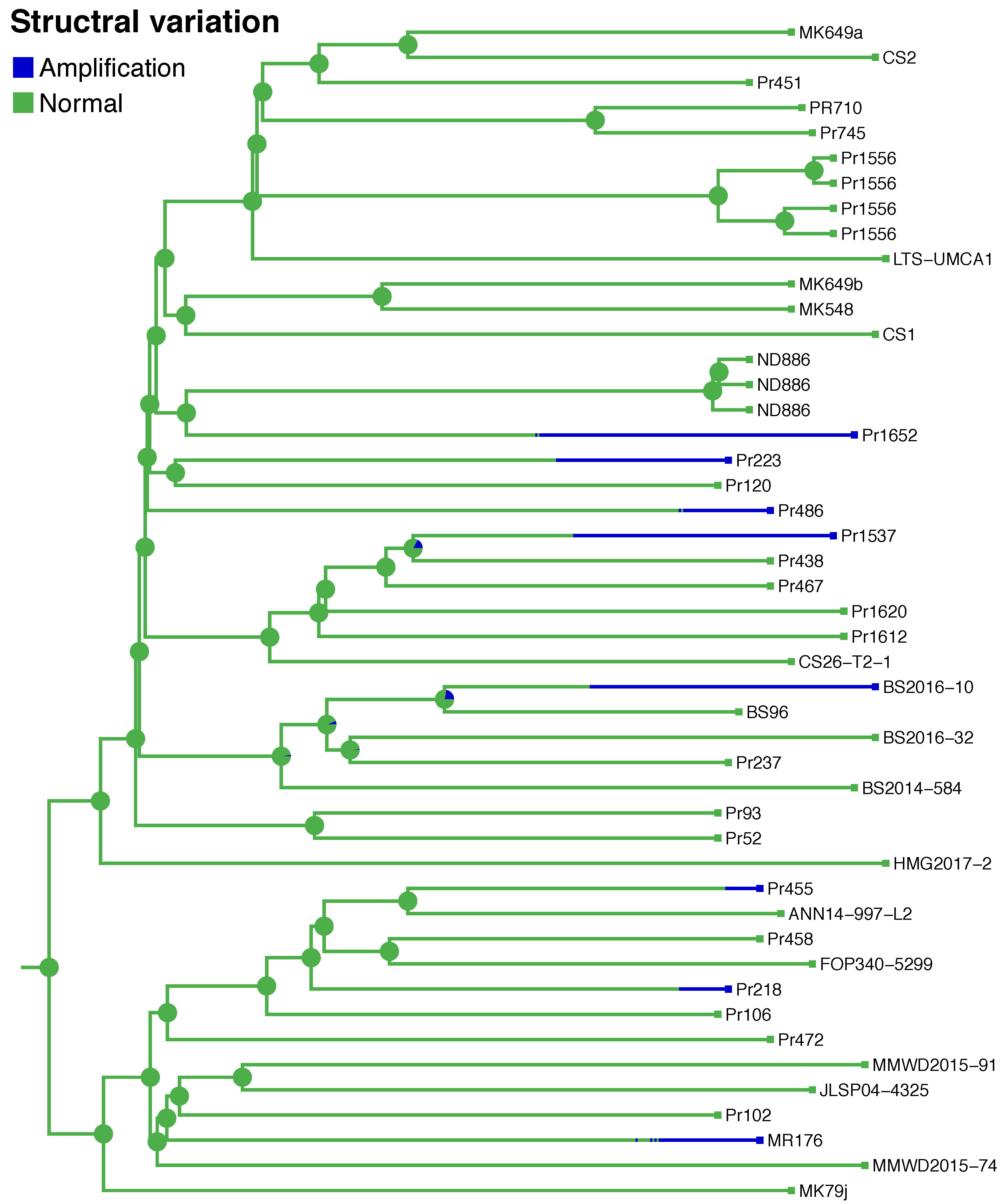
3.2. SNPs and SVs Support Asexual Evolution
3.3. Persistence of SVs in the Population is Associated with Host and Mutation Load
3.4. Stress-Related Environmental Conditions Drive SV Parallelism
3.5. SVs are Associated with Genes Involved in Pathogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martin, F.N.; Tooley, P.W. Phylogenetic relationships of Phytophthora ramorum, P. nemorosa, and P. pseudosyringae, three species recovered from areas in California with sudden oak death. Mycol. Res. 2003, 107, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Omilian, A.R.; Cristescu, M.E.A.; Dudycha, J.L.; Lynch, M. Ameiotic recombination in asexual lineages of Daphnia. Proc. Natl. Acad. Sci. USA 2006, 103, 18638–18643. [Google Scholar] [CrossRef] [PubMed]
- Schwander, T.; Crespi, B.J. Multiple Direct Transitions from Sexual Reproduction to Apomictic Parthenogenesis in Timema Stick Insects. Evolution 2009, 63, 84–103. [Google Scholar] [CrossRef]
- Oxley, P.R.; Ji, L.; Fetter-Pruneda, I.; McKenzie, S.K.; Li, C.; Hu, H.; Zhang, G.; Kronauer, D.J.C. The genome of the clonal raider ant. Cerapachys biroi. Curr. Biol. CB 2014, 24, 451–458. [Google Scholar] [CrossRef]
- Seidl, M.F.; Thomma, B.P.H.J. Sex or no sex: Evolutionary adaptation occurs regardless. BioEssays 2014, 36, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Weir, W.; Capewell, P.; Foth, B.; Clucas, C.; Pountain, A.; Steketee, P.; Veitch, N.; Koffi, M.; Meeûs, T.D.; Kaboré, J.; et al. Population Genomics Reveals the Origin and Asexual Evolution of Human Infective Trypanosomes. Available online: https://elifesciences.org/articles/11473 (accessed on 26 July 2018).
- García-Cuenca, A.F.; Dumas, Z.; Schwander, T. Extreme genetic diversity in asexual grass thrips populations. J. Evol. Biol. 2016, 29, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Hann-Soden, C.; Branco, S.; Sylvain, I.; Ellison, C.E. Clonal reproduction in fungi. Proc. Natl. Acad. Sci. USA 2015, 112, 8901–8908. [Google Scholar] [CrossRef]
- Debortoli, N.; Li, X.; Eyres, I.; Fontaneto, D.; Hespeels, B.; Tang, C.Q.; Flot, J.-F.; Van Doninck, K. Genetic Exchange among Bdelloid Rotifers Is More Likely Due to Horizontal Gene Transfer Than to Meiotic Sex. Curr. Biol. 2016, 26, 723–732. [Google Scholar] [CrossRef]
- Kasuga, T.; Bui, M.; Bernhardt, E.; Swiecki, T.; Aram, K.; Cano, L.M.; Webber, J.; Brasier, C.; Press, C.; Grünwald, N.J.; et al. Host-induced aneuploidy and phenotypic diversification in the Sudden Oak Death pathogen Phytophthora ramorum. BMC Genom. 2016, 17, 385. [Google Scholar] [CrossRef]
- Elliott, M.; Yuzon, J.; Malar, C.M.; Tripathy, S.; Bui, M.; Chastagner, G.A.; Coats, K.; Rizzo, D.M.; Garbelotto, M.; Kasuga, T. Characterization of phenotypic variation and genome aberrations observed among Phytophthora ramorum isolates from diverse hosts. BMC Genom. 2018, 19, 320. [Google Scholar] [CrossRef]
- Garbelotto, M.; Davidson, J.; Ivors, K.; Maloney, P.; Hüberli, D.; Koike, S.; Rizzo, D. Non-oak native plants are main hosts for sudden oak death pathogen in California. Calif. Agric. 2003, 57, 18–23. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Garbelotto, M. Sudden oak death: Endangering California and Oregon forest ecosystems. Front. Ecol. Environ. 2003, 1, 197–204. [Google Scholar] [CrossRef]
- Garbelotto, M.; Rizzo, D.M. A California-Based Chronological Review (1995–2004) of Research on Phytophora ramorum, the Causal Agent of Sudden Oak Death. Phytopathol. Mediterr. 2005, 44, 127–143. [Google Scholar] [CrossRef]
- Goss, E.M.; Carbone, I.; Grünwald, N.J. Ancient isolation and independent evolution of the three clonal lineages of the exotic sudden oak death pathogen Phytophthora ramorum. Mol. Ecol. 2009, 18, 1161–1174. [Google Scholar] [CrossRef]
- Grünwald, N.J.; Garbelotto, M.; Goss, E.M.; Heungens, K.; Prospero, S. Emergence of the sudden oak death pathogen Phytophthora ramorum. Trends Microbiol. 2012, 20, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Van Poucke, K.; Franceschini, S.; Webber, J.F.; Vercauteren, A.; Turner, J.A.; McCracken, A.R.; Heungens, K.; Brasier, C.M. Discovery of a fourth evolutionary lineage of Phytophthora ramorum: EU2. Fungal Biol. 2012, 116, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Brasier, C.; Kirk, S. Production of gametangia by Phytophthora ramorum in vitro. Mycol. Res. 2004, 108, 823–827. [Google Scholar] [CrossRef]
- Davidson, J.M.; Wickland, A.C.; Patterson, H.A.; Falk, K.R.; Rizzo, D.M. Transmission of Phytophthora ramorum in Mixed-Evergreen Forest in California. Phytopathology 2005, 95, 587–596. [Google Scholar] [CrossRef]
- Ivors, K.; Garbelotto, M.; Vries, I.D.E.; Ruyter-Spira, C.; Hekkert, B.T.; Rosenzweig, N.; Bonants, P. Microsatellite markers identify three lineages of Phytophthora ramorum in US nurseries, yet single lineages in US forest and European nursery populations. Mol. Ecol. 2006, 15, 1493–1505. [Google Scholar] [CrossRef]
- Kasuga, T.; Kozanitas, M.; Bui, M.; Hüberli, D.; Rizzo, D.M.; Garbelotto, M. Phenotypic Diversification Is Associated with Host-Induced Transposon Derepression in the Sudden Oak Death Pathogen Phytophthora ramorum. PLoS ONE 2012, 7, e34728. [Google Scholar] [CrossRef]
- Dale, A.L.; Feau, N.; Everhart, S.E.; Dhillon, B.; Wong, B.; Sheppard, J.; Bilodeau, G.J.; Brar, A.; Tabima, J.F.; Shen, D.; et al. Mitotic Recombination and Rapid Genome Evolution in the Invasive Forest Pathogen Phytophthora ramorum. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Anonymous APHIS List of Regulated Hosts and Plants Proven or Associated with Phytophthora ramorum. 2013. Available online: https://www.aphis.usda.gov/plant_health/plant_pest_info/pram/downloads/pdf_files/usdaprlist.pdf (accessed on 22 June 2020).
- Davidson, J.M.; Patterson, H.A.; Rizzo, D.M. Sources of Inoculum for Phytophthora ramorum in a Redwood Forest. Phytopathology 2008, 98, 860–866. [Google Scholar] [CrossRef]
- Maloney, P.E.; Lynch, S.C.; Kane, S.F.; Jensen, C.E.; Rizzo, D.M. Establishment of an emerging generalist pathogen in redwood forest communities. J. Ecol. 2005, 93, 899–905. [Google Scholar] [CrossRef]
- Raffaele, S.; Farrer, R.A.; Cano, L.M.; Studholme, D.J.; MacLean, D.; Thines, M.; Jiang, R.H.Y.; Zody, M.C.; Kunjeti, S.G.; Donofrio, N.M.; et al. Genome Evolution Following Host Jumps in the Irish Potato Famine Pathogen Lineage. Science 2010, 330, 1540–1543. [Google Scholar] [CrossRef]
- Lamour, K.H.; Mudge, J.; Gobena, D.; Hurtado-Gonzales, O.P.; Schmutz, J.; Kuo, A.; Miller, N.A.; Rice, B.J.; Raffaele, S.; Cano, L.M.; et al. Genome Sequencing and Mapping Reveal Loss of Heterozygosity as a Mechanism for Rapid Adaptation in the Vegetable Pathogen Phytophthora capsici. Mol. Plant Microbe Interact. 2012, 25, 1350–1360. [Google Scholar] [CrossRef]
- Dobrowolski, M.P.; Tommerup, I.C.; Shearer, B.L.; O’Brien, P.A. Three Clonal Lineages of Phytophthora cinnamomi in Australia Revealed by Microsatellites. Phytopathology 2003, 93, 695–704. [Google Scholar] [CrossRef]
- Cobb, R.C.; Meentemeyer, R.K.; Rizzo, D.M. Apparent competition in canopy trees determined by pathogen transmission rather than susceptibility. Ecology 2010, 91, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Cunniffe, N.J.; Cobb, R.C.; Meentemeyer, R.K.; Rizzo, D.M.; Gilligan, C.A. Modeling when, where, and how to manage a forest epidemic, motivated by sudden oak death in California. Proc. Natl. Acad. Sci. USA 2016, 113, 5640–5645. [Google Scholar] [CrossRef] [PubMed]
- Davidson, J.M.; Patterson, H.A.; Wickland, A.C.; Fichtner, E.J.; Rizzo, D.M. Forest Type Influences Transmission of Phytophthora ramorum in California Oak Woodlands. Phytopathology 2011, 101, 492–501. [Google Scholar] [CrossRef] [PubMed]
- DiLeo, M.V.; Bostock, R.M.; Rizzo, D.M. Microclimate Impacts Survival and Prevalence of Phytophthora ramorum in Umbellularia californica, a Key Reservoir Host of Sudden Oak Death in Northern California Forests. PLoS ONE 2014, 9, e98195. [Google Scholar] [CrossRef]
- Garbelotto, M.; Schmidt, D.; Swain, S.; Hayden, K.; Lione, G. The ecology of infection between a transmissive and a dead-end host provides clues for the treatment of a plant disease. Ecosphere 2017, 8, e01815. [Google Scholar] [CrossRef]
- Mascheretti, S.; Croucher, P.J.P.; Vettraino, A.; Prospero, S.; Garbelotto, M. Reconstruction of the Sudden Oak Death epidemic in California through microsatellite analysis of the pathogen Phytophthora ramorum. Mol. Ecol. 2008, 17, 2755–2768. [Google Scholar] [CrossRef]
- Mascheretti, S.; Croucher, P.J.P.; Kozanitas, M.; Baker, L.; Garbelotto, M. Genetic epidemiology of the Sudden Oak Death pathogen Phytophthora ramorum in California. Mol. Ecol. 2009, 18, 4577–4590. [Google Scholar] [CrossRef] [PubMed]
- Croucher, P.J.P.; Mascheretti, S.; Garbelotto, M. Combining field epidemiological information and genetic data to comprehensively reconstruct the invasion history and the microevolution of the sudden oak death agent Phytophthora ramorum (Stramenopila: Oomycetes) in California. Biol. Invasions 2013, 15, 2281–2297. [Google Scholar] [CrossRef] [PubMed]
- Johnston, S.F.; Cohen, M.F.; Torok, T.; Meentemeyer, R.K.; Rank, N.E. Host Phenology and Leaf Effects on Susceptibility of California Bay Laurel to Phytophthora ramorum. Phytopathology 2015, 106, 47–55. [Google Scholar] [CrossRef]
- Malar, C.M.; Yuzon, J.D.; Das, S.; Das, A.; Panda, A.; Ghosh, S.; Tyler, B.M.; Kasuga, T.; Tripathy, S. Haplotype-phased genome assembly of virulent Phythophthora ramorum isolate ND886 facilitated by long-read sequencing reveals effector polymorphisms and copy number variation. Mol. Plant Microbe Interact. 2019, 32, 1047–1060. [Google Scholar] [CrossRef] [PubMed]
- Malar, C.M.; Yuzon, J.D.; Panda, A.; Kasuga, T.; Tripathy, S. Updated assembly resource of Phytophthora ramorum Pr102 isolate incorporating long reads from PacBio sequencing. Mol. Plant Microbe Interact. 2019. [Google Scholar] [CrossRef]
- Prospero, S.; Hansen, E.M.; Grünwald, N.J.; Winton, L.M. Population dynamics of the sudden oak death pathogen Phytophthora ramorum in Oregon from 2001 to 2004. Mol. Ecol. 2007, 16, 2958–2973. [Google Scholar] [CrossRef] [PubMed]
- Vercauteren, A.; De Dobbelaere, I.; Grünwald, N.J.; Bonants, P.; Van Bockstaele, E.; Maes, M.; Heungens, K. Clonal expansion of the Belgian Phytophthora ramorum populations based on new microsatellite markers. Mol. Ecol. 2010, 19, 92–107. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P. GENALEX 6: Genetic Analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Langella, O. Populations, 1.2.30. 1999. Available online: http://bioinformatics.org/~tryphon/populations/ (accessed on 22 June 2020).
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Ha, G.; Roth, A.; Khattra, J.; Ho, J.; Yap, D.; Prentice, L.M.; Melnyk, N.; McPherson, A.; Bashashati, A.; Laks, E.; et al. TITAN: Inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 2014, 24, 1881–1893. [Google Scholar] [CrossRef] [PubMed]
- Fradin, H.; Kiontke, K.; Zegar, C.; Gutwein, M.; Lucas, J.; Kovtun, M.; Corcoran, D.L.; Baugh, L.R.; Fitch, D.H.A.; Piano, F.; et al. Genome Architecture and Evolution of a Unichromosomal Asexual Nematode. Curr. Biol. 2017, 27. [Google Scholar] [CrossRef]
- Warren, W.C.; García-Pérez, R.; Xu, S.; Lampert, K.P.; Chalopin, D.; Stöck, M.; Loewe, L.; Lu, Y.; Kuderna, L.; Minx, P.; et al. Clonal polymorphism and high heterozygosity in the celibate genome of the Amazon molly. Nat. Ecol. Evol. 2018, 2, 669. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap. Available online: sourceforge.net/projects/bbmap/ (accessed on 22 June 2020).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl. 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl. 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinforma. Oxf. Engl. 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P. Confounding Asymmetries in Evolutionary Diversification and Character Change. Evolution 2006, 60, 1743–1746. [Google Scholar] [CrossRef]
- Stadler, T. Sampling-through-time in birth–death trees. J. Theor. Biol. 2010, 267, 396–404. [Google Scholar] [CrossRef]
- Bouckaert, R.R.; Drummond, A.J. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 2017, 17, 42. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Ayres, D.L.; Darling, A.; Zwickl, D.J.; Beerli, P.; Holder, M.T.; Lewis, P.O.; Huelsenbeck, J.P.; Ronquist, F.; Swofford, D.L.; Cummings, M.P.; et al. BEAGLE: An Application Programming Interface and High-Performance Computing Library for Statistical Phylogenetics. Syst. Biol. 2012, 61, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Minin, V.N.; Suchard, M.A. Fast, accurate and simulation-free stochastic mapping. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 3985–3995. [Google Scholar] [CrossRef]
- Lemey, P.; Minin, V.N.; Bielejec, F.; Pond, S.L.K.; Suchard, M.A. A counting renaissance: Combining stochastic mapping and empirical Bayes to quickly detect amino acid sites under positive selection. Bioinforma. Oxf. Engl. 2012, 28, 3248–3256. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Mächler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M. “Finding Groups in Data”: Cluster Analysis Extended Rousseeuw. Available online: https://cran.r-project.org/web/packages/cluster/index.html (accessed on 19 March 2017).
- Nielsen, R. Mapping Mutations on Phylogenies. Syst. Biol. 2002, 51, 729–739. [Google Scholar] [CrossRef]
- Pearson, R.G.; Stanton, J.C.; Shoemaker, K.T.; Aiello-Lammens, M.E.; Ersts, P.J.; Horning, N.; Fordham, D.A.; Raxworthy, C.J.; Ryu, H.Y.; McNees, J.; et al. Life history and spatial traits predict extinction risk due to climate change. Nat. Clim. Chang. 2014, 4, 217–221. [Google Scholar] [CrossRef]
- Drost, H.-G. Philentropy: Information Theory and Distance Quantification with R. J. Open Source Softw. 2018, 3, 765. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Falcon, S.; Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007, 23, 257–258. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kumar, S.; Subramanian, S. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. 2002, 99, 803–808. [Google Scholar] [CrossRef]
- Kasuga, T.; White, T.J.; Taylor, J.W. Estimation of Nucleotide Substitution Rates in Eurotiomycete Fungi. Mol. Biol. Evol. 2002, 19, 2318–2324. [Google Scholar] [CrossRef]
- Guillet-Claude, C.; Isabel, N.; Pelgas, B.; Bousquet, J. The Evolutionary Implications of knox-I Gene Duplications in Conifers: Correlated Evidence from Phylogeny, Gene Mapping, and Analysis of Functional Divergence. Mol. Biol. Evol. 2004, 21, 2232–2245. [Google Scholar] [CrossRef]
- Wicker, T.; Oberhaensli, S.; Parlange, F.; Buchmann, J.P.; Shatalina, M.; Roffler, S.; Ben-David, R.; Doležel, J.; Šimková, H.; Schulze-Lefert, P.; et al. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 2013, 45, 1092–1096. [Google Scholar] [CrossRef]
- Tatsumoto, S.; Go, Y.; Fukuta, K.; Noguchi, H.; Hayakawa, T.; Tomonaga, M.; Hirai, H.; Matsuzawa, T.; Agata, K.; Fujiyama, A. Direct estimation of de novo mutation rates in a chimpanzee parent-offspring trio by ultra-deep whole genome sequencing. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Kelly, M.; Meentemeyer, R.K. Landscape Dynamics of the Spread of Sudden Oak Death. Photogramm. Eng. Remote Sens. 2002. [Google Scholar]
- Whisson, S.C.; Boevink, P.C.; Moleleki, L.; Avrova, A.O.; Morales, J.G.; Gilroy, E.M.; Armstrong, M.R.; Grouffaud, S.; van West, P.; Chapman, S.; et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 2007, 450, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Kamoun, S.; Zody, M.C.; Jiang, R.H.Y.; Handsaker, R.E.; Cano, L.M.; Grabherr, M.; Kodira, C.D.; Raffaele, S.; Torto-Alalibo, T.; et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 2009, 461, 393–398. [Google Scholar] [CrossRef]
- Zhang, J.; Kumar, S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol. Biol. Evol. 1997, 14, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.E.; Burke, J.M.; Rieseberg, L.H. Parallel genotypic adaptation: When evolution repeats itself. Genetica 2005, 123, 157–170. [Google Scholar] [CrossRef]
- Bailey, S.F.; Blanquart, F.; Bataillon, T.; Kassen, R. What drives parallel evolution? BioEssays 2017, 39, e201600176. [Google Scholar] [CrossRef]
- Pickersgill, B. Parallel vs. Convergent Evolution in Domestication and Diversification of Crops in the Americas. Front. Ecol. Evol. 2018, 6. [Google Scholar] [CrossRef]
- Excoffier, L.; Ray, N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 2008, 23, 347–351. [Google Scholar] [CrossRef]
- Browning, M.; Englander, L.; Tooley, P.W.; Berner, D. Survival of Phytophthora ramorum hyphae after exposure to temperature extremes and various humidities. Mycologia 2008, 100, 236–245. [Google Scholar] [CrossRef]
- Morales-Cruz, A.; Amrine, K.C.H.; Blanco-Ulate, B.; Lawrence, D.P.; Travadon, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC Genom. 2015, 16, 469. [Google Scholar] [CrossRef]
- Walker, C.A.; van West, P. Zoospore development in the oomycetes. Fungal Biol. Rev. 2007, 21, 10–18. [Google Scholar] [CrossRef]
- Muller, H.J. The relation of recombination to mutational advance. Mutat. Res. Mol. Mech. Mutagen. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Englander, L.; Browning, M.; Tooley, P.W. Growth and sporulation of Phytophthora ramorum in vitro in response to temperature and light. Mycologia 2006, 98, 365–373. [Google Scholar] [CrossRef]
- Tooley, P.W.; Browning, M. Temperature Effects on the Onset of Sporulation by Phytophthora ramorum on Rhododendron ‘Cunningham’s White’. J. Phytopathol. 2015, 163, 908–914. [Google Scholar] [CrossRef]
| Host/Source | Common | Uncommon | |
|---|---|---|---|
| Cluster | |||
| 1 | 16 | 4 | |
| 2 | 1 | 4 | |
| Count by SV Type | Monterey Co.: Coef. Est. | Monterey Co.: 5% | Monterey Co.: 95% | Sonoma Co.: Coef. Est. | Sonoma Co.: 5% | Sonoma Co.: 95% | Bayes Factor Relative to Model/ AMPno. | Bayes Factor Relative to Model/ BNDno. | Bayes Factor Relative To Model/ DELno. | Bayes Factor Relative to Model/ SVno. | Bayes Factor Relative to Model/ Parallel SIG_SV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMPno | 0.08 | −0.01 | 0.22 | 0.10 | 0.03 | 0.21 | ⏤ | 27.86 | 0.06 | 0.82 | 9.19 × 1014 |
| BNDno | −0.39 | −1.97 | 0.80 | 0.31 | −0.48 | 1.47 | 0.04 | ⏤ | 0.00 | 0.03 | 3.10 × 1013 |
| DELno | 2.89 | 0.35 | 5.72 | 1.55 | 0.37 | 3.02 | 15.51 | 489.51 | ⏤ | 13.05 | 1.47 × 1016 |
| SVno | 0.07 | −0.01 | 0.21 | 0.10 | 0.03 | 0.21 | 1.19 | 35.03 | 0.08 | ⏤ | 1.21 × 1015 |
| Parallel SIG_SV | 0.35 | −0.25 | 1.08 | 0.35 | −0.25 | 1.07 | 0.00 | 0.00 | 0.00 | 0.00 | ⏤ |
| Genomic Feature | Intersect SV and Genomic Feature | Unique to Genomic Feature | Unique to SV | Neither SV Nor Genomic Feature | Odds Ratio | p-Value |
|---|---|---|---|---|---|---|
| N. densiflorus RNAseq | 58534 | 1095221 | 2635487 | 56523374 | 1.15 | <2.2 × 10−16 |
| RXLR | 12691 | 243686 | 2681330 | 57374909 | 1.11 | <2.2 × 10−16 |
| CRN | 1850 | 20319 | 2692171 | 57598276 | 1.95 | <2.2 × 10−16 |
| Genes | 2200663 | 46065733 | 493358 | 11552862 | 1.12 | <2.2 × 10−16 |
| Repeats | 459449 | 14743843 | 2234572 | 42874752 | 0.60 | <2.2 × 10−16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuzon, J.D.; Travadon, R.; Malar C, M.; Tripathy, S.; Rank, N.; Mehl, H.K.; Rizzo, D.M.; Cobb, R.; Small, C.; Tang, T.; et al. Asexual Evolution and Forest Conditions Drive Genetic Parallelism in Phytophthora ramorum. Microorganisms 2020, 8, 940. https://doi.org/10.3390/microorganisms8060940
Yuzon JD, Travadon R, Malar C M, Tripathy S, Rank N, Mehl HK, Rizzo DM, Cobb R, Small C, Tang T, et al. Asexual Evolution and Forest Conditions Drive Genetic Parallelism in Phytophthora ramorum. Microorganisms. 2020; 8(6):940. https://doi.org/10.3390/microorganisms8060940
Chicago/Turabian StyleYuzon, Jennifer David, Renaud Travadon, Mathu Malar C, Sucheta Tripathy, Nathan Rank, Heather K. Mehl, David M. Rizzo, Richard Cobb, Corinn Small, Tiffany Tang, and et al. 2020. "Asexual Evolution and Forest Conditions Drive Genetic Parallelism in Phytophthora ramorum" Microorganisms 8, no. 6: 940. https://doi.org/10.3390/microorganisms8060940
APA StyleYuzon, J. D., Travadon, R., Malar C, M., Tripathy, S., Rank, N., Mehl, H. K., Rizzo, D. M., Cobb, R., Small, C., Tang, T., McCown, H. E., Garbelotto, M., & Kasuga, T. (2020). Asexual Evolution and Forest Conditions Drive Genetic Parallelism in Phytophthora ramorum. Microorganisms, 8(6), 940. https://doi.org/10.3390/microorganisms8060940





